Abstract
DNA helicases are ubiquitous enzymes that catalyze unwinding of duplex DNA and function in all metabolic processes in which access to single-stranded DNA is required, including DNA replication, repair, recombination and RNA transcription. RecQ helicases are a conserved family of DNA helicases that display highly specialized and vital roles in the maintenance of genome stability. Mutations in three of the five human RecQ helicases, BLM, WRN and RECQL4 are associated with the genetic disorders Bloom syndrome, Werner syndrome and Rothmund-Thomson syndrome that are characterized by chromosomal instability, premature aging and predisposition to cancer. The biological role of human RECQL5 is only partially understood and RECQL5 has not yet been associated with any human disease. Illegitimate recombination and replication stress are hallmarks of human cancers and common instigators for genomic instability and cell death. Recql5 knockout mice are cancer prone and show increased chromosomal instability. Recql5-deficient mouse embryonic fibroblasts are sensitive to camptothecin and display elevated levels of sister chromatid exchanges. Unlike other human RecQ helicases, RECQL5 is recruited to single-stranded DNA breaks and is also proposed to play an essential role in RNA transcription. Here, we review the established roles of RECQL5 at the cross roads of DNA replication, recombination and transcription, and propose that human RECQL5 provides important backup functions in the absence of other DNA helicases.
Keywords: Base excision repair, DNA replication, homologous recombination, RecQ helicases, transcription
Biochemical and cellular characterization of human RECQL5
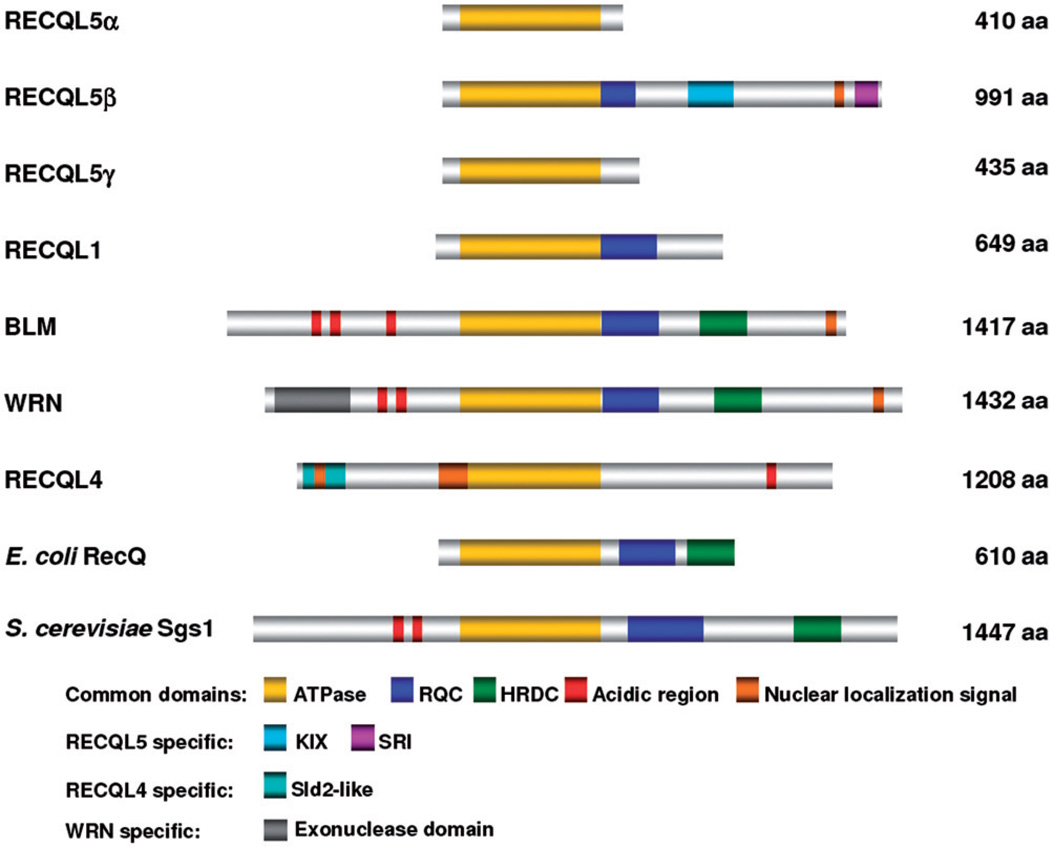
RecQ helicases play several critical functions in promoting genomic stability and are often called “caretakers of the genome” (Bohr, 2008; Chu & Hickson, 2009). Human cells express five RecQ helicases, while lower organisms such as Escherichia coli and Saccharomyces cerevisiae express only one member per species. RECQL5 was first identified and cloned from human cells in 1998, based on its homology with the other human RecQ homologs (Kitao et al., 1998). Unlike other RecQ helicases, RECQL5 exists in three alternatively-spliced isoforms, RECQL5α, RECQL5β and RECQL5γ (Shimamoto et al., 2000). RECQL5α and RECQL5γ are cytoplasmic in localization, and contain 410 and 435 amino acids, respectively (Figure 1). RECQL5β is nuclear and contains 991 amino acids including an extended C-terminal region that is not homologous to that of the other human RecQ homologs (Figure 1) (Shimamoto et al., 2000). RECQL5α is truncated upstream of the putative Zn2+-binding motif of RecQ C-terminal domain, whereas RECQL5γ contains 25 additional amino acids at the C-terminus that are not present in RECQL5α and RECQL5β (Figure 1).
Figure 1.
Schematic representation of RecQ helicases from various organisms. The common domains and the protein-specific domains are listed at the bottom. RECQL5 has three isoforms, RECQL5α, RECQL5β and RECQL5γ. The length of the proteins indicated at right in terms of number of aminoacids (aa). (see colour version of this figure at www.informahealthcare.com/bmg).
All three RECQL5 isoforms contain the conserved helicase domain; however, only RECQL5β possesses ATPase and/or helicase activity (Ren et al., 2008). RECQL5β can unwind forked duplexes and M13mpl8-based partial duplex DNA structures, whereas it does not efficiently unwind D-loop, G-quadruplex and Holliday junction (HJ) substrates (Garcia et al., 2004; Ghosh et al., 2009) that are preferentially unwound by BLM and WRN. RECQL5β requires the single-stranded DNA binding protein Replication protein A (RPA) for efficient DNA unwinding activity, while RPA binding inhibits its strand annealing activity (Garcia et al., 2004). It should also be noted that RECQL5β lacks the winged-helix motif that plays a critical role in DNA unwinding activity of RECQL1 and WRN (Pike et al., 2009; Tadokoro et al., 2012). RECQL5α possesses neither ATPase nor DNA unwinding activities, but surprisingly displays a strong strand annealing activity (Ren et al., 2008). Although there are no reported studies on RECQL5γ, its enzymatic properties may be similar to that of the a isoform. It is not known if RECQL5α and RECQL5γ isoforms possess any redundant roles with that of the relatively well characterized RECQL5β. The larger RECQL5β isoform is significantly expressed at a higher level than the other isoforms in both mice and humans and is the focus of this review and will be referred to as RECQL5 in the remainder of the text.
The relative expression of human RecQ helicases in different tissues and their modulation at different stages of the cell cycle in replicating cells helps us discern a putative biological function for RECQL5. While the expression of BLM, WRN and RECQL4 appears to be tissue specific, RECQL5 is expressed ubiquitously in all tissues, independent of their cell cycle phase (Kawabe et al., 2000; Kitao et al., 1998). Further, agents that stimulate B-cell transformation or proliferation cause significant up regulation of all human RecQ helicases except RECQL5 (Kawabe et al., 2000). These observations indicate a general and cell cycle-independent role for RECQL5 and a possible role in G0 and G1 in resting cells, which is also compatible with its recent implications in transcription. RECQL5 differs from other human RecQ helicases in its possible role as a transcriptional modulator (Aygun et al., 2009) and in its recruitment to single-strand breaks (SSBs) (Tadokoro et al., 2012). Although not associated with any disease, RECQL5 was proposed to be a tumor suppressor and Recql5 deletion in mice results in higher cancer susceptibility (Hu et al., 2007). Hence, it is important to understand its biological role in the maintenance of genomic stability.
RCQ-5, the RECQL5 homologue in Caenorhabditis elegans, is expressed in all developmental phases and its perturbation reduced the life span of the organism and slightly increased its sensitivity to ionizing radiation (Jeong et al., 2003). Genetic studies with Drosophila melanogaster RECQ5/QE, the closest fly homologue of human RECQL5, also provided some insights into RECQL5 function in genome stability (Jeong et al., 2000; Nakayama et al., 2004, 2006). Flies carrying homozygous deletion of the RECQ5/QE gene are viable and fertile but exhibit mitotic defects and chromosomal abnormalities in larval neuroblasts (Nakayama et al., 2009). Interestingly, RECQ5/QE was able to rescue several phenotypes such as higher frequencies of homologous recombination (HR), sister chromatid exchanges (SCEs), and increased sensitivity to methyl methanesulfonate (MMS) and hydroxyurea (HU) in the cells lacking the single budding yeast RecQ helicase Sgs1 (Nakayama et al., 2004). Moreover, RECQ5/QE also complements the growth defects in sgs1, srs2 and sgs1 top3 yeast, suggesting a possible role in HR (Nakayama et al., 2004). Recql5-deficient mice exhibit elevated frequencies of SCEs and chromosomal translocations in response to replicative stress (Hu et al., 2007).
At least 10 human proteins have been shown to interact with RECQL5, as summarized in Table 1. These protein interacting partners suggest the involvement of RECQL5 in various DNA metabolic pathways including DNA replication, DNA double strand break (DSB) repair, base excision repair (BER) and transcription. Cellular studies are consistent with this notion, and the functional significance of RECQL5 protein interactions will be discussed in more detail in the following sections.
Table 1.
Proposed interaction partners of RECQL5 till date.
| Protein | Location | Methods | Region | Function | References |
|---|---|---|---|---|---|
| FEN1 | Nuc | CBD-PD | ND | RECQL5 stimulates flap endonuclease activity |
Speina et al. (2010) |
| TOPO IIa | Nuc | CoIP | ND | RECQL5 stimulates DNA decatenation activity |
Ramamoorthy et al. (2012) |
| TOPO IIIa/b | Nuc | CoIP | ND | ND | Shimamoto et al. (2000) |
| PCNA | Nuc | GST-PD | 542–991 | ND | Kanagaraj et al. (2006) |
| WRN | Nuc | CoIP | ND | RECQL5 stimulates WRN helicase activity |
Popuri et al. (2012) |
| Rad51 | Nuc | CoIP, CBD-PD | 515–653 | RECQL5 disrupts Rad51 filament formation |
Schwendener et al. (2010) |
| Mrell | Nuc | MS, CoIP | ND | RECQL5 inhibits Mrell exonuclease activity |
Zheng et al. (2009) |
| NBS1 | Nuc | MS, CoIP | ND | ND | Zheng et al. (2009) |
| RAD50 | Nuc | MS, CoIP | ND | ND | Zheng et al. (2009) |
| RNAP II | Nuc | MS, CoIP | KIX, SRI | RECQL5 inhibits RNA Pol II transcription |
Aygun et al. (2008), Islam et al. (2010) |
| SWI/SNF comp | Nuc | MS, CoIP | ND | ND | Zhou et al. (2010) |
| PARP-1 | Nuc | CoIP | ND | PARP-1 regulates | Tadokoro et al. (2012) |
| RECQL5 helicase activity |
The interaction partners of RECQL5 in various DNA metabolic processes such as DNA replication, transcription and HR. The modes of functional interaction and the interacting domains involved, with the appropriate references are also listed.
Role of RECQL5 in DNA replication and chromosomal segregation
Recent studies suggest that RECQL5 plays a role in DNA replication. Recql5-deficient mouse embryonic stem (ES) cells and mouse embryonic fibroblasts (MEFs) are hypersensitive to camptothecin (CPT), a drug that inhibits Topoisomerase I and blocks DNA replication (Hu et al., 2009). Specifically, a higher proportion of Recql5-deficient MEFs failed to incorporate bromodeoxyuridine when compared to their wild-type counterparts after treatment with CPT. Moreover, the loss of replication capability upon CPT treatment was also accompanied by a rapid increase in DSB accumulation, gross chromosomal rearrangements and replication-dependent cell death (Hu et al., 2009). Co-treatment with CPT and aphidicolin, an inhibitor of DNA replication (Ishii & Bender, 1980), can alleviate the cytotoxic effects in both wild-type and Recql5-deficient ES cells, suggesting that the enhanced cytotoxicity of CPT in Recql5-deficient cells is in part due to a defect in DNA replication (Hu et al., 2009). The amino acid composition of the mouse and human RECQL5 proteins is 78% identical (Kawabe et al., 2000; Shimamoto et al., 2000) and expression of human RECQL5 in Recql5−/− ES cells can partially rescue sensitivity to CPT, indicating that the function of RECQL5 is partially conserved in mammals (Hu et al., 2009).
Mouse Recql5−/− ES cells also show an increase in spontaneous γH2AX foci that are positive for proliferating cell nuclear antigen (PCNA), indicating that these DNA lesions might be associated with DNA replication (Hu et al., 2007). Spontaneous DNA strand breaks also accumulate in D. melanogaster RECQ5/QE mutant larvae (Nakayama et al., 2009) and in human RECQL5 knockdown cells (Li et al., 2011). Such an elevated level of DSBs might not be associated with a defect in DSB repair, as these cells are proficient in HR (Hu et al., 2007, 2009). However, it is possible that RECQL5 plays an important role in maintaining active DNA replication to prevent replication fork collapse and generation of DSBs. This implies possible roles during normal DNA replication and also in cells experiencing replicative stress. For instance, RECQL5 physically interacts with human Flap Endonuclease 1 (FEN1), which plays a critical role in the processing of Okazaki fragments during DNA replication and dramatically stimulates the rate of FEN1 cleavage on both 5′ flap and nicked DNA substrates (Speina et al., 2010). This raises the intriguing possibility that RECQL5, via stimulation of FEN1, could also coordinate the cleavage event needed to reset an uncoupled replication fork (Speina et al., 2010). Furthermore, RECQL5 can promote strand exchange on synthetic DNA structures that resemble a stalled replication fork. In particular, RECQL5 can overcome a leading strand gap and can promote the displacement of both arms of the fork and subsequently anneal the displaced leading and lagging strands, suggesting that it can mediate regression of stalled replication forks to facilitate bypass of DNA damage via template-switching (Kanagaraj et al., 2006). RECQL5 associates with DNA replication factories in S phase nuclei, persists at the sites of stalled replication forks, and physically interacts with PCNA both in vivo and in vitro (Kanagaraj et al., 2006). In thymidine-treated cells, RECQL5 relocates to stalled replication forks, suppresses thymidine-induced RPA foci, and inhibits CHK1 signaling and γH2AX activation and has been proposed to be involved in replication fork stabilization (Blundred et al., 2010). Furthermore, the amount of chromatin-bound RECQL5 co-localizing with CldU also increases in thymidine- and HU-treated cells (Blundred et al., 2010).
It was reported that CPT-induced Topo I–DNA complex formation can result in retardation of DNA supercoiling which is also associated with an impediment of transcription and replication in vivo (Koster et al., 2007). This may also suggest a possible involvement of RECQL5 in DNA super-coiling and in DNA decatenation and/or chromosomal separation. Our recent observations indicate a physical and functional interaction of Topoisomerase IIα with RECQL5 and this direct interaction stimulates the DNA decatenation activity of Topoisomerase IIα (Ramamoorthy et al., 2012). Stable depletion of RECQL5 compromises cell proliferation, induces late S-phase cycling defects in cells and activates a G2/M decatenation checkpoint leading to apoptosis (Ramamoorthy et al., 2012). These effects are similar to those observed after depletion of Topoisomerase IIα (Bower et al., 2010; Gonzalez et al., 2011). Moreover, stable RECQL5 knockdown cells are also characterized by the presence of under-condensed and entangled chromosomes (Ramamoorthy et al., 2012). In separate studies, loss of maternal RECQL5 in Drosophila embryos was also reported to induce formation of DNA bridge-like structures in anaphase chromosomes, indicating chromosomal lagging or defects in chromosomal segregation (Sakurai et al., 2011). Together, these observations link RECQL5 with topoisomerases and suggest that it participates in chromosomal segregation.
Role of RECQL5 in recombination and DNA repair
Role of RECQL5 in HR
In mice, Recql5-deficiency leads to genome instability (Hu et al., 2007, 2009). Specifically, Recql5−/− ES cells and primary MEFs show a high level of SCEs and an increased rate of chromosomal rearrangements, an end result of RAD51-dependent HR-mediated DSB repair. The elevated SCE frequency in Recql5−/− cells is reminiscent of the phenotype of BLM deficient cells, suggesting that RECQL5 and BLM share an anti-recombination function (Hu et al., 2005). Indeed, both BLM and RECQL5 can dissociate RAD51 from ssDNA in vitro (Bugreev et al., 2007, 2009; Hu et al., 2007). Mouse Recql5−/− Blm−/− double mutant cells exhibit an even greater SCE frequency than the Recql5−/− or Blm−/− single mutant cells (Hu et al., 2005), suggesting non-redundant roles for RECQL5 and BLM in suppressing SCEs. Interestingly, RECQL5 interacts with topoisomerase IIIα (Shimamoto et al., 2000), but does not substitute for BLM in double HJ dissolution (Raynard et al., 2006; Wu et al., 2005).
A physical interaction between RECQL5 and RAD51 is essential for the anti-recombinase activity of RECQL5 in vitro (Schwendener et al., 2010). RECQL5 contains a single RAD51-interacting domain located between amino acids 654 and 725 (Schwendener et al., 2010). Deletion of amino acids 652–674 or alanine substitution of F666 (F666A) completely abolished RECQL5 and RAD51 binding and inhibited the anti-recombinase activity of RECQL5. RECQL5 contains a BRC variant motif (BRCv) in the RAD51 binding domain, similar to the BRC motif in BRCA2, and was shown to regulate its anti-recombinase activity in vitro (Islam et al., 2012). Mutations in the BRCv repeat disrupts the major functions of RECQL5, association with RAD51 and its ability to block RAD51-mediated D-loop formation, suppression of SCEs, and resistance to CPT-induced replication stress (Islam et al., 2012). In this context, it would be interesting to evaluate any functional interactions and/or involvement of RECQL5 in the BRCA2-mediated recombination pathway.
Role of RECQL5 in DSB repair
Our recent studies indicate that RECQL5-depleted cells accumulate spontaneous DNA DSBs and exhibit slower DNA repair capacity followed by γ-irradiation (Popuri et al., 2012). We and others have shown direct recruitment of RECQL5 to DSBs (Popuri et al., 2012; Zheng et al., 2009) and constitutive association between RECQL5 and the MRE11-RAD50-NBS1 (MRN) complex (Zheng et al., 2009), a primary sensor of DSBs. The MRN complex promotes DSB repair and regulates DNA damage signaling via activation of ATM kinase. RECQL5 interacts with the MRN complex through the MRE11 and NBS1 subunits and specifically inhibits MRE11 exonuclease activity (Zheng et al., 2009). RECQL5 co-localizes with the MRN complex in cells exposed to HU or ultraviolet light (UVC), both of which stall replication fork progression (Zheng et al., 2009). More importantly, MRE11 plays an essential role in recruiting RECQL5 to laser-induced DNA DSBs and other types of DNA damage associated with replication arrest (Zheng et al., 2009).
In living cells, visualized using confocal laser microscopy, RECQL5 is recruited to DNA DSBs with similar kinetics as other human RecQ helicases such as BLM, WRN and RECQL4 (Popuri et al., 2012). However, RECQL5 remains at DSBs for a shorter period of time than BLM and WRN but for a longer period of time than RECQL4 (Popuri et al., 2012). These studies might reflect the differential involvement of RecQ helicases in DNA DSB repair. Mapping of domains within RECQL5 that are necessary for recruitment to DSBs revealed that both the helicase and KIX domains of RECQL5 are required for DNA damage recognition and for stable association to the DSB sites (Popuri et al., 2012). Although both these domains are recruited independently, they disassociate faster than the full length RECQL5 protein, indicating that RECQL5 requires both domains in order to be retained efficiently at DSBs and to facilitate efficient DNA repair (Popuri et al., 2012). In a separate study, Islam et al. showed that RECQL5 requires both the helicase and KIX domains to correct SCEs and to confer cellular resistance towards CPT (Islam et al., 2010).
Consistent with its role in DSB repair, deletion of RCQ-5 in Caenorhabditis elegans also results in germ line sensitivity to ionizing radiation (Jeong et al., 2003). Drosophila RECQ5/ QE is also proposed to be involved in DSB repair and interestingly, the protein is up-regulated in drosophila S2 cells exposed to MMS (Nakayama et al., 2006). Loss of function of RECQ5/QE leads to genome instability and loss of heterozygosity (Chen et al., 2010). RECQL5 was also proposed to function in the resolution of knot-like DNA structures during mitosis and can play an important role in DSB repair during interphase in syncytial Drosophila embryos (Nakayama et al., 2009). Loss of maternally derived RECQ5/QE leads to accumulation of spontaneous DSBs during mitosis in syncytial embryos (Nakayama et al., 2009).
Role of RECQL5 in single-strand break repair and BER
BER is the most prominent pathway for repair of endogenous DNA damage. Endogenous reactive oxygen species and exogenous xenobiotic compounds can induce DNA base damages. SSBs generated by exogenous DNA-damaging agents or during processing of other types of DNA damage can also be repaired by BER. SSB repair (SSBR) requires several components of the BER pathway such as FEN1, PCNA and poly(ADP-ribose) polymerase 1 (PARP1). Since both FEN1 and PCNA form part of a core complex shared in replication and in BER and SSBR pathways (especially long patch BER sub-pathway), the interaction of RECQL5 with these proteins (Kanagaraj et al., 2010; Speina et al., 2010) implicates RECQL5 in these processes. Consistent with this hypothesis, our recent studies suggest that RECQL5 is involved in the repair of endogenous DNA damage (Tadokoro et al., 2012). Specifically, the dissociation kinetics of XRCC1 foci was delayed in RECQL5-knockdown cells, suggesting inefficient SSBR in these cells (Tadokoro et al., 2012). A large load of unrepaired SSBs contributes to genetic instability and can lead to accumulation of DNA DSBs and eventually cell death (Caldecott, 2008; Heeres & Hergenrother, 2007). Depletion of RECQL5 in human cells sensitizes them to oxidative stress, leads to accumulation of endogenous DNA damage and increases cellular PAR levels (Tadokoro et al., 2012). In particular, DNA strand breaks and alkaline/Fpg𠈓sensitive sites, including Fapy and 8-oxo-dG, as well as abasic sites are more abundant in RECQL5-knock-down cells than in control cells (Tadokoro et al., 2012). Consistent with these results, depletion of RECQ5/QE in Drosophila also results in the accumulation of DNA strand breaks, including SSBs and DSBs (Nakayama et al., 2009). Moreover, Recql5 deletion in mouse cells resulted in elevated SCEs, a phenomenon frequently associated with increased SSBs (Hu et al., 2009). Involvement of RECQL5 in the BER and/or SSBR pathway is not unique among RecQ helicases, as BLM, WRN, RECQL4 and RECQL1 are all proposed to be involved in this pathway (Harrigan et al., 2006; Schurman et al., 2009; Sharma et al., 2012; Szekely et al., 2005). However, in contrast to the other human RecQ helicases, only RECQL5 accumulates at laser-induced DNA SSBs, suggesting a unique role of RECQL5 in this pathway (Singh et al., 2010; Tadokoro et al., 2012). Interestingly, XRCC1 and PARP1 are expressed at a lower level in RECQL5-depleted cells, suggesting that RECQL5 might have a possible role as a transcription modulator in relation to BER/SSBR genes (Tadokoro et al., 2012). Collectively, these observations indicate that RECQL5 could play a non-redundant and unique role in BER, specifically in modulating and/or directly participating in BER-mediated repair of endogenous DNA damage.
Role of RECQL5 in transcription
A number of recent proteomic studies have revealed that RECQL5 forms a stable complex with RNA polymerase II (RNA Pol II) in human cells (Aygun et al., 2008; Islam et al., 2010; Kanagaraj et al., 2010). The RECQL5–RNA Pol II interaction is direct and is mediated by the largest subunit of RNA Pol II, RPB1 (Aygun et al., 2008). Knockdown of RECQL5 in human cells increases transcription of several genes (Izumikawa et al., 2008). Further, in vitro transcription assays and small interfering RNA (siRNA) studies have shown that the RECQL5 inhibits RNA Pol II-catalyzed transcriptional initiation and elongation (Aygun et al., 2009; Izumikawa et al., 2008), while other RecQ helicases do not (Aygun et al., 2008; Islam et al., 2010), suggesting a specific role for RECQL5 in RNA metabolism. In agreement with this, several biochemical and bioinformatic analyses identified two regions in RECQL5 that could interact with RNA Pol II (Islam et al., 2010): aa 540 to 620, homologous to the KIX domain in several Pol II transcriptional regulators (Parker et al., 1996); and aa 909 to 991, homologous to the SRI (Set2 Rpb 1-interacting) domain in histone methyltransferase SetD2, which also regulates transcription (Kizer et al., 2005). Interestingly, the SetD2-SRI domain interacts with the phosphorylated C-terminal domain (CTD) (pCTD) of Pol IIO, which is in complete agreement with the function of the predicted SRI domain in RECQL5 (Islam et al., 2010; Li et al., 2011). RECQL5 also co-purifies with subunits of the RNA Pol II-associated initiation complex (MED4, MED1, MED24 and MED 15) as well as with the elongation factors SPT5 and SPT6, indicating that RECQL5 likely plays an important role in the initiation and elongation phases of RNA Pol II-dependent transcription (reviewed in Aygun & Svejstrup, 2010).
Several studies have been carried out to elucidate the role of RECQL5 in transcription. It was proposed that RECQL5 associates with hypophosphorylated RNA Pol II, involved in transcription initiation (Pol IIa, via the KIX domain) and with hyperphosphorylated RNA Pol II, involved in the productive/ elongation phase of transcription (Pol IIo, via the SRI domain) (Islam et al., 2010). It was speculated that RECQL5 might regulate RNA Pol II association with chromatin and possibly prevent spontaneous DNA breaks (Li et al., 2011). A significant increase in the levels of chromatin bound RNA Pol II was found in cells lacking RECQL5 and more importantly, increased numbers of spontaneous DNA breaks were associated with RNA Pol II-dependent transcribed loci (Li et al., 2011), indicating that the DSBs were transcription-associated. Furthermore, overex-pression of RECQL5 in human 293 T cells reduced the amount of the elongation subunit RNA Pol IIo associated with DNA. In this context, the SRI domain of RECQL5 prevents the accumulation of RNA Pol IIo on chromatin, while overexpression of a RECQL5 fragment lacking the SRI domain in turn leads to an accumulation of the transcriptionally active form of RNA Pol II on chromatin and induces a p53-dependent transcriptional stress response (Li et al., 2011). Collectively, these observations suggest that RECQL5 negatively regulates elongation by RNA Pol II in vivo. Interestingly, our previous observations indicate that WRN could be a potential transcriptional activator and can stimulate RNA Pol II-dependent transcription in vitro (Balajee et al., 1999). Consistently, Werner syndrome cells exhibit reduced levels of RNA Pol II-mediated transcription (Balajee et al., 1999). These observations indicate differential involvement of these RecQ helicases in transcription.
Chromatin-remodeling factors also play important roles during transcription and can reportedly enhance the recruitment of RNA polymerase to the promoter-proximal region immediately upstream of the transcription start site, which is a prerequisite for the establishment of transcriptional machinery and the formation of the pre-initiation complex (PIC) (Thomas & Chiang, 2006). Chromatin modulators, such as the SWI/SNF chromatin-remodeling complex facilitate assembly of a bulky PIC complex at a promoter site (Li et al., 2007; Venters & Pugh, 2009). In addition, chromatin-remodeling factors play important roles in other aspects of RNA Pol II transcription, such as transcription elongation and/or pre-mRNA processing (Li et al., 2007). Recent mass spectrometry analyses demonstrate that the SWI/SNF complex (BRG1, BAF170, SNF5) interacts with RECQL5, providing a clue for the presence of RECQL5 in the SWI/SNF-RNAPII complex probably during the assembly of the PIC complex (Zhou et al., 2010). In this context, RECQL5 could affect the recruitment of SWI/SNF chromatin-remodeling complex, thereby modulating assembly of the RNA Pol II transcription machinery.
RECQL5 safeguards the intersection of DNA replication and transcription
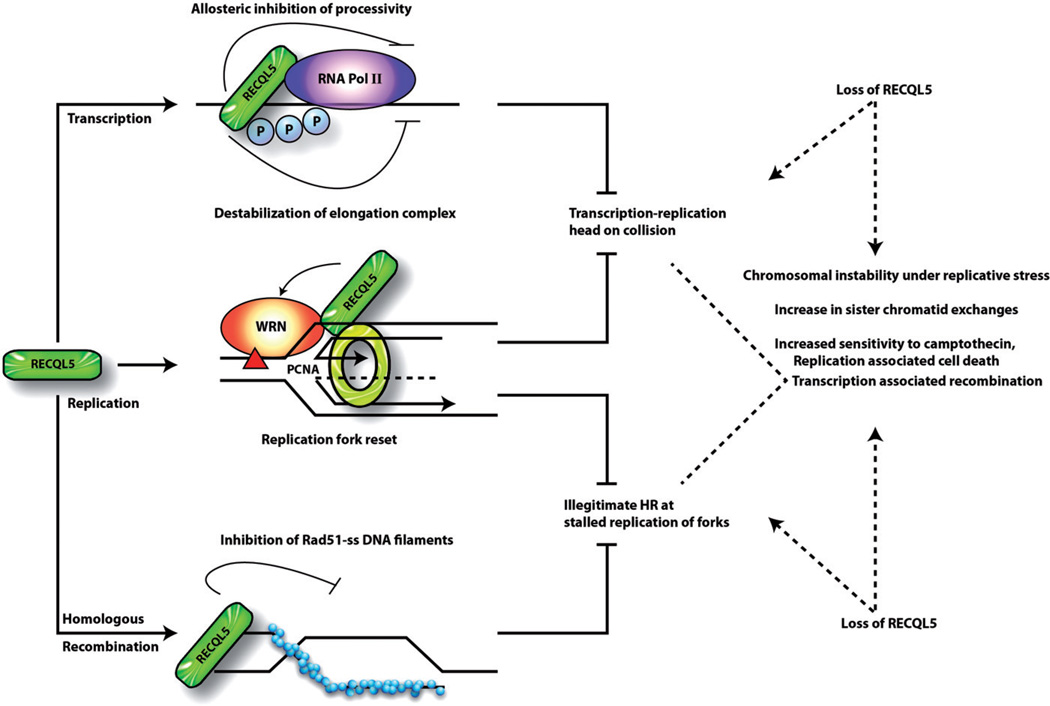
While RECQL5 may have separate roles in transcription and replication, recent studies link its potential involvement in regulating RNA Pol II with its role in suppressing DSBs during DNA replication. Interestingly, the interaction of RECQL5 with RNA Pol II was reportedly enhanced during S-phase and this increase may be required for regulating the levels of RNA Pol IIo molecules on DNA during S-phase (Li et al., 2011). In RECQL5-depleted cells, spontaneous DSBs accumulate during replication, but only in association with RNA Pol II transcription (Li et al., 2011). Transient incubation with the transcription inhibitor 5,6-dichloro-beta-D-ribofuranosyl benzimidazole (DRB) not only eliminated DNA-bound RNA Pol IIo, but also inhibited DNA strand breaks and γH2AX foci in RECQL5-depleted cells (Li et al., 2011). Even though transcription is required for cell viability, this essential cellular process is known to trigger DNA lesions in various organisms (Aguilera, 2002; Azvolinsky et al., 2009; Gottipati & Helleday, 2009). Current models suggest that transcription-associated genome instability is intimately linked to DNA replication (Azvolinsky et al., 2009; Gottipati & Helleday, 2009; Tuduri et al., 2009). For example, a head-on collision between the transcription machinery and a replication fork is likely to cause fork stalling and/or collapse, leading to accumulation of spontaneous DSBs (Figure 2). RNA polymerase and the DNA replisome share the same DNA template for their respective functions. The rate of replication is typically 20 times faster than transcription and six or more replication forks may be present at the same time on the chromosome (Gotta et al., 1991; Hirose et al., 1983). Interference between replication and transcription represents a major source of spontaneous replication stress, which could drive genomic instability during the early stages of tumorigenesis (Tuduri et al., 2009). Recent studies indicate that collisions of the transcription machinery with a replication fork are inevitable on long human genes and can result in the formation of fragile sites (Helmrich et al., 2011). Hence, adaptation of the molecular machinery is absolutely necessary for the resolution of conflicts between transcription and replication to ensure genomic stability. Unlike other human RecQ helicases, RECQL5 stands out uniquely with its strong inherent association with RNA Pol II and participates at the intersection of DNA replication and transcription. It has been proposed that RECQL5 promotes genome stabilization through two mechanisms: (1) by interacting with RNA polymerase IIa (via the KIX domain) and (2) via its catalytic/helicase activity (Islam et al., 2010). Consistent with this notion, we observed that both the KIX and the helicase domain of RECQL5 are required for its recruitment and stable retention at DNA DSB sites (Popuri et al., 2012). RECQL5 may inhibit Pol II transcription elongation to reduce the frequency of head-on collisions between transcription and replication machineries that would lead to stalled/collapsed replication forks (Rudolph et al., 2007). RECQL5 may then directly stabilize and restart blocked replication forks (Kanagaraj et al., 2006) by its catalytic activity (Figure 2).
Figure 2.
Human RECQL5 at the intersection of DNA replication, transcription and recombination. RECQL5 inhibits transcription elongation by interaction with the hyperphosphorylated form of RNA Pol II. RECQL5 promotes strand exchange at stalled replication forks. Collision of a replication fork with the transcription machinery might result in stalled/collapsed replication forks. Alternatively, if a fork encounters a blocking lesion (red triangle) in the leading-strand template, the fork might regress and the nascent DNA strands might anneal to form a four-way junction. Replication past the lesion might be facilitated by the strand exchange and uncoupling of leading- and lagging-strand synthesis. The leading strand could be extended using the longer lagging strand as a template. We speculate that the interactions of RECQL5 with WRN and PCNA at these stalled replication forks might then reset the fork and the leading strand would now be able to extend beyond the lesion. RECQL5 acts as an anti-recombination factor by inhibiting RAD51 nucleo-protein filaments. Transcription associated recombination might also occur when transcription machinery collides with a replication fork as an alternative for fork-reset. Loss of RECQL5 might result in collision of transcription and replication machinery that would result in stalled/collapsed replication forks and eventual elevation of recombination events, (see colour version of this figure at www.informahealthcare.com/bmg).
Transcription can also enhance recombination events in all organisms ranging from prokaryotes to higher eukaryotes (Aguilera, 2002), which is a ubiquitous phenomenon referred to as transcription-associated recombination (TAR). The collision between transcription and replication machineries may result in replication fork stalling and recombination is the major mechanism involved in resolving the stalled forks (Figure 2) (Aguilera & Gomez-Gonzalez, 2008; Gottipati et al., 2008). TAR may involve transient RNA: DNA hybrids (R-loops) that would also impair replication fork progression (Aguilera & Gomez-Gonzalez, 2008). RECQL5 cannot disrupt R-loops, but interestingly, depletion of RECQL5 reduces the cellular sensitivity to diospyrin, a plant-derived bisnaphthoquinonoid, which interferes with spliceosome assembly that also leads to the formation of R-loops (Kanagaraj et al., 2010). These data suggest that RECQL5 negatively affects cell viability under conditions leading to the formation of R-loops. The ability of RECQL5 to regulate recombination, replication and transcription makes it a crucial guardian of genome integrity.
Interestingly, our preliminary observations indicate that depletion of RECQL5 may have more deleterious consequences in primary fibroblasts than in Simian virus (SV40)-transformed fibroblasts (Popuri et al., 2012). This could be because SV40 T-antigen can activate transcription from certain RNA polymerase II and III promoters (Contreras & Fiers, 1981; Loeken et al., 1988; Rajan et al., 1991). It would be interesting in future studies to explore the role of RECQL5 in primary cells in comparison with the transformed cells. Consistent to our observations, it has been previously speculated that RECQL5 would have a critical function in resting/primary cells than the transformed cells based on its expression levels and due to its involvement in transcription (Kawabe et al., 2000).
RECQL5 provides essential backup functions
Recent studies indicate that both BLM and RECQL5 are important anti-recombination factors, similar to Srs2 in yeast that blocks invasion of RAD51-ssDNA filaments onto duplex DNA to form D-loops (Bugreev et al., 2007; Burgess et al., 2009; Hu et al., 2007). Mouse Recql5−/− ES cells have an elevated frequency of SCEs and an increased incidence of multi-radial structures, similar to Blm−/− cells (Hu et al., 2005), but the overall frequency of SCEs in double-knockout ES cells (Recql5−/− and Blm−/− ) is significantly higher than in Bln−/− or Recql5−/− ES cells (Hu et al., 2005). Similar studies from Wang et al. (2003) indicate that double depletion of RECQL5 and BLM in chicken DT40 cells results in a higher frequency of SCEs than in BLM−/− cells, indicating that RECQL5 suppresses SCEs in the absence of BLM. Specifically, it is possible that RECQL5, perhaps in conjunction with BLM, plays an early role in preventing premature or unnecessary engagement of HR-mediated restart of stalled/ collapsed replication forks and this engagement could be more significant in the absence of BLM (Lu et al., 2011). In this context, our studies indicate that RECQL5 is retained longer at DSB sites in BLM-deficient cells (Popuri et al., 2012).
RTEL1 is a human functional homolog of Srs2 that suppresses HR and disrupts preformed D-loops (Barber et al., 2008). RTEL1 can also promote synthesis-dependent DNA strand annealing to direct non-crossover outcomes during mitotic (or meiotic) repair of DSBs (Youds et al., 2010). Recent studies also indicate that RTEL1 may suppress recombination in telomeres as well (Uringa et al., 2011). Specifically, RTEL1 disassembles telomeric loops and counteracts telomeric G4-DNA to ensure telomere stability and is also required for telomere elongation (Vannier et al., 2012). Interestingly, rte1–1, a C. elegans homolog of human RTEL1 is synthetically lethal with C. elegans recql5, with 100% lethality at the embryonic stage (Barber et al., 2008). Double mutant germ lines are characterized by massive accumulation of recombination events: accumulation of RAD51 foci and high frequencies of meiotic recombination, leading to cross-over products (Barber et al., 2008). Although it is still unclear why these double germ lines are synthetically lethal, this phenomenon has been attributed to the inability of cells to reverse nonproductive HR intermediates (Barber et al., 2008). Collectively, these results indicate that RECQL5’s backup anti-recombination activity becomes more important in the absence of other recombination suppressors.
Although WRN is not thought to suppress HR in the same manner as BLM and RECQL5, it has been proposed that WRN also suppresses illegitimate recombination (Lebel, 2002; Saunders et al., 2008; Yamagata et al., 1998). WS cells exhibit variegated chromosomal translocations (Salk et al., 1981), elevated recombination levels between micro-homology plasmids (Cheng et al., 1990) and an increase in spontaneous formation of RAD51 foci (Pichierri et al., 2001). Although WRN is involved in end-resection during HR (Chen et al., 2003; Saintigny et al., 2002), it also plays an important role in the repair of stalled/collapsed replication forks by suppressing illegitimate recombination (Dhillon et al., 2007; Pirzio et al., 2008; Rodriguez-Lopez et al., 2007). Consistent with this notion, ectopic expression of a HJ resolvase in WS cells enhances viability and proliferation, suggesting that HR intermediates might accumulate during DNA replication in cells lacking WRN (Franchitto et al., 2008; Rodriguez-Lopez et al., 2002). Consistently, a two-fold increase in the frequency of spontaneous HR was reported in mice with helicase domain deficient WRN protein (Lebel, 2002). Furthermore, in agreement with its potential role in telomeric replication (Crabbe et al., 2004), WRN also suppresses telomeric SCEs (Laud et al., 2005). Interestingly, our recent studies demonstrate that RECQL5 could be synthetically lethal with WRN. We observed that RECQL5 plays both co-operative and complementary roles with WRN helicase (Popuri et al., 2012). RECQL5 specifically interacts with WRN (but not BLM) both physically and functionally and the interaction is maximal during S-phase after replicative stress (Popuri et al., 2012).
RECQL5 co-operates with WRN on synthetic stalled replication fork-like structures and specifically stimulates its helicase activity (but not BLM) on DNA forked duplexes. Both RECQL5 and WRN re-localize from the nucleolus into the nucleus after replicative stress and significantly associate with each other during S-phase (Popuri et al., 2012). Moreover RECQL5 is essential for cell survival in the absence of WRN (Popuri et al., 2012). Loss of both RECQL5 and WRN severely compromises DNA replication and elevates RAD51 foci formation, indicating increased illegitimate recombination. Furthermore, the retention of RECQL5 at DNA DSBs was also extended in the absence of both WRN and BLM (Popuri et al., 2012). These observations suggest that RECQL5 provides an essential function in cells lacking BLM and WRN and would further implicate the divergent and complementary roles that these RecQ helicases might play in the cell. We speculate that RECQL5 responds to stress differently in BLM- and WRN-deficient cells. In BLM-deficient cells, the stress likely arises from elevated SCEs and deposition of RAD51 nucleo-protein filaments. In WRN-deficient cells and in normal cells exposed to replication blocking DNA damage, RECQL5 is probably responding to stalled or collapsed replication forks. In the latter case, WRN’s catalytic activities may be insufficient to handle the total cellular burden and therefore RECQL5 may provide assistance and stimulation (Popuri et al., 2012). This functional co-operation could also be enhanced by their mutual interactions with other major players such as FEN1, MRN complex or PCNA. For instance, RECQL5 interacts with PCNA and co-localizes with the MRN complex and this association increases upon replicative stress (Kanagaraj et al., 2006). Similarly, previous studies also indicate a physical interaction of WRN with PCNA (Rodriguez-Lopez et al., 2003) and its potential interplay with the MRN complex in cells experiencing replicative stress (Franchitto & Pichierri, 2004). We speculate that although RECQL5 is less processive than WRN, it cooperates with WRN to facilitate proper replication fork progression in vivo.
Interestingly, loss of both RECQL5 and WRN results in abnormal or defective separation of the two sister chromatids (Popuri et al., 2012). Efficient sister chromatid pairing or cohesion between the two sister chromatids is established during DNA replication and is fundamental for cell viability (Carson & Christman, 2001; Skibbens, 2009). Upon depletion of RECQL5 in the absence of WRN, we observed severe defects in DNA replication with very few cells in S-phase (Popuri et al., 2012). Further, loss of checkpoint (CHK1) activation would drive cells into mitosis with defective DNA replication and results in loss of cell viability (Castedo et al., 2004; Yin et al., 2008). Our previous studies indicate that WRN plays both upstream and downstream roles to ATM and ATR. In particular, WRN-depleted cells fail to activate pATM, pATR and pCHK1 in response to replicative stress (Cheng et al., 2008; Patro et al., 2011). WRN-depleted cells also progress through S-phase at a faster rate in response to replicative stress, suggesting a defect in the intra S-phase checkpoint (Cheng et al., 2008). Defects in DNA replication can also trigger mitotic arrest and we observe that most of the RECQL5/WRN double mutant cells were arrested at G2/M phase with the onset of apoptosis and polyploidy (Popuri et al., 2012) that are also hallmarks of mitotic cell death (Castedo et al., 2004; Pflumm & Botchan, 2001). Collectively, these observations highlight a novel backup role for RECQL5 in DNA replication and recombination.
Future perspectives
Moving forward, it will be important to determine if the basic subcellular localization of RECQL5 might be important in differentiating its various roles. RECQL5 predominantly localizes to the nucleolus but can re-localize to the nucleus during S-phase after replicative stress (Popuri et al., 2012). On the other hand, we observe that RECQL5’s localization to the nucleolus is altered in ATM- and ATR-deficient cells (unpublished results). We speculate that subcellular localization may depend on specific post-translational modification of RECQL5 and/or its interacting partners which would affect its cellular functions and its coordinated involvement in various DNA metabolic processes (Figure 2). It would be interesting to investigate whether post-translational modifications of RECQL5 modulate its role in transcription and/or other cellular metabolic pathways. Our previous studies indicate that depletion of RECQL5 induces a moderate G2/M cell cycle arrest (Ramamoorthy et al., 2012); in this context, it would also be interesting to investigate involvement of RECQL5 with cyclindependent kinases and cell cycle checkpoints.
Kecgl5-deficient MEFs exhibit elevated frequencies of spontaneous DNA double-strand breaks and HR events and are prone to gross chromosomal rearrangements (Hu et al., 2007). More importantly, deletion of Recql5 in mice results in cancer susceptibility. Detailed phenotypic analysis of Recql5−/− mice uncovered an age-dependent increase in the incidence of multiple types of sporadic cancers (Hu et al., 2007), but most prominently with gastro-intestinal and colonic tumors (Hu et al., 2010). Human RECQL5 haplotypes were also shown associated with colon cancer in a population-based case control study (Heather et al., 2010). RECQL5 confers CPT-tolerance to human colorectal cancer cells and both RECQL5-deficient human colorectal cancer cells and Recql5−/− MEFs are hypersensitive to CPT, a prototype of irinotecan, a drug approved by the FDA for treating colon cancer (Hu et al., 2010; Wang et al., 2011). Collectively, these observations indicate that RECQL5 may suppress colorectal cancer and that RECQL5 may also be used as a potential biomarker for the disease (Hu et al., 2010).
Altered telomere length is linked to a number of diseases, including cancer. Single nucleotide polymorphisms in the RECQL5 gene were inversely associated with telomere length in DNA from immortalized colon cancers cells (Mirabello et al., 2010; Pellatt et al., 2012). Shorter telomere length results in greater likelihood of developing colon cancer (Pellatt et al., 2012). Incorrect HR and defective DSB repair are also associated with short telomeres (Royle et al., 2009; Wu et al., 2003). A missense mutation could result in loss of function of RECQL5 that might impair proper stabilization of stalled/collapsed replication forks at telomeres and would eventually result in telomere shortening (Pellatt et al., 2012). Moreover, it has also been shown that C. elegans RECQL5 is synthetically lethal with the telomeric helicase RTEL1 (Barber et al., 2008). Future studies are essential to explore any functional implications of RECQL5 in telomere maintenance, although our previous studies indicate that RECQL5 cannot efficiently disrupt telomeric D-loops (Ghosh et al., 2009).
Loss of RECQL5 impacts both DNA replication and transcription and can lead to excessive recombination events that might eventually result in gross chromosomal rearrangements and higher tumor incidence. With the observation of increased cancer susceptibility in Recql5-deficient mice, it may be possible that defects in RECQL5 could also be associated with a disease in humans, with a rare patient yet to be identified, similar to the first reported patient with ERCC1 deficiency (Jaspers et al., 2007). Thus, we speculate that loss of RECQL5 would thus be characterized by higher levels of transcription that would hinder DNA replication fork progression and eventually elevate recombination events that would be responsible for higher tumor incidence and increased risk of chromosomal abnormalities (Figure 2). This would most likely occur in highly replicative tissues like skin, gut, bone marrow and hematopoietic tissue.
Acknowledgements
We thank Drs Morten Scheibye-Knudsen and Raghavendra Shamanna for critical reading of the review.
This work was supported by funds from the Intramural Research Program of the National Institute on Aging, NIH, AG000726–20.
Footnotes
Declaration of interest
The authors declare no conflict of interest.
References
- Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- Aygun O, Svejstrup JQ. RECQL5 helicase: connections to DNA recombination and RNA polymerase II transcription. DNA Repair (Amst) 2010;9:345–353. doi: 10.1016/j.dnarep.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci USA. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygun O, Xu X, Liu Y, et al. Direct inhibition of RNA polymerase II transcription by RECQL5. J Biol Chem. 2009;284:23197–23203. doi: 10.1074/jbc.M109.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae . Mol Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee AS, Machwe A, May A, et al. The Werner syndrome protein is involved in RNA polymerase II transcription. Mol Biol Cell. 1999;10:2655–2668. doi: 10.1091/mbc.10.8.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Youds JL, Ward JD, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundred R, Myers K, Helleday T, et al. Human RECQL5 overcomes thymidine-induced replication stress. DNA Repair (Amst) 2010;9:964–975. doi: 10.1016/j.dnarep.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower JJ, Karaca GF, Zhou Y, et al. Topoisomerase Ilalpha maintains genomic stability through decatenation G(2) checkpoint signaling. Oncogene. 2010;29:4787–4799. doi: 10.1038/onc.2010.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Mazina OM, Mazin AV. Bloom syndrome helicase stimulates RAD51 DNA strand exchange activity through a novel mechanism. J Biol Chem. 2009;284:26349–26359. doi: 10.1074/jbc.M109.029371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RC, Lisby M, Altmannova V, et al. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J Cell Biol. 2009;185:969–981. doi: 10.1083/jcb.200810055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- Carson DR, Christman ME. Evidence that replication fork components catalyze establishment of cohesion between sister chromatids. Proc Natl Acad Sci USA. 2001;98:8270–8275. doi: 10.1073/pnas.131022798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M, Perfettini JL, Roumier T, et al. Cell death by mitotic catastrophe: a molecular definition. Oncogene. 2004;23:2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- Chen Y, Dui W, Yu Z, et al. Drosophila RecQ5 is required for efficient SSA repair and suppression of LOH in vivo. Protein Cell. 2010;1:478–490. doi: 10.1007/s13238-010-0058-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Huang S, Lee L, et al. WRN, the protein deficient in Werner syndrome, plays a critical structural role in optimizing DNA repair. Aging Cell. 2003;2:191–199. doi: 10.1046/j.1474-9728.2003.00052.x. [DOI] [PubMed] [Google Scholar]
- Cheng WH, Muftic D, Muftuoglu M, et al. WRN is required for ATM activation and the S-phase checkpoint in response to interstrand cross-link-induced DNA double-strand breaks. Mol Biol Cell. 2008;19:3923–3933. doi: 10.1091/mbc.E07-07-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng RZ, Murano S, Kurz B, Shmookler Reis RJ. Homologous recombination is elevated in some Werner-like syndromes but not during normal in vitro or in vivo senescence of mammalian cells. Mutat Res. 1990;237:259–269. doi: 10.1016/0921-8734(90)90008-f. [DOI] [PubMed] [Google Scholar]
- Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- Contreras R, Fiers W. Initiation of transcription by RNA polymerase II in permeable, SV40-infected or noninfected, CVI cells; evidence for multiple promoters of SV40 late transcription. Nucleic Acids Res. 1981;9:215–236. doi: 10.1093/nar/9.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- Dhillon KK, Sidorova J, Saintigny Y, et al. Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell. 2007;6:53–61. doi: 10.1111/j.1474-9726.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- Franchitto A, Pichierri P. Werner syndrome protein and the MRE11 complex are involved in a common pathway of replication fork recovery. Cell Cycle. 2004;3:1331–1339. doi: 10.4161/cc.3.10.1185. [DOI] [PubMed] [Google Scholar]
- Franchitto A, Pirzio LM, Prosperi E, et al. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J Cell Biol. 2008;183:241–252. doi: 10.1083/jcb.200803173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia PL, Liu Y, Jiricny J, et al. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Rossi ML, Aulds J, et al. Telomeric D-loops containing 8-oxo-2’-deoxyguanosine are preferred substrates for Werner and Bloom syndrome helicases and are bound by POT1. J Biol Chem. 2009;284:31074–31084. doi: 10.1074/jbc.M109.027532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez RE, Lim CU, Cole K, et al. Effects of conditional depletion of topoisomerase II on cell cycle progression in mammalian cells. Cell Cycle. 2011;10:3505–3514. doi: 10.4161/cc.10.20.17778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta SL, Miller OL, Jr, French SL. rRNA transcription rate in Escherichia coli . J Bacteriol. 1991;173:6647–6649. doi: 10.1128/jb.173.20.6647-6649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottipati P, Cassel TN, Savolainen L, Helleday T. Transcription-associated recombination is dependent on replication in mammalian cells. Mol Cell Biol. 2008;28:154–164. doi: 10.1128/MCB.00816-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottipati P, Helleday T. Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination. Mutagenesis. 2009;24:203–210. doi: 10.1093/mutage/gen072. [DOI] [PubMed] [Google Scholar]
- Harrigan JA, Wilson DM, III, Prasad R, et al. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase beta. Nucleic Acids Res. 2006;34:745–754. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heather MO-B, Cheryl LT, Sarah P, et al. A RecQ protein-like 5 haplotype is associated with colon cancer. Gastroenterol Res. 2010;3:101–105. doi: 10.4021/gr2010.06.214w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeres JT, Hergenrother PJ. Poly(ADP-ribose) makes a date with death. Curr Opin Chem Biol. 2007;11:644–653. doi: 10.1016/j.cbpa.2007.08.038. [DOI] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Hirose S, Hiraga S, Okazaki T. Initiation site of deoxyribonu-cleotide polymerization at the replication origin of the Escherichia coli chromosome. Mol Gen Genet. 1983;189:422–431. doi: 10.1007/BF00325904. [DOI] [PubMed] [Google Scholar]
- Hu Y, Lu X, Barnes E, et al. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol Cell Biol. 2005;25:3431–3412. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Lu X, Luo G. Effect of Recql5 deficiency on the intestinal tumor susceptibility of Apc(min) mice. World J Gastroenterol. 2010;16:1482–1486. doi: 10.3748/wjg.v16.i12.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Lu X, Zhou G, et al. Recql5 plays an important role in DNA replication and cell survival after camptothecin treatment. Mol Biol Cell. 2009;20:114–123. doi: 10.1091/mbc.E08-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Raynard S, Sehorn MG, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, Bender MA. Effects of inhibitors of DNA synthesis on spontaneous and ultraviolet light-induced sister-chromatid exchanges in Chinese hamster cells. Mutat Res. 1980;79:19–32. doi: 10.1016/0165-1218(80)90144-5. [DOI] [PubMed] [Google Scholar]
- Islam MN, Fox D, III, Guo R, et al. RecQL5 promotes genome stabilization through two parallel mechanisms - interacting with RNA polymerase II and acting as a helicase. Mol Cell Biol. 2010;30:2460–2472. doi: 10.1128/MCB.01583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MN, Paquet N, Fox D, III, et al. A variant of the breast cancer type 2 susceptibility protein (BRC) repeat is essential for the RECQL5 helicase to interact with RAD51 recombinase for genome stabilization. J Biol Chem. 2012;287:23808–23818. doi: 10.1074/jbc.M112.375014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa K, Yanagida M, Hayano T, et al. Association of human DNA helicase RecQ5beta with RNA polymerase II and its possible role in transcription. Biochem J. 2008;413:505–516. doi: 10.1042/BJ20071392. [DOI] [PubMed] [Google Scholar]
- Jaspers NG, Raams A, Silengo MC, et al. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am J Hum Genet. 2007;80:457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YS, Kang Y, Lim KH, et al. Deficiency of Caenorhabditis elegans RecQ5 homologue reduces life span and increases sensitivity to ionizing radiation. DNA Repair (Amst) 2003;2:1309–1319. doi: 10.1016/j.dnarep.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Jeong SM, Kawasaki K, Juni N, Shibata T. Identification of Drosophila melanogaster RECQE as a member of a new family of RecQ homologues that is preferentially expressed in early embryos. Mol Gen Genet. 2000;263:183–193. doi: 10.1007/s004380051159. [DOI] [PubMed] [Google Scholar]
- Kanagaraj R, Huehn D, MacKellar A, et al. RECQ5 helicase associates with the C-terminal repeat domain of RNA polymerase II during productive elongation phase of transcription. Nucleic Acids Res. 2010;38:8131–8110. doi: 10.1093/nar/gkq697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagaraj R, Saydam N, Garcia PL, et al. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe T, Tsuyama N, Kitao S, et al. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene. 2000;19:4764–4772. doi: 10.1038/sj.onc.1203841. [DOI] [PubMed] [Google Scholar]
- Kitao S, Ohsugi I, Ichikawa K, et al. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- Kizer KO, Phatnani HP, Shibata Y, et al. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster DA, Palle K, Bot ES, et al. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- Laud PR, Multani AS, Bailey SM, et al. Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev. 2005;19:2560–2570. doi: 10.1101/gad.1321305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel M. Increased frequency of DNA deletions in pink-eyed unstable mice carrying a mutation in the Werner syndrome gene homologue. Carcinogenesis. 2002;23:213–216. doi: 10.1093/carcin/23.1.213. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li M, Xu X, Liu Y. The SET2-RPB1 interaction domain of human RECQ5 is important for transcription-associated genome stability. Mol Cell Biol. 2011;31:2090–2099. doi: 10.1128/MCB.01137-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeken M, Bikel I, Livingston DM, Brady J. trans-Activation of RNA polymerase II and III promoters by SV40 small t antigen. Cell. 1988;55:1171–1177. doi: 10.1016/0092-8674(88)90261-9. [DOI] [PubMed] [Google Scholar]
- Lu X, Lou H, Luo G. A Blm-Recql5 partnership in replication stress response. J Mol Cell Biol. 2011;3:31–38. doi: 10.1093/jmcb/mjq056. [DOI] [PubMed] [Google Scholar]
- Mirabello L, Yu K, Kraft P, et al. The association of telomere length and genetic variation in telomere biology genes. Hum Mutat. 2010;31:1050–1058. doi: 10.1002/humu.21314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, Kawasaki K, Matsumoto K, Shibata T. The possible roles of the DNA helicase and C-terminal domains in RECQ5/QE: complementation study in yeast. DNA Repair (Amst) 2004;3:369–378. doi: 10.1016/j.dnarep.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Maruyama S, Kanda H, et al. Relationships of Drosophila melanogaster RECQ5/QE to cell-cycle progression and DNA damage. FEBS Lett. 2006;580:6938–6912. doi: 10.1016/j.febslet.2006.11.059. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Yamaguchi S, Sagisu Y, et al. Loss of RecQ5 leads to spontaneous mitotic defects and chromosomal aberrations in Drosophila melanogaster . DNA Repair (Amst) 2009;8:232–231. doi: 10.1016/j.dnarep.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Parker D, Ferreri K, Nakajima T, et al. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro BS, Frohlich R, Bohr VA, Stevnsner T. WRN helicase regulates the ATR-CHKl-induced S-phase checkpoint pathway in response to topoisomerase-I-DNA covalent complexes. J Cell Sci. 2011;124:3967–3979. doi: 10.1242/jcs.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellatt AJ, Wolff RK, Lundgreen A, et al. Genetic and lifestyle influence on telomere length and subsequent risk of colon cancer in a case control study. Int J Mol Epidemiol Genet. 2012;3:184–194. [PMC free article] [PubMed] [Google Scholar]
- Pflumm MF, Botchan MR. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development. 2001;128:1697–1707. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]
- Pichierri P, Franchitto A, Mosesso P, Palitti F. Werner’s syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol Biol Cell. 2001;12:2412–2421. doi: 10.1091/mbc.12.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike AC, Shrestha B, Popuri V, et al. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc Natl Acad Sci USA. 2009;106:1039–1014. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirzio LM, Pichierri P, Bignami M, Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J Cell Biol. 2008;180:305–314. doi: 10.1083/jcb.200705126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popuri V, Huang J, Ramamoorthy M, et al. RECQL5 plays cooperative and complementary roles with WRN syndrome helicase. Nucleic Acids Res. 2012;41:881–899. doi: 10.1093/nar/gks1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popuri V, Ramamoorthy M, Tadokoro T, et al. Recruitment and retention dynamics of RECQL5 at DNA double strand break sites. DNA Repair (Amst) 2012;11:624–635. doi: 10.1016/j.dnarep.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan P, Dhamankar V, Rundell K, Thimmapaya B. Simian virus 40 small-t does not transactivate RNA polymerase II promoters in virus infections. J Virol. 1991;65:6553–6561. doi: 10.1128/jvi.65.12.6553-6561.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy M, Tadokoro T, Rybanska I, et al. RECQL5 cooperates with Topoisomerase II alpha in DNA decatenation and cell cycle progression. Nucleic Acids Res. 2012;40:1621–1635. doi: 10.1093/nar/gkr844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- Ren H, Dou SX, Zhang XD, et al. The zinc-binding motif of human RECQ5beta suppresses the intrinsic strand-annealing activity of its DExH helicase domain and is essential for the helicase activity of the enzyme. Biochem J. 2008;412:425–433. doi: 10.1042/BJ20071150. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Jackson DA, Iborra F, Cox LS. Asymmetry of DNA replication fork progression in Werner’s syndrome. Aging Cell. 2002;1:30–39. doi: 10.1046/j.1474-9728.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Jackson DA, Nehlin JO, et al. Characterisation of the interaction between WRN, the helicase/ exonuclease defective in progeroid Werner’s syndrome, and an essential replication factor, PCNA. Mech Ageing Dev. 2003;124:167–174. doi: 10.1016/s0047-6374(02)00131-8. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Whitby MC, Borer CM, et al. Correction of proliferation and drug sensitivity defects in the progeroid Werner’s Syndrome by Holliday junction resolution. Rejuvenation Res. 2007;10:27–10. doi: 10.1089/rej.2006.0503. [DOI] [PubMed] [Google Scholar]
- Royle NJ, Mendez-Bermudez A, Gravani A, et al. The role of recombination in telomere length maintenance. Biochem Soc Trans. 2009;37:589–595. doi: 10.1042/BST0370589. [DOI] [PubMed] [Google Scholar]
- Rudolph CJ, Dhillon P, Moore T, Lloyd RG. Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair (Amst) 2007;6:981–993. doi: 10.1016/j.dnarep.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Saintigny Y, Makienko K, Swanson C, et al. Homologous recombination resolution defect in werner syndrome. Mol Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H, Okado M, Ito F, Kawasaki K. Anaphase DNA bridges induced by lack of RecQ5 in Drosophila syncytial embryos. FEBS Lett. 2011;585:1923–1928. doi: 10.1016/j.febslet.2011.04.074. [DOI] [PubMed] [Google Scholar]
- Salk D, Au K, Hoehn H, Martin GM. Cytogenetics of Werner’s syndrome cultured skin fibroblasts: variegated translocation mosaicism. Cytogenet Cell Genet. 1981;30:92–107. doi: 10.1159/000131596. [DOI] [PubMed] [Google Scholar]
- Saunders RD, Boubriak I, Clancy DJ, Cox LS. Identification and characterization of a Drosophila ortholog of WRN exonuclease that is required to maintain genome integrity. Aging Cell. 2008;7:418–425. doi: 10.1111/j.1474-9726.2008.00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurman SH, Hedayati M, Wang Z, et al. Direct and indirect roles of RECQL4 in modulating base excision repair capacity. Hum Mol Genet. 2009;18:3470–3483. doi: 10.1093/hmg/ddp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendener S, Raynard S, Paliwal S, et al. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J Biol Chem. 2010;285:15739–15715. doi: 10.1074/jbc.M110.110478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Phatak P, Stortchevoi A, et al. RECQ1 plays a distinct role in cellular response to oxidative DNA damage. DNA Repair (Amst) 2012;11:537–549. doi: 10.1016/j.dnarep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto A, Nishikawa K, Kitao S, Furuichi Y. Human RecQ5beta, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3alpha and 3beta. Nucleic Acids Res. 2000;28:1647–1655. doi: 10.1093/nar/28.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Karmakar P, Aamann M, et al. The involvement of human RECQL4 in DNA double-strand break repair. Aging Cell. 2010;9:358–371. doi: 10.1111/j.1474-9726.2010.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV. Establishment of sister chromatid cohesion. Curr Biol. 2009;19:R1126–R1132. doi: 10.1016/j.cub.2009.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speina E, Dawut L, Hedayati M, et al. Human RECQL5beta stimulates flap endonuclease 1. Nucleic Acids Res. 2010;38:2904–2916. doi: 10.1093/nar/gkp1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely AM, Bleichert F, Numann A, et al. Werner protein protects nonproliferating cells from oxidative DNA damage. Mol Cell Biol. 2005;25:10492–10506. doi: 10.1128/MCB.25.23.10492-10506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T, Ramamoorthy M, Popuri V, et al. Human RECQL5 participates in the removal of endogenous DNA damage. Mol Biol Cell. 2012;23:4273–4285. doi: 10.1091/mbc.E12-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uringa EJ, Youds JL, Lisaingo K, et al. RTEL1: an essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res. 2011;39:1647–1655. doi: 10.1093/nar/gkq1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, et al. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- Venters BJ, Pugh BE. How eukaryotic genes are transcribed. Crit Rev Biochem Mol Biol. 2009;44:117–111. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu X, Zhou G, et al. RECQL5 is an important determinant for camptothecin tolerance in human colorectal cancer cells. Biosci Rep. 2011;31:363–369. doi: 10.1042/BSR20100108. [DOI] [PubMed] [Google Scholar]
- Wang W, Seki M, Narita Y, et al. Functional relation among RecQ family helicases RecQLl, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol Cell Biol. 2003;23:3527–3535. doi: 10.1128/MCB.23.10.3527-3535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Chan KL, Ralf C, et al. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Jiang X, Lee WH, Chen PL. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen Breakage Syndrome 1. Cancer Res. 2003;63:2589–2595. [PubMed] [Google Scholar]
- Yamagata K, Kato J, Shimamoto A, et al. Bloom’s and Werner’s syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc Natl Acad Sci USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Locovei AM, D’Urso G. Activation of the DNA damage checkpoint in mutants defective in DNA replication initiation. Mol Biol Cell. 2008;19:4374–4382. doi: 10.1091/mbc.E08-01-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Mets DG, Mcllwraith MJ, et al. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327:1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Kanagaraj R, Mihaljevic B, et al. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 2009;37:2645–2657. doi: 10.1093/nar/gkp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Liu Y, Wu SY, et al. Purification of a novel RECQL5-SWI/SNF-RNAPII super complex. Int J Biochem Mol Biol. 2010;1:101–111. [PMC free article] [PubMed] [Google Scholar]