Abstract
Brain metastasis is a major complication of non-small cell lung cancer (NSCLC) and leads to most of the mortality of this disease. However, the biological mechanisms and molecular features in brain metastasis of NSCLC are poorly understood. In the present study, we compared whole-genome copy number variations (CNVs) between a primary lung adenocarcinoma and secondary metastatic brain lesion from the same patient using array comparative genomic hybridization (aCGH). The number of CNV regions was markedly higher in the secondary metastatic tumor than the primary tumor in the lung. In detail, the common CNVs in both tumors included gains of 7p22, 7p12-p11, 7q11, 7q22, 21q22, and 19q13; gains of 1p33-p34, 1q22, 5p13 and 14q11 whereas losses of 3p, 4q31, 5q, 11p15, Xp21-p22 and Xq21 were identified only in the secondary lesion. Gene Ontology enrichment analysis revealed that the genes with amplified copy numbers in both tumors were related to such processes as DNA replication and mismatch repair. Genes only amplified in the metastatic tumor were enriched in processes that include leukocyte migration and organ development, and genes with a lower copy number in the secondary tumor included the processes of proteolysis regulation, negative regulation of cell proliferation and cell adhesion. These findings provided new insight into the genomic mechanism of the spread of lung adenocarcinoma to the brain, and the candidate genes identified serve as novel indicators or putative targets in NSCLC brain metastasis.
Keywords: non-small cell lung cancer, brain metastasis, copy number variation, array comparative genomic hybridization
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide (1), with a majority of the histologic subtype of non-small cell lung cancer (NSCLC). Approximately 33% of all NSCLC patients suffer from brain metastasis (2), which is a major complication accounting for the mortality of patients (3). Indeed, the incidence proportion (IP) percentage of brain metastases from single primary lung cancer was reported to be 19.9%, the highest propensity of all malignant tumors (4). The median time to brain relapse after surgical resection of locally advanced NSCLC is 7.5–9.3 months (5–7). The prognosis of patients with brain metastases of NSCLC is remarkably poor, with a median survival time of 1–2 months for untreated patients and 6 months for those receiving surgery, radiotherapy and chemotherapy (8,9). Therefore, an understanding of the biological characteristics and molecular mechanisms of NSCLC metastasis to the brain is urgently needed for the better prevention or treatment of this disease.
Several genes or genetic alterations have been reported to be involved in brain metastasis of NSCLC. The expression of miR-145 was found to be downregulated in brain metastatic tumors compared to its primary lung adenocarcinoma (10), and chemokine CXCL12 and its receptor CXCR4 appear to play a role in the development of NSCLC brain metastasis (11). An immunohistochemistry analysis revealed that the expression of integrin αvβ3 was significantly increased in brain metastases compared to primary NSCLC (12). The S100B (13) and matrix metalloproteinase-2 (MMP-2) (14) proteins are also likely involved in NSCLC brain metastasis.
Furthermore, DNA copy number variations (CNVs) have also been found to be significant in NSCLC brain metastasis. A recent study reported that FGFR1 amplification in brain metastasis of lung adenocarcinoma (ADC) is 15.3%, 5-fold more frequent than primary tumors (15). Using Molecular Inversion Probe (MIP) technology, Lee et al (16) identified that amplifications of 5q35, 10q23 and 17q23-24 in primary lung ADC are significantly correlated with brain metastasis. To the best of our knowledge, CNV scanning based on a high-resolution methodology such as array comparative genomic hybridization (aCGH) in brain metastasis and primary NSCLC has not yet been reported.
In the present study, we compared the CNV characteristics of a primary lung tumor and brain metastasis in the same patient diagnosed with lung ADC utilizing the aCGH method.
Patient and methods
Patient
The patient was a 48-year-old female diagnosed with middle lobe of the right lung adenocarcinoma in 2010 January at China-Japan Union Hospital, Jilin University, Changchun, China. She then underwent lung tumor resection surgery. The tumor dimensions were 4.0×3.5×3.0 cm and TNM stage was T2N1M0, IIB. Chemotherapy with the combination of carboplatin and docetaxel was administered at three months after right lobectomy, when the patient condition was suitable for adjuvant treatment. After chemotherapy, the patient received prophylactic granulocyte colony-stimulating factor 2 (G-CSF) supportive therapy at each cycle on days 2, 3 and 4.
In December 2010, this patient was admitted again to hospital for a headache and was diagnosed as 'brain metastasis of lung cancer' by Magnetic Resonance Imaging (MRI). She underwent brain tumor resection; the tumor dimensions were 3.5×2.5×2 cm. Radiotherapy was also administered after the brain tumor resection. The patient's condition to date is optimal. Regular radiological examination and blood testing were performed, with confirmed evidence indicating a progression-free stage.
DNA extraction and aCGH microarray analysis
Samples were collected from 5-μm thick formalin-fixed paraffin-embedded (FFPE) sections from both the primary and secondary lesions. Genomic DNA (gDNA) was extracted from the cells according to the G4410-90020_CGH_Protocol_FFPE1 from Agilent. The concentrations and purities were precisely measured using the NanoDrop ND-1000 UV-VIS spectrophotometer.
Microarray comparative genomic hybridization was performed using the human 4×44k Agilent Oligo aCGH microarray according to the manufacturer's protocol. In brief, Agilent Oligo aCGH Labeling kit was used to label the tumor DNA and reference DNA with Cy-5 and Cy-3, respectively. The labeled DNA samples were mixed and hybridized with the aCGH microarray at 65°C and 20 rpm for 40 h. The microarray slide was scanned with an Agilent scanner, and Feature Extraction software was used to extract and normalize the raw image data. The final data of log10 ratio (tumor/reference) was generated for the subsequent analysis.
aCGH microarray data analysis
The human genome data, hg18 version was used for annotation mapping. The whole-genome (except chromosome Y) log10 ratio (tumor/reference) value was smoothed, and abnormal copy number regions were detected using the circular binary segmentation algorithm (17,18) with the R package 'DNAcopy' from Bioconductor. The segments with average segmentation values of 0.2/−0.2 were defined as gain/loss copy number regions, respectively. 'Circos' software (19) or R package 'SNPchip' was used to plot the chromosome cytoband and corresponding DNA copy number data. The genes in the gain/loss copy number regions were compared between the primary and secondary lesions. The test for gene overlap between different gene sets was performed by the hypergeometric distribution test. The Gene Ontology (GO) term enrichment significance of the gene sets of interest was estimated by the hypergeometric distribution test, and the false discovery rate (FDR) was calculated using the Benjamini and Hochberg method (20); FDR<0.05 was the significance cut-off.
Immunohistochemistry (IHC)
IHC staining was performed using a standard streptavidin-biotin-peroxidase complex method. The sections (5 mm-thick) were incubated at 4°C in a humid chamber overnight with mouse monoclonal antibody against human epidermal growth factor receptor (EGFR; ready to use, catalog MAB-0196; Maixin Fuzhou, China), rabbit monoclonal antibody against short in-frame deletions in exon 19 (E746-A750del Specific) of EGFR (1:100, catalog #5747), and rabbit monoclonal antibody against L858R mutant in 21 exon of EGFR (1:100, catalog #3197) (both from Cell Signaling Technology, Boston, MA, USA). Phosphate-buffered saline (PBS) instead of primary antibody was used as negative control. The nuclei were counterstained with hematoxylin. IHC results were reviewed by two experienced pathologists, independently.
Results
Overall DNA CNVs of primary and secondary tumor lesions
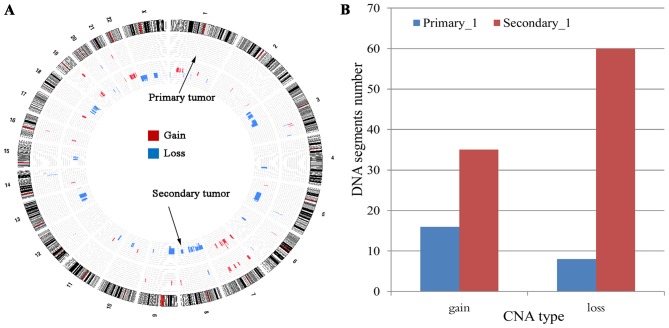
As shown in Fig. 1A, the DNA copy number alteration pattern of the secondary tumor lesion was quite different from that of the primary tumor. Many DNA copy number alteration regions (gain or loss) present in the secondary tumor were not found in the primary tumor. Specifically, 16 gains in DNA copy number variation regions (segments) and 8 loss regions were detected in the primary tumor. In contrast, the number of gains and losses of CNV regions found in the secondary tumor were 35 and 60, respectively (Fig. 1B). The increased CNV region number suggests a much greater genomic instability or dysregulation of biological functions in the secondary lesion.
Figure 1.
Global view of DNA copy number variation patterns in primary and secondary tumors. (A) Genomic visualization of CNV patterns in the two tumor lesions. The outer circle indicates chromosome 1 to chromosome X, the middle circle represents the DNA copy number alteration of the primary tumor, and the inner circle represents the DNA copy number alteration of the secondary tumor. The colored blocks (red for gain, blue for loss) indicate the average log10 ratio value (only showing those reaching the cut-off 0.2) after segmentation by the circular binary segmentation algorithm. (B) Statistics of altered DNA regions (segments) between the primary and secondary tumors. The x-axis presents the CNV type, and the y-axis indicates the altered DNA region numbers. CNV, copy number variations.
The CNV gain regions in the primary tumor include 3q, 6p21, 7p, 7q, 8q, 9p, 9q, 14q32, 16q12, 19q and 21q22. Such DNA regions as 3q22-q24, 5q32, 6q25, 8p22 12q13, 14q24 and 20q13 were found to be lost in the primary tumor. With regard to the secondary tumor, the gain regions include 1p33-p34, 1q22, 2p11, 2q35, 5p13, 6q15, 7p, 7q, 9q22, 10q11, 11p12-p11, 14q11, 16p12-p11, 17q, 19q and 22q; and the loss regions include 1p31.1, 1q41, 3p, 4q31, 5q, 6q12, 8q, 8q, 9p, 10q21-q24, 11p15, 13q, 15q14, 15q21, 18q, 20q11 and 20q13. The common gain regions found in both the primary and secondary tumors include 7p22, 7p12-p11.2, 7q11, 7q22, 21q22 and 19q13.3.
Genes in CNV regions from the two tumors
We also analyzed the genes in the CNV regions in both the primary and secondary tumors (Fig. 2A). For the primary tumor, the 16 gained CNV regions covered 351 genes. In addition, 1,198 genes were found in the CNV gain regions of the secondary tumor, with 249 genes overlapping with those of the primary tumor (hypergeometric test, P<0.001). Similarly, 27 genes were found in the lost CNV regions of the primary tumor, with 561 genes in those of the secondary tumor. The 12 overlapping genes between the lost regions from the two tumor lesions were not random (hypergeometric test, P<0.001). Importantly, the EGFR gene, located at 7p12 was amplified in both the primary and secondary tumors (Fig. 2B).
Figure 2.
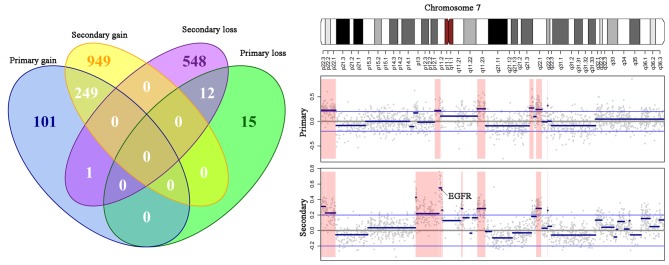
Gene comparison in DNA copy number variation regions. (A) Venn diagram presenting gene numbers of different gene sets derived from the gain/loss regions of the primary/secondary tumors. (B) DNA copy number alteration patterns for chromosome 7. The upper panel presents the cytogenetic bands of chromosome 7. The middle and lower panels indicate the CNV data of the primary and secondary tumors, respectively. For the CNV data, the x-axis indicates the genomic positions, y-axis indicates the log10 ratio value, grey dots indicate the raw value of each microarray probe, blue thick indicates the DNA segments detected by the circular binary segmentation algorithm, blue thin lines indicate the cut-off 0.2, and red shadows indicate the DNA regions with altered copy numbers. CNV, copy number variations.
According to the results of the GO functional terms enrichment analysis, the 249 genes overlapping between the gain regions of the primary and secondary tumors were significantly (FDR <0.05) related to DNA replication, mismatch and nucleotide excision repairs, and regulation of apoptosis. The overlapping gene set in the loss regions did not reach statistical significance in the GO analysis, which may be due to the small gene number (only 12 genes). Importantly, the genes showing gain in copy number in the secondary but not in the primary tumor were significantly (FDR <0.05) enriched in GO processes as leukocyte migration, lipoprotein metabolic and organ development. In contrast, genes related to processes such as proteolysis and negative regulation of cell proliferation, cell adhesion and cell-matrix adhesion exhibited a loss in copy number only in the primary tumor.
EGFR expression in primary tumor in lung and secondary metastasis in brain
EGFR staining was predominantly located in cytoplasm and membrane of tumor cells. Overexpression of EGFR was detected both in primary tumor in lung and secondary metastasis tumor in brain (Fig. 3B and F). Furthermore, secondary tumor showed stronger staining of E746-A750del specific short in-frame deletions in 19 exon of EGFR than in primary tumor (Fig. 3C and G). L858R mutant in 21 exon of EGFR was not detectable in either the primary or the secondary tumors (Fig. 3D and H).
Figure 3.
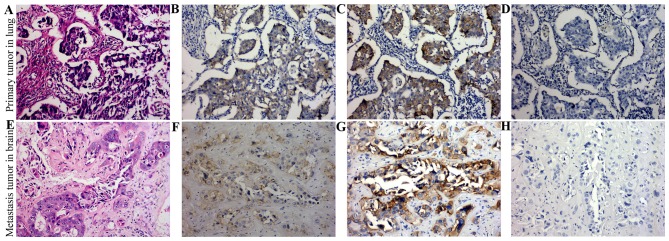
Expression of EGFR protein in formalin-fixed and paraffin-embedded tissues by IHC staining. (A and E) H&E staining of primary tumor in lung and secondary metastasis tumor in the brain. (B and F) Overexpression of EGFR was detected both in primary tumor in lung and secondary metastasis tumor in the brain. (C and G) Secondary tumor showed stronger staining of E746-A750del specific short in-frame deletions in 19 exon of EGFR than in primary tumor. (D and H) L858R mutant in 21 exon of EGFR was not detectable in either the primary or the secondary tumors. EGFR, epidermal growth factor receptor; IHC, immunohistochemistry.
Discussion
Brain metastasis is one of the most frequent complications of NSCLC, accounting for the major mortality in this disease. Nonetheless, the complex molecular events involved in this process are poorly understood. In the present study, we compared DNA copy number variations (CNVs) between a tumor from the primary site and a secondary brain metastasis lesion from the same patient diagnosed with lung ADC using aCGH technology. Compared with the primary tumor, an increase in the number of DNA CNV regions was observed in the secondary tumor, indicating enhanced genome instability during the evolution of brain metastasis. Our analysis of Gene Ontology (GO) terms revealed that the CNV regions identified only in the secondary tumor included genes related to leukocyte migration, organ development, cell adhesion, and extracellular matrix compounds.
CNV is a common genomic alteration that may contribute to tumor initiation and the progression in a variety of cancers, including NSCLC. In the lung ADC case of the present study, the primary tumor and the heterochronous brain relapse lesion showed several common amplified CNV regions, including 7p22, 7p12-p11.2, 7q11, 7q22, 19q13.3 and 21q22. This observation suggests that the primary and secondary tumors likely shared the same ancestor lineage during progression of the tumor. Importantly, the amplification of 7p12 in NSCLC was previously reported, and a gain of copy number of the EGFR gene in this region was a predictor for a patient to benefit from treatment with tyrosine kinase inhibitors (TKIs) (21). Our IHC results showed that the EGFR protein was upregulated in both primary and secondary tumor. We further investigated the expression condition of two major EGFR mutation proteins, E746-A750del specific short in-frame deletions in 19 exon and L858R mutant in 21 exon. For this patient, the 19 exon E746-A750del mutation protein was found significantly overexpression in the metastatic tumor.
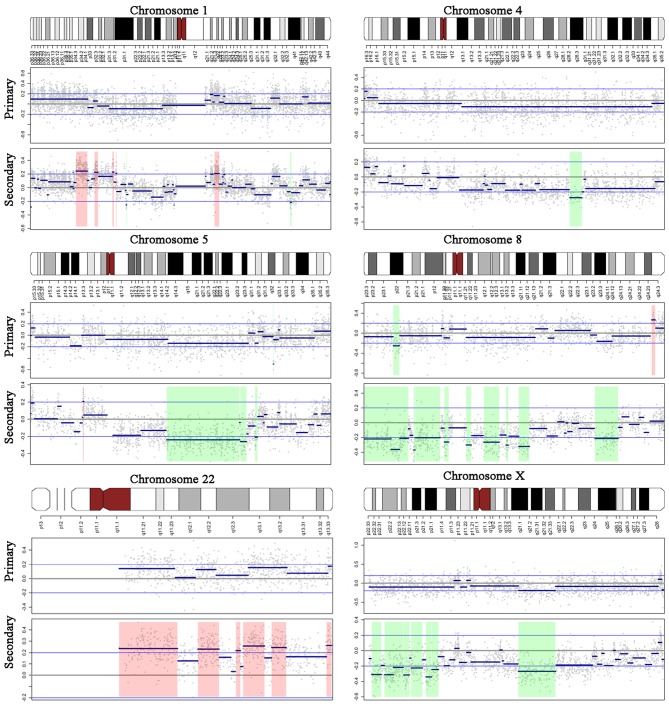
The amplified regions only present in the secondary tumor included 1p33-p34, 1q22, 5p13 and 14q11. Our data showed 4q28-q31 loss in the secondary lesion but not in the primary tumor (Fig. 4), which was consistent with a previous study indicating that 4q deletion is associated with the distant metastatic propensity of lung cancer (22). In addition, the secondary tumor exhibited Xp21-22 and Xq21 loss compared with the primary tumor, which may be an explanation for the observation of more frequent brain metastasis in female patients with lung cancer than males (4).
Figure 4.
Different DNA copy number variations in the primary and secondary tumors. DNA copy number alteration patterns for chromosome 1, 4, 5, 8, 22 and X. The upper panel represents the cytogenetic bands of each chromosome. The middle and lower panels indicate the CNV data of the primary and secondary tumors, respectively. For the CNV data, the x-axis indicates genomic positions, y-axis indicates the log10 ratio value, grey dots indicate the raw value of each microarray probe, blue thick lines indicate the DNA segments detected by the circular binary segmentation algorithm, blue thin lines indicate the cut-off 0.2, and red shadows represent the DNA regions with altered copy numbers. CNV, copy number variations.
According to our data, leukocyte migration-related genes were enriched in the regions showing gain in DNA copy number only in the metastatic tumor, indicating the involvement of leukocyte-related molecules in brain dissemination of NSCLC. Cytokines and chemokines play significant roles in regulating the tumor-leukocyte interaction (23,24), which contributes to the migration and invasion of cancer cells. In addition, the upregulation of the chemokine-receptor of CXCL12/CXCR4 was observed in NSCLC brain metastatic tumors compared with the primary lesion (11). The percentage of TH17 cells and IL-17 levels in peripheral blood was found to be increased in patients with brain metastases of lung cancer, suggesting a potential involvement of TH17 cells in lung metastasis to the brain (25).
Degradation and remodeling of the extracellular matrix play important roles in the invasiveness and metastatic capacity of cancer cells to metastasize (26). For example, high expression of matrix metalloproteinase-2 (MMP-2) was significantly correlated with the central nervous system metastasis of lung cancer (14). In the present study, a loss in DNA copy number of serpin family members, which are negative regulators of proteolysis occurred in the brain metastatic tumor. Consistent with our findings, a recent study (11) reported that SERPINB2 (of the serpin family) inhibited the metastatic capacity of cancer cells (27). Accordingly, cell adhesion genes (such as collagens of COL9A1, COL19A1 and protocadherins of PCDH10 and PCDH20) also showed loss in DNA copy number in the secondary tumor in the brain. These observations indicate that the promotion of extracellular degradation and suppression of the expression of cell adherin molecules are critical steps for the development of brain metastasis of lung cancer. However, the decrease in copy number of CDH2 and POSTN in the metastatic tumor of our case is not in agreement with other studies showing that CDH2 (N-cadherin) is a positive marker for metastatic spread to the brain (28) and that POSTIN upregulation is associated with the progression and poor outcome of NSCLC (29). The exact mechanism of the extracellular balance of NSCLC metastasis to the brain needs to be studied further.
The dysregulation of rapid cell proliferation is one of the characteristics of cancer. Indeed, a high level of cell division contributes to poor differentiation, aggressive progression and local or distant relapse of NSCLC. According to our results, genes related to the negative regulation of cell proliferation (APC, PTEN, CDKN2A and CDKN2B) and induction of apoptosis (FAS and IFNA2) were identified in the lost regions of the secondary tumor in the brain. APC (30) and PTEN (31) are well-known tumor suppressor genes that regulate the cell cycle to prevent cancer cells from uncontrolled rapid division. Consistently, loss of heterozygosity (LOH) of the APC gene has been observed in brain metastases originating from the lung or colon (32). The loss of PTEN DNA copy number was reported to be associated with the poor prognosis of NSCLC, with or without gefitinib treatment (33). Collectively, these data suggest that the suppression of cell cycle inhibitors and apoptosis inducers is involved in the brain metastasis process of NSCLC.
In summary, the present study compared CNVs of a brain metastasis and primary lung adenocarcinoma from the same patient. Several CNVs were identified only in the metastatic lesion, including gains of 1p33-p34, 1q22, 5p13 and 14q11 and; loss of 3p, 4q31, 5q, 11p15, Xp21-22 and Xq21. These findings provide new insight into the genomic mechanism of lung adenocarcinoma spread to the brain, and these candidate genes may serve as novel indicators or putative targets in the brain metastasis of NSCLC.
References
- 1.Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N Engl J Med. 2004;350:379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 2.Quint LE, Tummala S, Brisson LJ, Francis IR, Krupnick AS, Kazerooni EA, Iannettoni MD, Whyte RI, Orringer MB. Distribution of distant metastases from newly diagnosed non-small cell lung cancer. Ann Thorac Surg. 1996;62:246–250. doi: 10.1016/0003-4975(96)00220-2. [DOI] [PubMed] [Google Scholar]
- 3.Han L, Liang XH, Chen LX, Bao SM, Yan ZQ. SIRT1 is highly expressed in brain metastasis tissues of non-small cell lung cancer (NSCLC) and in positive regulation of NSCLC cell migration. Int J Clin Exp Pathol. 2013;6:2357–2365. [PMC free article] [PubMed] [Google Scholar]
- 4.Barnholtz-Sloan JS, Sloan AE, Davis FG, Vigneau FD, Lai P, Sawaya RE. Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the Metropolitan Detroit Cancer Surveillance System. J Clin Oncol. 2004;22:2865–2872. doi: 10.1200/JCO.2004.12.149. [DOI] [PubMed] [Google Scholar]
- 5.Stuschke M, Eberhardt W, Pöttgen C, Stamatis G, Wilke H, Stüben G, Stöblen F, Wilhelm HH, Menker H, Teschler H, et al. Prophylactic cranial irradiation in locally advanced non-small-cell lung cancer after multimodality treatment: Long-term follow-up and investigations of late neuropsychologic effects. J Clin Oncol. 1999;17:2700–2709. doi: 10.1200/JCO.1999.17.9.2700. [DOI] [PubMed] [Google Scholar]
- 6.Wang SY, Ye X, Ou W, Lin YB, Zhang BB, Yang H. Risk of cerebral metastases for postoperative locally advanced non-small-cell lung cancer. Lung Cancer. 2009;64:238–243. doi: 10.1016/j.lungcan.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Eberhardt W, Wilke H, Stamatis G, Stuschke M, Harstrick A, Menker H, Krause B, Müeller MR, Stahl M, Flasshove M, et al. Preoperative chemotherapy followed by concurrent chemoradiation therapy based on hyperfractionated accelerated radiotherapy and definitive surgery in locally advanced non-small-cell lung cancer: Mature results of a phase II trial. J Clin Oncol. 1998;16:622–634. doi: 10.1200/JCO.1998.16.2.622. [DOI] [PubMed] [Google Scholar]
- 8.Knisely JP, Berkey B, Chakravarti A, Yung AW, Curran WJ, Jr, Robins HI, Movsas B, Brachman DG, Henderson RH, Mehta MP. A phase III study of conventional radiation therapy plus thalidomide versus conventional radiation therapy for multiple brain metastases (RTOG 0118) Int J Radiat Oncol Biol Phys. 2008;71:79–86. doi: 10.1016/j.ijrobp.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Fidler IJ. The role of the organ microenvironment in brain metastasis. Semin Cancer Biol. 2011;21:107–112. doi: 10.1016/j.semcancer.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Zhao C, Xu Y, Zhang Y, Tan W, Xue J, Yang Z, Zhang Y, Lu Y, Hu X. Downregulation of miR-145 contributes to lung adenocarcinoma cell growth to form brain metastases. Oncol Rep. 2013;30:2027–2034. doi: 10.3892/or.2013.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavallaro S. CXCR4/CXCL12 in non-small-cell lung cancer metastasis to the brain. Int J Mol Sci. 2013;14:1713–1727. doi: 10.3390/ijms14011713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogetseder A, Thies S, Ingold B, Roth P, Weller M, Schraml P, Goodman SL, Moch H. αv-Integrin isoform expression in primary human tumors and brain metastases. Int J Cancer. 2013;133:2362–2371. doi: 10.1002/ijc.28267. [DOI] [PubMed] [Google Scholar]
- 13.Pang X, Min J, Liu L, Liu Y, Ma N, Zhang H. S100B protein as a possible participant in the brain metastasis of NSCLC. Med Oncol. 2012;29:2626–2632. doi: 10.1007/s12032-012-0169-0. [DOI] [PubMed] [Google Scholar]
- 14.Rojiani MV, Alidina J, Esposito N, Rojiani AM. Expression of MMP-2 correlates with increased angiogenesis in CNS metastasis of lung carcinoma. Int J Clin Exp Pathol. 2010;3:775–781. [PMC free article] [PubMed] [Google Scholar]
- 15.Preusser M, Berghoff AS, Berger W, Ilhan-Mutlu A, Dinhof C, Widhalm G, Dieckmann K, Wöhrer A, Hackl M, von Deimling A, et al. High rate of FGFR1 amplifications in brain metastases of squamous and non-squamous lung cancer. Lung Cancer. 2014;83:83–89. doi: 10.1016/j.lungcan.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Lee HW, Seol HJ, Choi YL, Ju HJ, Joo KM, Ko YH, Lee JI, Nam DH. Genomic copy number alterations associated with the early brain metastasis of non-small cell lung cancer. Int J Oncol. 2012;41:2013–2020. doi: 10.3892/ijo.2012.1663. [DOI] [PubMed] [Google Scholar]
- 17.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 18.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 19.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series. 1995;57:289–300. [Google Scholar]
- 21.Casorzo L, Corigliano M, Ferrero P, Venesio T, Risio M. Evaluation of 7q31 region improves the accuracy of EGFR FISH assay in non small cell lung cancer. Diagn Pathol. 2009;4:36. doi: 10.1186/1746-1596-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wrage M, Ruosaari S, Eijk PP, Kaifi JT, Hollmén J, Yekebas EF, Izbicki JR, Brakenhoff RH, Streichert T, Riethdorf S, et al. Genomic profiles associated with early micrometastasis in lung cancer: Relevance of 4q deletion. Clin Cancer Res. 2009;15:1566–1574. doi: 10.1158/1078-0432.CCR-08-2188. [DOI] [PubMed] [Google Scholar]
- 23.Lee HJ, Song IC, Yun HJ, Jo DY, Kim S. CXC chemokines and chemokine receptors in gastric cancer: From basic findings towards therapeutic targeting. World J Gastroenterol. 2014;20:1681–1693. doi: 10.3748/wjg.v20.i7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarvaiya PJ, Guo D, Ulasov I, Gabikian P, Lesniak MS. Chemokines in tumor progression and metastasis. Oncotarget. 2013;4:2171–2185. doi: 10.18632/oncotarget.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He G, Zhang B, Zhang B, Qiao L, Tian Z, Zhai G, Xin X, Yang C, Liu P, Zhang Y, et al. Th17 cells and IL-17 are increased in patients with brain metastases from the primary lung cancer. Zhongguo Fei Ai Za Zhi. 2013;16:476–481. doi: 10.3779/j.issn.1009-3419.2013.09.07. In Chinese. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: Drivers of tumour metastasis. Nat Rev Cancer. 2014;14:430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroder WA, Major LD, Le TT, Gardner J, Sweet MJ, Janciauskiene S, Suhrbier A. Tumor cell-expressed SerpinB2 is present on microparticles and inhibits metastasis. Cancer Med. 2014;3:500–513. doi: 10.1002/cam4.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grinberg-Rashi H, Ofek E, Perelman M, Skarda J, Yaron P, Hajdúch M, Jacob-Hirsch J, Amariglio N, Krupsky M, Simansky DA, et al. The expression of three genes in primary non-small cell lung cancer is associated with metastatic spread to the brain. Clin Cancer Res. 2009;15:1755–1761. doi: 10.1158/1078-0432.CCR-08-2124. [DOI] [PubMed] [Google Scholar]
- 29.Hong LZ, Wei XW, Chen JF, Shi Y. Overexpression of periostin predicts poor prognosis in non-small cell lung cancer. Oncol Lett. 2013;6:1595–1603. doi: 10.3892/ol.2013.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su LK, Vogelstein B, Kinzler KW. Association of the APC tumor suppressor protein with catenins. Science. 1993;262:1734–1737. doi: 10.1126/science.8259519. [DOI] [PubMed] [Google Scholar]
- 31.Chu EC, Tarnawski AS. PTEN regulatory functions in tumor suppression and cell biology. Med Sci Monit. 2004;10:RA235–RA241. [PubMed] [Google Scholar]
- 32.Pećina-Šlaus N, Nikuševa Martić T, Zeljko M, Bulat S. Brain metastases exhibit gross deletions of the APC gene. Brain Tumor Pathol. 2011;28:223–228. doi: 10.1007/s10014-011-0030-8. [DOI] [PubMed] [Google Scholar]
- 33.Fidler MJ, Morrison LE, Basu S, Buckingham L, Walters K, Batus M, Jacobson KK, Jewell SS, Coon J, IV, Bonomi PD. PTEN and PIK3CA gene copy numbers and poor outcomes in non-small cell lung cancer patients with gefitinib therapy. Br J Cancer. 2011;105:1920–1926. doi: 10.1038/bjc.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]