Abstract
Biologically related processes operate across multiple spatiotemporal scales. For computational modeling methodologies to mimic this biological complexity, individual scale models must be linked in ways that allow for dynamic exchange of information across scales. A powerful methodology is to combine a discrete modeling approach, agent-based models (ABMs), with continuum models to form hybrid models. Hybrid multi-scale ABMs have been used to simulate emergent responses of biological systems. Here, we review two aspects of hybrid multi-scale ABMs: linking individual scale models and efficiently solving the resulting model. We discuss the computational choices associated with aspects of linking individual scale models while simultaneously maintaining model tractability. We demonstrate implementations of existing numerical methods in the context of hybrid multi-scale ABMs. Using an example model describing Mycobacterium tuberculosis infection, we show relative computational speeds of various combinations of numerical methods. Efficient linking and solution of hybrid multi-scale ABMs is key to model portability, modularity, and their use in understanding biological phenomena at a systems level.
Key Terms: Multi-Scale Modeling, Hybrid Modeling, Agent-Based Modeling, Numerical Implementation, Linking Models, Tuneable Resolution
Introduction
Computational models are used in systems biology for understanding, predicting, and translating a wealth of experimentally generated data into a realization of systems behavior. Multi-scale computational models in particular have provided valuable insights for application to areas as diverse as infectious disease 18,26,27, inflammation 3, cancer 24,82,87, angiogenesis 67, and disease treatment 38,44,58,86. A defining feature of multi-scale computational models is a description of biological mechanisms that operate over different spatiotemporal scales 4,75,76,86,89. When building multi-scale models four different areas must be considered (Figure 1): (1) constructing models – how to create a mathematical formulation that is able to recapitulate the dynamics of a biological system at an individual scale, (2) linking models – how to join individual scale models to allow for exchange of information, (3) solving models – determining the most efficient way to solve the underlying mathematics, and (4) analyzing models – how to calibrate and validate the model and then make and understand model predictions. In this paper, we review strategies to link individual scale models and methods to efficiently solve the resulting multi-scale model.
Figure 1. Considerations for building multi-scale models.
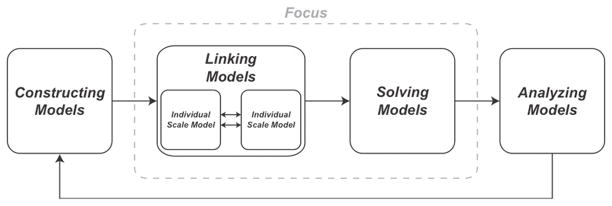
(1) Constructing models: how to create mathematical formulations that accurately represent individual scale dynamics of a biological system, (2) Linking models: how to connect mathematical formulations of individual scale models to create multi-scale models, (3) Solving models: implementing efficient methods to solve multi-scale models, and (4) Analyzing models: Understanding and translating model predictions. Model analysis commonly iterates back to model construction in order to include new biological mechanisms of interest/relevance. In this work, we focus on how to link individual scale models and efficiently solve the resultant multi-scale model.
We typically link descriptions of biological phenomena operating at an individual scale together to form a multi-scale model 48,86. One approach to linking individual scale models, referred to as hierarchical linking, is to solve the lowest scale (i.e. the smallest length or time scale) model to completion and subsequently feed the output, for example in the form of an initial condition or parameter value, to the model at the next higher scale 20,48,86. However, this type of linking methodology is inappropriate when information across scales needs to be continually exchanged and in both directions. For instance, vascular endothelial growth factor (VEGF) can induce an endothelial cell to divide which may elongate a blood vessel in a new direction. The new endothelial cells will produce more VEGF, leading to cellular movement and continued growth of the blood vessel in the new direction 66,67. Without exchange of information in both directions (molecular to cellular and cellular to molecular), sprouting of new blood vessels would likely not occur. The nature of biological systems, with constant exchange of information across scales, necessitates multi-scale models that mimic this dynamic exchange of information. The resulting multi-scale model is more complex than the individual-scale parts, making it more difficult to link, solve, and analyze than traditional mathematical or computational models. Therefore, efficient strategies and approaches that balance model complexity, computational time, and ease of implementation to link and solve multi-scale models are necessary.
Agent-based models (ABMs, sometimes called individual based models – IBMs) are stochastic, discrete models that utilize individual entities known as agents, here representing individual biological cells (Figure 2). Each agent is autonomous and behaves based on decisions from the set of rules, interactions, and states given to it, leading to heterogeneity between agents. ABMs can generate complex system-level emergent behavior from simple rule-based descriptions of each individual 3,11,70,73,86,89. Agents can receive inputs from the environment, influencing their decision making, and can also have the ability to alter their environment. Models that combine aspects of both continuous and discrete model constructs are commonly referred to as hybrid models. Hybrid ABMs arise when continuum models are used to describe part of the overall system, such as the environment and parts of the agent decision-making processes 4,18,26,38,50,89. A hybrid ABM is also termed multi-scale when a portion of the model, such as the continuum models, describes behaviors occurring at a different spatiotemporal scale than that of the ABM.
Figure 2. Mathematical representations of biological processes acting across different spatiotemporal scales.

Individual scale models are combined with exchange of information across scales. (1) An agent-based model (ABM) represents tissue and cellular scales (e.g. cell activation states). (2) Ordinary differential equation models represent molecular scale behaviors associated with cells (e.g. receptor-ligand trafficking and intracellular signaling). (3) Partial differential equation models represent molecular scale behaviors of the environment (e.g. extracellular molecule diffusion). Together these integrated individual scale models form the basis of a hybrid multi-scale agent-based model.

We focus this review on providing strategies, guidelines, and a general framework for developing hybrid multi-scale agent-based models where an agent-based model (discrete), is informed by differential equation models (continuous) operating at a different scale. This review can function as a guide for new modeling efforts (e.g. How are individual scale models be linked? What solution methods should I consider?) and also as a framework for extending existing agent-based models into hybrid multi-scale agent-based models (e.g. What needs to be incorporated into the existing model? What is the easiest way to get it working?).
We focus on the use of continuum models to describe the dynamics of the environment and agent-associated reactions that occur at a smaller spatial and faster time scale and influence agent decision-making processes (Figure 2, Figure 3). These hybrid multi-scale ABMs use a temporally separated approach in which the continuum models are solved using conventional numerical methods on a faster time scale than the ABM; syncing between scales is required to reconcile information exchange 22,85. While many platforms exist for developing ABMs (e.g. NetLogo, Repast, Swarm, SPARK, CHASTE, MASON, and FLAME 5,8,18,22,29,46,72), we have found the flexibility of an in-house platform (written in an object-oriented programming language – C++) necessary to link and solve hybrid multi-scale ABMs. After introducing key modeling concepts, we review how to link individual scale ABMs to differential equation models and efficient implementations of numerical solvers that allow frequent exchange of information across models. Finally, we present a case study in order to demonstrate the trade-off between computational speeds and model complexity using an established hybrid multi-scale ABM of Mycobacterium tuberculosis infection.
Figure 3. Example of how information is exchanged across scales in a hybrid multi-scale agent-based model.

Extracellular molecules in the environment (with diffusion and degradation described using partial differential equations) interact with agents through agent-associated reactions (ordinary differential equations). Based on relative levels of agent-associated species (species A, green and species B, blue) agents make different decisions: (1) if both species A and B are above specified thresholds the agent will die, (2) if only species A (green) is above the specified threshold the agent will proliferate, (3) if only species B (blue) is above the specified threshold the agent will change state, (4) if both species A and B are below specified thresholds the agent will be quiescent. Agent decision logic using thresholds is only one example of how agent-associated reactions can be linked to various dynamics. Other examples include Poisson processes based on agent-associated quantities and rate of change of agent-associated species 26,28,89. Figure style partially adapted from 89.

Examples of Hybrid Multi-Scale ABMs
Hybrid multi-scale ABMs (Figure 2) are being used to describe many biological systems. To orient the reader, we briefly review three such systems: epithelial restitution, growth of brain tumors, and immune responses to bacterial pathogens.
Epithelial restitution
The wound healing response of damaged epithelial cells involves restitution (resealing the epithelial layer), proliferation, and differentiation 78. The cytokines transforming-growth-factor-β (TGF-β) and epidermal growth factor (EGF) are necessary for beginning restitution processes, diffusing through the extracellular matrix (Figure 2C), binding to TGF-β and EGF receptors on endothelial cells, and signaling through SMAD and ERK pathways (Figure 2B) 78. In the ABM, endothelial cells are represented as individual agents (Figure 2A) whose behavior, such as migration and adherence, depends on SMAD and ERK signaling (Figure 3). At the tissue scale, the reconnection of damaged epithelium (restitution) with healthy endothelial is critical to wound healing and constitutes much of the early wound healing response to damaged tissue. This hybrid multi-scale ABM qualitatively matches temporal experimental data and predicts the importance of environmental interactions on the dynamics of epithelial restitution.
Growth patterns of brain tumors
The expression of EGF receptors in brain tumors is associated with rapid growth and invasion. Yet, in growing tumors, cells only display a single phenotype of either migration or proliferation. Transforming-growth-factor-α (TGF-α) diffuses within the extracellular environment (Figure 2C) and binds and dimerizes with EGF receptors, initiating downstream signaling through PLC (Figure 2B). These downstream signaling processes mediate the phenotype of a tumor cell (Figure 3). In the ABM, tumor cells are represented as individual agents (Figure 2A) with both proliferative and migratory potentials determined by levels of PLC and bound EGFR (Figure 3). The proliferative and migratory nature of tumor cells leads to tumor growth and expansion 6,7,88. These hybrid multi-scale ABMs of tumor growth have shown that increased EGF receptor density correlates with tumor expansion based on early phenotypic switching driven by TGF-α autocrine signaling 6,7,88.
Immune response to Mycobacterium tuberculosis
During M. tuberculosis infection the immune system relies on a variety of cells and molecules to coordinate an effective immune response 18,26,28,30,57,70. Two extracellular diffusing molecules of interest are the pro-inflammatory cytokine tumor necrosis factor-α (TNF-α) and the anti-inflammatory cytokine interleukin-10 (IL-10). These cytokines diffuse through the lung tissue (Figure 2C), bind to cell-associated receptors (TNFR1, TNFR2, and IL-10R), and signal through pathways such as NFκB and STAT3 (Figure 2B). Macrophages and T cells are key immune cells, modeled as agents (Figure 2A), with many states (e.g. resting, activated, deactivated) and functions (e.g. bactericidal ability) driven by levels of NFκB and STAT3 (Figure 3). Control of infection relies on the formation of an organized structure of immune cells, known as a granuloma, and its function over the long timescale of infection. Hybrid multi-scale ABMs of M. tuberculosis infection are able to reproduce the emergent phenomenon of granuloma formation and demonstrate a critical balance between TNF-α and IL-10 in controlling granuloma function 18,26,28. In addition, the effects of two first-line antibiotics, rifampicin (RIF) and isoniazid (INH), on bacterial burden have been simulated in a hybrid multi-scale ABM (Pienaar et al. accepted with revision). Antibiotics diffuse through the lung environment (Figure 2C), are taken up by immune cells (Figure 2A), and are able to kill bacteria. We use this hybrid multi-scale ABM describing M. tuberculosis infection as a case study in a later section to illustrate the principles and numerical methods described in this review.
Central Concepts For Hybrid Multi-Scale ABMs
Mathematical Framework and Linking
We describe the elements of a 2-dimensional hybrid multi-scale ABM (Figure 2), but the methodology presented is easily adapted to any dimensionality required. An ABM describes relevant biological cells (or any unit) as individual agents. Each agent (A) has an associated state (V) and position (x, y) that can change with time. Examples of cell agent states include activated, proliferating, infected, and cancerous. Changes in state are based on a set of stochastic agent rules and interactions, and are also influenced by extracellular diffusing molecules in the environment (e.g. ligands that bind to surface receptors on the cell). The construction of an ABM is beyond the scope of this article but is well-described in the literature 1,11,17,29,36,66,69,72,73,78. The concept of agents in an ABM is naturally analogous to objects in an object-oriented programming language, and therefore constructing hybrid multi-scale ABM in a language such as C++ or Java is a logical choice.
For each individual agent, a set of agent-associated reactions can occur:
| (Eqn. 1) |
Eqn. 1 represents the agent-associated species (R total species), where Yr is an agent-associated species, and t is time. L is an extracellular diffusing molecule (e.g. a ligand that binds to a surface receptor) that is modeled on a continuum scale, and β is a vector of rate parameters independent of t and Yr. fr is a function of agent-associated species, diffusing molecules, and rate parameters (e.g as necessary for zeroth, first, or second order reaction kinetics). β is dependent upon the state of the agent (V), and thus rate parameters can change when an agent changes state. Multiple diffusing molecules can be included, although in our examples we will only include one for simplicity. The agent-associated reactions describe reactions occurring in agents, such as cell-cycle proteins controlling proliferation or actin remodeling controlling cellular movement 2,92. More common is a description of receptor-ligand binding and trafficking and ensuing intracellular signaling processes, as in the three examples given above 10,14,28,49,83. These reactions are typically based on mass-action kinetics 51. A simple example of receptor-ligand binding, trafficking, and intracellular signaling reactions for a single agent is given in Table 1.
Table 1.
Example of Agent-Associated Reactions: Receptor-Ligand Binding, Trafficking, and Intracellular Signaling
| Description | Equation | |
|---|---|---|
| Diffusing Molecule – [L] (M) |
|
|
| Free Molecule – [Y1] (#/cell) |
|
|
| Bound Receptor – [Y2] (#/cell) |
|
|
| Internalized Receptor – [Y3] (#/cell) |
|
|
| Signaling Molecule – [Y4] (#/cell) |
|
|
| Response Factor – [Y5] (#/cell) |
|
kf – association rate constant (1/M*s), kr – dissociation rate constant (1/s), kint – internalization rate constant (1/s), krec – recycling rate constant (1/s), ksynth – synthesis rate constant (#/cell*s), ksig – signaling rate constant (1/s), kdec – signal decay rate constant (1/s), kresp – signal response rate constant (1/s), kd – response decay rate constant (1/s), ρ (cells/L), NAV – Avogadro’s constant (#/mol)
Agent-associated reactions are dependent upon the local concentration of extracellular diffusing molecules (L) (Eqn. 1). These molecules diffuse and degrade in the extracellular environment, and are typically described by continuum equations. The linked mathematical representation is a diffusion-reaction equation:
| (Eqn. 2) |
where D is the isotropic diffusion coefficient, kdeg is the extracellular degradation rate constant, and g(L, Y1, Y2, …, YR, β) is the effect agent-associated reactions (Eqn. 1) on the concentration of extracellular diffusing molecules 13,51. For instance, secretion of extracellular ligand or dissociation of ligand from cell surface receptors will increase the concentration of extracellular diffusing molecules, while binding will decrease it. Conversions to correct units are necessary as agent-associated quantities are usually given on a per cell basis (e.g. #/cell), while extracellular diffusing molecules are typically described by concentration in the extracellular space (e.g. nM) (see Table 1). This mathematical formulation allows each agent to interact with extracellular diffusing molecules in the environment through agent-associated species unique to each agent, A(x,y,V). This gives the dynamic exchange of information between differential equation models and the ABM, as Eqn. 1 and Eqn. 2 are dependent upon both agents and extracellular diffusing molecules. Agents are moving, interacting, and changing state in the ABM with a simultaneous and direct interface with extracellular diffusing molecules. Changes to concentrations of extracellular diffusing molecules in the environment factor into the agent decision-making processes, while changes in agent-associated reactions can influence agent states (Figure 3).
Operator Splitting
We can use temporal operator splitting to decouple Eqn. 2 into simpler and more tractable systems for numerical solution 16,38,54,59,79,81. Here, we split Eqn. 2 into three equations: (1) extracellular molecule diffusion (operator Θ1), (2) agent-associated reactions (operator Θ2), and (3) extracellular molecule degradation (operator Θ3). Therefore, Eqn. 2 becomes:
| (Eqn. 3) |
| (Eqn. 4) |
| (Eqn. 5) |
The agent-associated reactions, fr, (Eqn. 1) and the effect of agent-associated reactions, g, (Eqn. 2) on the extracellular diffusing molecule must be solved together as the equations depend on quantities from both equations (Eqn. 4). This reduces the problem to solving a partial differential equation (PDE) (Eqn. 3), a set of non-linear ordinary differential equations (ODEs) for each agent (Eqn. 4), and a simple linear first order ODE (Eqn. 5). Eqn. 5 has an analytical solution:
| (Eqn. 6) |
The remaining equations, Eqn. 3 and Eqn. 4 (such as those shown in Table 1), can now be solved using existing numerical methods and discrete time steps. The overall numerical approximation to Eqn. 2 is then obtained by mathematically combining the solution to each individual equation (Eqn. 3, 4, and 5). The splitting method determines the accuracy of the overall solution and also determines relative time steps used for the individual equations.
Lie splitting (also known as first-order splitting) is the most commonly used method in multi- scale ABMs. As shown in Figure 4, the solution to each individual equation is estimated in sequence using the same time step (Δt), with the solution of each operator as the input to the next operator 26,38,59:
| (Eqn. 7) |
Figure 4. Operator splitting algorithms.

The top panel represents Lie Splitting, where each operator (Θ1, Θ2, and Θ3) is advanced in time one after the other. The bottom panel represents Strang splitting, where one operator (Θ2) is advanced halfway in time, followed by the other operators being advanced all the way in time (Θ1 and Θ3), and then the first operator (Θ2) is advanced another half-step in time.
Lie splitting has first order accuracy with solution error due to splitting of Eqn. 2 being proportional to the discrete time step O(Δt) 26,38,59.
A simple improvement over Lie splitting is Strang splitting (Eqns. 8–9), shown in Figure 4, which is second order accurate O(Δt2) 79:
| (Eqn. 8) |
| (Eqn. 9) |
The most computationally intensive operator is solved using the full time step (Δt), while the less computationally intensive operator is solved using a half time step (Δt/2). With three operators, two are grouped together (in this case Θ1 and Θ3) and the splitting method is used on the combined operator (Θ1 and Θ3). The splitting between the Θ1 and Θ3 operators remains Lie splitting. We group the molecular diffusion operator (Θ1) and the molecular degradation operator (Θ3) together in all further sections and refer to the combined operator as Θ4. We advocate the use of Strang splitting as the increased accuracy provides increased flexibility in choosing appropriate time steps. We base all further analysis on implementation of Strang splitting.
While operator splitting makes Eqn. 2 easier to solve, if too large of a time step (Δt) is chosen, one is essentially considering the system to be mathematically decoupled. This can lead to non-phenomenological behavior of the system. Additional operator splitting techniques have been developed and include higher order methods such as Yoshida splitting (4th and 6th order), Kahan splitting, and Zassenhaus products 21,34,45,93. Although the accuracy of the splitting method increases with these methods, additional function evaluations (some requiring steps backwards in time) make them more complicated approaches.
Model Layers and Discretization
Hybrid multi-scale ABMs are implemented using multiple super-imposed layers of information38. We follow this methodology and describe two layers: an environment layer and an agent layer. The environment layer holds information for each extracellular diffusing molecule (L) and is discretized into grid points of uniform spacing, Δx and Δy (Figure 5). The discretized grid is described using lattice parameters; i increases in the x-dimension and j increases in the y-dimension. Thus the local concentration of an extracellular molecule is given by Li,j. The agent layer, also a discretized grid, holds positional information of the agents, providing a framework for agent movement, behavior, and interaction. Agents in the agent layer interact with the environment layer at their corresponding positions. We prefer to maintain the same discretization size for both agent and environment layer, due to the simplicity in mapping between the two layers. Different discretization sizes of the environment and agent layers (Δx, Δy) have been used in the context of a hybrid multi-scale ABM but require interpolation between agent and environment layers 38.
Figure 5. Model layers and discretization.
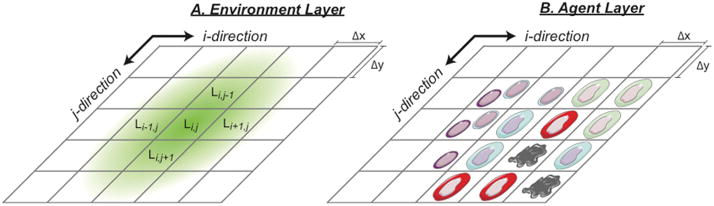
Implementation of multiple layers holding different types of information (extracellular molecule concentration, L) discretized into grid of spacing Δx and Δy represented by i,j coordinates. (A) The environment layer represents the extracellular space of the hybrid agent-based model and holds extracellular molecule concentrations. (B) The agent layer holds positional information of agents. Agents in the agent layer interact with the environment layer at their corresponding positions.
Tuneable Resolution
As more agent-associated reactions and species are included in Eqn. 4 (see Table 1), the computational cost of solving the equations grows. Each additional reaction or species must be solved for every agent in the system; in published models the number of agents ranges from a few hundred to as many as 100,000. Tuneable resolution is an approach that advocates reducing the complexity of a system by ‘coarse-graining’ a detailed model (or aspects of that model) to save computational resources while preserving key mechanisms and behaviors 49. For instance, an initial and fairly simple or ‘coarse-grained’ model is developed and the computational cost associated with solving the model is acceptable. Spurred by more biological data or additional questions, a more detailed or ‘fine-grained’ model containing many more agent-associated reactions is formulated, but it requires significant computational resources to solve. For subsequent biological questions, however, all of the detail of this ‘fine-grained’ model may not be needed. The goal is to use the ‘fine-grained’ model to build a better ‘coarse-grained’ model that estimates key mechanisms and behaviors from the ‘fine-grained’ model while simultaneously alleviating computational burdens (and facilitating model sharing and analysis). For example, it may be reasonable to assume pseudo steady-state for some agent-associated reactions or to use an apparent or lumped rate constant to approximate a set of reactions; sensitivity analysis and consideration of time scales may aid in these decisions (Pienaar et al. accepted with revision) 35,49,55.
Numerical Methods for PDE Sub-Models
To solve the two-dimensional form of Eqn. 3 in Cartesian coordinates (Eqn.10) one can utilize either explicit (the solution at a later time point can directly be computed from the current state of the system) or implicit (the solution at a later time point is calculated from both the current state of the system and the solution at the later time point) methods.
| (Eqn. 10) |
As explicit methods are more straightforward and also do not require linear algebra solvers, these methods are relatively easy to incorporate into a hybrid multi-scale ABM. Thus, we describe the basis and central features of three explicit methods: Forward-Time Central-Space, Alternating-Direction Explicit, and Spectral.
Forward-Time Central-Space Method
The simplest and most frequently used numerical method for solving diffusion equations in the context of a hybrid multi-scale ABM is the forward-time central-space (FTCS) approximation 9,26,38. FTCS is an explicit method (the concentration at the next time step can be directly calculated from concentrations at the current time step) that uses a first order forward approximation of the time derivative and a second order central difference approximation of the spatial derivatives 9,65. FTCS requires the discretized grid concentrations (Li,j) in the environment layer to be stored for both the current time point (t) and the next time point (t+Δt). Insulating boundary conditions are applied by ensuring the flux across the boundary is zero, while Dirichlet boundary conditions are applied by setting the appropriate concentration at the boundary.
The FTCS method is O(Δt) accurate in time and O(Δx2, Δy2) in space, while the computational cost is O(n2) per time step. The FTCS method is numerically stable only if the following criterion is met:
| (Eqn. 11) |
Therefore, the time step is directly dependent upon the diffusivity of the extracellular molecule in the environment (e.g. larger diffusivities mean lower time steps).
Alternating-Direction Explicit Method
The alternating-direction explicit (ADE) numerical method is an extension of the FTCS method built upon the Peaceman-Rachford alternating direction concept 9,62. Let both ui,j and vi,j be finite difference approximations of the extracellular molecule concentration, L(x,y,t).
| (Eqn. 12) |
| (Eqn. 13) |
In the case of u, iteration proceeds in both the forward i and j directions. Thus, the values in Eqn. 12 of and are known from previous calculations when iterating forward through the grid. In the case of v, iteration proceeds in the reverse i and j directions (i.e. iterating backwards through the grid). Thus, the values in Eqn. 13 at and are known from previous calculations when iterating backwards through the grid. Boundary conditions are set in the same manner as the FTCS method. The extracellular molecule concentration at the next time point, , is calculated by averaging and . The ADE method requires values to be stored for ui,j, vi,j, and Li,j for both the current time point (t) and the next time point (t+Δt) in the environment layer.
The ADE method is O(Δt2) accurate in time and O(Δx2, Δy2) in space, i.e. it is more accurate than the FTCS method, but the computational cost remains O(n2) per time step. Furthermore, the ADE method is an explicit method that is unconditionally numerically stable (typically seen with implicit methods), which does not place a restriction on Δt 9. Thus, Δt can be chosen based solely on the accuracy of the solution needed. A maximum Δt of approximately 4–6 times the Δt predicted by the conditional stability criterion of the FTCS method (Eqn. 11) can be used for the ADE method while maintaining acceptable accuracy 9,18.
Spectral Methods – Discrete Sine and Cosine Transforms
Spectral methods solve PDEs by assuming the solution is a sum of basis functions and choosing basis coefficients in order to best satisfy the solution 19,31,84. Spectral methods reduce PDEs to ODEs, greatly diminishing the difficulty of computation 60. The two-dimensional discrete sine transform (DST) and discrete cosine transform (DCT) are defined as:
| (Eqn. 14) |
| (Eqn. 15) |
Where k and l are the spectrally transformed i and j discretization, M and N are the lengths of k and l, and Āk,l are the spectral coefficients. The DST is an even-valued function at k = −1, k = M, l = −1, and l = N which naturally applies a Dirichlet boundary condition (constant zero concentration). The DCT is an odd-valued function at k = −1, k = M, l = −1, and l = N which naturally applies a Neumann boundary condition (insulating).
The appropriate transform (depending on the boundary condition for the modeled situation) is applied to Eqn. 10 and the resulting ODE is solved analytically, given below 19.
| (Eqn. 16) |
In the case of the DST (Eqn. 17) and DCT (Eqn. 18):
| (Eqn. 17) |
| (Eqn. 18) |
Pi,j is invariant and can be calculated from the discretization of the simulation space and the diffusion coefficient of the extracellular molecule. The spectral coefficients can be determined using a Fast Fourier Transform (FFT) and advanced forward in time using Eqn. 16. The spectral coefficients can be converted back into extracellular molecule concentrations (Li,j) using an inverse Fast Fourier Transform (FFT) 25. The FFTw library provides a simple C++ interface for computing spectral coefficients and their inverses 19,33. Using FFTw algorithms the computational cost for the forward and inverse FFTs are O(n log n), however multiplication of with Pi,j is O(n3) per time step 33. Therefore, the overall cost of the spectral method is dominated by the multiplication cost of O(n3) per time step.
Spectral methods require the extracellular molecule concentrations to be stored at (t) and (t+Δt) in the environment layer. Stability requirements are typically determined by examining the solution with different combinations of time step and discretization size 19,31. The accuracy of spectral methods is difficult to relate to O notation, but numerical errors tend to decay exponentially leading to much greater accuracy than purely finite difference methods 19,31,84. However, spectral methods have difficulty handling discontinuities or shock-like behaviors in input matrices and produce artifacts (or aliasing) at jump discontinuities, known as the ‘Gibb’s Phenomenon’ 19,37,84. As the input concentration field into spectral methods can be fairly discontinuous a technique known as smoothing (or anti-aliasing) is applied to alleviate these issues 43. The most common smoothing method is the ‘2/3’ rule, yet its computational cost can be large 43. We have successfully implemented a simple smoothing method in hybrid multi-scale ABMs by using the FTCS method presented above (Pienaar et al. accepted with revision). We take between 2–5 FTCS algorithm steps before solving Eqn. 10 using spectral methods. The number of FTCS steps is estimated by comparing the differences in the concentration field solution from a different method (e.g. FTCS or ADE) to the smoothed spectral method concentration field solution.
Other Available Numerical Methods
Implicit and semi-implicit algorithms (e.g. Crank-Nicholson) have also been used in hybrid multi-scale ABMs due to their stability characteristics 32,91,94. While increased stability allows for larger time steps, the need to assemble and determine a Jacobian matrix along with incorporating linear algebra solvers for systems of equations can be a complex task. Many libraries are available (e.g. LAPACK, LINPACK, PETSc, and GSL), but adapting existing code to libraries can be difficult. In the context of building a new hybrid multi-scale ABM or extending an existing ABM, in our experience it is much easier to use explicit schemas. Other advancements in solving PDEs are the multigrid and discrete wavelet transform (DWT) methods. The multigrid method has been demonstrated in the context of hybrid multi-scale ABMs 46,90,91. Implementation of the algorithm without libraries could be challenging, and available libraries (PETSc, Dune, Trilinos, FETK) may be difficult to interface with existing code. The DWT captures information in both the frequency and time domain, unlike the DCT and DST which capture only frequency information 23. Thus, the DWT can handle local discontinuities better than spectral methods while still using similar methods to compute wavelet coefficients at an O (n log n) or even O(n) computational cost. DWTs are still relatively new and have yet to be applied in the context of a hybrid multi-scale ABM. Additionally, only limited libraries exist presenting a barrier to efficient usage. We are currently evaluating the possible benefits of both the multigrid and DWT algorithms in the context of solving extracellular molecule diffusion in hybrid multi-scale ABMs.
Numerical Methods for ODE Sub-Models
Eqn. 4 is a description of any agent-associated reactions and must be solved for each agent in the simulation. For ease of explanation we present the numerical methods in the context of a single agent in vector notation (Eqn. 19):
| (Eqn. 19) |
Forward Euler
The simplest and easiest algorithm to implement is the forward Euler (FE) method, which uses a forward finite difference estimation of the first order derivative in Eqn. 19 65. FE is an explicit method and only requires the concentrations of each species at the current time point (t) and an estimate of the derivative at the current time point. FE is O(Δt) accurate in time and its computational cost is O(n) per time step. The FE method is conditionally stable and is numerically unstable for stiff equations and large time steps. The criteria for numerical stability of the solution of a set of linear ODEs is given by:
| (Eqn. 20) |
where λ is the set of eigenvalues for the system 71. For non-linear systems of ODEs (as in Table 1) the equations can be linearized and the behavior of the linearized system analyzed for stability, giving an approximate local stability criteria for the non-linear system 52. In practice this can be difficult for large systems of ODEs, and hence the stability limit for a particular set of ODEs is typically determined by trial-and-error 68. Numerical stability requirements of the FE method in no way guarantee accuracy of the solution.
4th Order Runge-Kutta
Runge-Kutta methods use higher-order terms from the Taylor-series expansion of the first derivative. The higher order terms are evaluated at distinct points and subsequently combined to give a better approximation to the first derivative 65,71. Most commonly used is the 4th order Runge-Kutta (RK4) method that requires concentrations of each species at the current time point (t) along with four estimates of the derivative. The RK4 method is O(Δt4) accurate in time, a significant improvement on the FE method. The computational cost of RK4 remains O(n) per time step and is a conditionally stable method. The criteria for numerical stability of linear ODEs is shown below, where λ is the set of eigenvalues for the system 52:
| (Eqn. 21) |
As mentioned above, biological systems are in general non-linear; therefore in practice the stability limit is again determined by trial-and-error 68.
Other Available Numerical Methods
A simple extension of the RK4 method is an adaptive step size Runge-Kutta method, known as RK4–5 65,76,77,95. This method determines the necessary time step for a given accuracy on-the-fly based on estimates of local error by calculating both the 4th order and 5th order Runge-Kutta solutions. The additional computational cost in calculating both 4th and 5th order solutions, estimating error, and determining the correct time step could be outweighed by significantly larger time steps. Implicit algorithms for solving ODEs can also be used 16,32,41,58,59,61,91. Similar to PDE algorithms, implicit algorithms require determining a Jacobian matrix and implementation of linear algebra solvers. Considering the large number of agents per simulation (anywhere from 100s to 100,000s), assembly and solution of the Jacobian matrix can be an overwhelming task. Recently, some have suggested the use of in situ adaptive tabulation (ISAT) to minimize the number of numerical method calls 15,22,39,40,64,74. ISAT functions by tabulating existing solutions and determining appropriate regions of solution space where use of existing solutions or simple interpolation can accurately represent the solution without ever calling the numerical method 64,74. ISAT has not been demonstrated in the context of hybrid multi-scale ABMs but the theoretical reduction in computational cost is enticing. Lastly, many external programs and frameworks for solving systems of ODEs, such as MATLAB, COPASI, and CVODE 1,22,42,80,86, exist and could be linked to a hybrid multi-scale ABM. In our experience linking external programs to existing code can be difficult and may confer a large computational cost associated with module communication.
Syncing Numerical Methods in Hybrid Multi-Scale ABMs
It is critical to maintain sync points where differing time steps used to solve the PDEs, ODEs, and ABM are reconciled, thus linking the mathematics, exchanging information across scales, and allowing the model to correctly proceed forward in time. A challenge of hybrid multi-scale ABMs is to determine the largest time step for each numerical method that maintains easily identifiable sync points. For instance, a numerical method that solves extracellular molecule diffusion with a large time step requires fewer sync points with the agent-associated reactions (Figure 6A) than a numerical method that solves extracellular molecule diffusion with a smaller time steps (Figure 6B). A simple procedure for determining time steps that maintain sync points is given below. We assume Strang splitting and a previously determined time step for the agent-based model, Δtagent:
Figure 6. Syncing time steps across hybrid multi-scale agent-based models.
Example of two different combinations of time steps for extracellular molecule diffusion and agent-associated reactions. (A) A large time step for extracellular molecule diffusion requires few sync points with the agent-associated reactions. (B) A small time step for extracellular molecule diffusion requires many sync points with the agent-associated reactions.
Estimate the maximum time step to solve the extracellular molecule diffusion and degradation (Θ4) for the chosen numerical method. A good starting point for all numerical methods is Eqn. 11. Reduce the estimated time step to a number divisible by Δtagent for syncing and set this value as Δtpde.
Estimate the maximum time step, Δtode, to solve the ODE model of agent-associated reactions (Θ2) for the chosen numerical method. This can be accomplished by linearizing the equations and using Eqn. 20 or Eqn. 21 or by using trial-and-error in a ‘test-bed’ environment such as a standalone implementation of the numerical method in C++ or MATLAB (The Mathworks Inc. – Natick, MA). Frequently, the maximal time step for numerical accuracy and stability will be significantly smaller than Δtpde/2. Choose Δtode such that stability and accuracy requirements are satisfied. Additionally reduce Δtode to a number that is evenly divisible by Δtpde/2 for syncing.
Using the estimated time steps (Δtagent, Δtpde, and Δtode) solve the hybrid ABM.
Reduce all time steps by a factor of 2 and re-solve the system.
Compare the model solutions. If the solutions are inconsistent the time steps are too large. Reduce all time steps (Δtpde and Δtode) and re-verify that each is able to ‘sync’. Repeat steps 3–5 until the model solutions are within the desired tolerance.
A full hybrid multi-scale ABM algorithm is shown in Figure 7 and includes: the agent time step, Δtagent (agents move, change state, etc.), the ODE time step, Δtode (agent-associated reactions), and the PDE time step, Δtpde (diffusion and degradation). Although not the focus of this review, we note that successfully linking and solving a hybrid multi-scale ABM does not of course guarantee fidelity to data or its usefulness. Extensive model validation and testing are necessary using methods such as uncertainty and sensitivity analysis or non-linear least squares 56; individual scale models may function differently in the context of the integrated model. Examples of model validation can be found in particular applications 8,18,44 and a broader discussion of some points can be found in 44,49.
Figure 7. Diagram of a solution algorithm for a hybrid multi-scale agent-based.

(1) Update agents (movement, states, proliferation, etc). (2) Solve a single time step (Δtode) for agent-associated reactions (Θ2). Increment a counter N. (3) If the total time step (N×Δtode) is equal to (Δtpde/2) then move on to extracellular molecule diffusion and degradation. If not, take another single time step for agent-associated reactions (Θ2) and check again. (4) Solve a single time step (Δtpde) for extracellular molecule diffusion and degradation. Increment a counter M. (5) Solve a single time step (Δtode) for agent-associated reactions (Θ2). Increment a counter N. (6) If the total time step (N×Δtode) is equal to (Δtpde/2) move on to the final check. If not, take another single time step for agent-associated reactions (Θ2) and check again. (7) If the total time step (M×Δtpde) is equal to (Δtagent) then a full time step has been completed. Continue by updating agents as indicated in (1). If not, continue solving with step (2).
Case Study: An Example Hybrid Multi-Scale ABM
Choosing numerical methods that maintain computational tractability of a hybrid multi-scale ABM is essential. Considerations affecting the choice of numerical methods include: number of agents, number of agent-associated reactions, and model dimensionality (i.e. 2D or 3D). In addition, uncertainty and sensitivity analysis is commonly used to understand how variations in parameter values affect model results and necessitates large numbers of model simulations (~103) 56. In practice, hybrid multi-scale ABMs that require over a day to run severely limit model usefulness.
As a case study of the application of numerical methods described above, we ran comparison simulations with a hybrid multi-scale ABM of M. tuberculosis infection discussed earlier 18,26,28 (Pienaar et al. accepted with revision) (Tables 2 and 3). Depending on the computational environment (i.e. computing cluster vs. laptop) and the processes modeled (e.g. number of extracellular diffusing molecules modeled on a continuum scale) the computational cost will vary greatly. However, this case study functions to illustrate the principles presented above and demonstrate the trade-offs between complexity of numerical algorithms (and implementation) and computational cost. The hybrid multi-scale ABM is constructed using the C++ programming language, Boost libraries (distributed under the Boost Software License – www.boost.org), FFTw libraries (distributed under GPL – www.fftw.org), and the Qt framework for visualization (distributed under GPL – www.qt.digia.com). The ABM is cross-platform (Macintosh, Windows, Unix) and can be run with or without our visualization software. Simulations were performed on the Flux high performance computing cluster, provided by Advanced Research Computing at the University of Michigan (http://goo.gl/WQIyCX). Computational accuracy was assessed by comparing output generated with different numerical methods, different platforms, and over different model resolutions. We also test individual sub-models for numerical convergence outside of the multi-scale model (e.g. solving the ODEs or PDEs in MATLAB). However, methods to validate numerical convergence inside hybrid multi-scale ABMs need to be developed.
Table 2.
Relative Computational Speeds for Case Study 1
| 100 × 100 Simulation Grid ~5,000 Agents
|
200 × 200 Simulation Grid ~10,000 Agents
|
|||||
|---|---|---|---|---|---|---|
| ODE Numerical Method | ODE Numerical Method | |||||
|
| ||||||
| PDE Numerical Method | FE (Δt = 0.6s) | RK4 (Δt = 6s) | TR (Δt = 6s) | FE (Δt = 0.6s) | RK4 (Δt = 6s) | TR (Δt = 6s) |
| FTCS (Δt = 6s) | 1.0 | 2.3 | 4.6 | 1.0 | 2.6 | 5.4 |
| ADE (Δt = 30s) | 1.8 | 4.3 | 12.8 | 1.2 | 3.9 | 7.7 |
| SM (Δt = 120s) | 1.8 | 5.5 | 22.7 | 1.2 | 5.2 | 23.6 |
FTCS – Forward Time Central Space, ADE – Alternating Direction Explicit, SM – Spectral Method, FE – Forward Euler, RK4 – Runge-Kutta 4th Order, TR – Tuneable Resolution
N=3 for all simulations. 100×100 – Relative Speed of 1.0 corresponds to 7589 s of simulation time, 200×200 – Relative Speed of 1.0 corresponds to 46907 s of simulation time
Table 3.
Relative Computational Speeds for Case Study 1
| 100 × 100 Simulation Grid ~5,000 Agents
|
200 × 200 Simulation Grid ~10,000 Agents
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ODE Numerical Method | ODE Numerical Method | |||||||||||
|
| ||||||||||||
| PDE Numerical Method | FE | RK4 | TR | FE | RK4 | TR | ||||||
| FTCS | (0.6/0.6) | 1.4 | (0.6/0.6) | 1.0 | (0.6/0.6) | 1.5 | (0.6/0.6) | 1.4 | (0.6/0.6) | 1.0 | (0.6/0.6) | 1.2 |
| ADE | (3.0/0.6) | 2.8 | (3.0/3.0) | 3.2 | (3.0/3.0) | 3.6 | (3.0/0.6) | 3.1 | (3.0/3.0) | 4.0 | (3.0/3.0) | 4.9 |
| SM | (60/0.6) | 6.8 | (60/6.0) | 15.8 | (60/6.0) | 43.8 | (60/0.6) | 8.3 | (60/6.0) | 26.3 | (60/6.0) | 63.5 |
(xx/xx) → (Soluble Molecule Diffusion Time Step/Agent-Associated Reactions Time Step)
FTCS – Forward Time Central Space, ADE – Alternating Direction Explicit, SM – Spectral Method, FE – Forward Euler, RK4 – Runge-Kutta 4th Order, TR – Tuneable Resolution
N=3 for all simulations. 100×100 – Relative Speed of 1.0 corresponds to 26968 s of simulation time, 200×200 – Relative Speed of 1.0 corresponds to 259150 s of simulation time
In Case Study 1 (see Table 2), we simulate 100 days of the immune response following an initial infection with M. tuberculosis. Diffusion and degradation of three extracellular molecules (two cytokines and one chemokine) are tracked, and thirteen agent-associated differential equations describe receptor-ligand binding, trafficking and signaling of TNF and IL-10 18. Extracellular molecule diffusivities are ~10−8 cm2/sec for cytokines and chemokines. In Case Study 2 (see Table 3), we simulate 50 days of the immune response following an initial infection with M. tuberculosis plus an additional 50 days of antibiotic treatment (Pienaar et al. accepted with revision). Diffusion and degradation of five extracellular molecules (two cytokines, one chemokine, and two antibiotics) are tracked, and thirteen agent-associated differential equations describe receptor-ligand binding, trafficking and signaling of TNF and IL-10. In addition, the model in Case Study 2 tracks the effects of antibiotics against bacteria. Extracellular molecule diffusivities for these antibiotics are ~10−7–10−6 cm2/sec. As antibiotics diffuse much faster through tissue than cytokines and chemokines (they are much smaller molecules), smaller time steps must be used in the numerical methods.
We show relative computational speeds for both a 100×100 (4 mm2) and 200×200 (16 mm2) simulation grid. Model discretization length (Δx, Δy) of the environment and agent layers is 20 μm. For the larger grid, the number of calculations is increased due to ~2-fold more agents (the number of agents present is not an input to the simulations but rather is generated from the stochastic nature of the simulations) and 4-fold more simulation space. Additionally, we employed a tuneable resolution approach (assuming a pseudo steady state for multiple reactions and using apparent rate constants) to reduce the thirteen agent-associated differential equations to two agent-associated differential equations (Pienaar et al. accepted with revision). For both Case Studies 1 and 2, we show the mean computational time from 3 simulations for each of the 38 different combinations of numerical methods (including tuneable resolution). Tables 2 and 3 depict fold-changes in computational speed normalized to the slowest value for the specific grid size (i.e., higher values indicate shorter computation times). The standard deviations for simulations range from ±1–30% of the mean computational time.
Our results in Tables 2 and 3 demonstrate that implementation of appropriate numerical methods and time steps can dramatically improve overall computational speed in hybrid multi-scale ABMs. Moving to more sophisticated numerical methods for solution both ODEs and PDEs increases computational speed by up to 5-fold in Case Study 1 or 25-fold in Case Study 2 (Table 2 and Table 3). The benefits of a tuneable resolution approach are easily observed, with an additional 2-fold to 5-fold increase in the computational speed beyond increases due to choice of numerical method (Table 2 and Table 3). Although Case Study 1 does benefit from spectral methods, the diffusion time step cannot be increased (beyond ~120 seconds) as the mathematics begin to decouple at larger time steps (see Operator Splitting section). The diffusion time steps used in Case Study 2 are restricted to smaller values as antibiotics diffuse much faster than cytokines and chemokines in the environment. With these restrictions on diffusion time steps, decoupling is less of a concern in Case Study 2 and spectral methods are more advantageous than in Case Study 1. In both Case Studies a combination of tuneable resolution and spectral methods increases computational speeds more than 20-fold.
There are caveats to implementation of faster and more efficient numerical algorithms in the context of hybrid multi-scale ABMs. Increasing the efficiency of solving one operator (Eqn. 3–5) is eventually limited by the efficiency of solving a different operator. For instance, implementing a spectral-based algorithm for solving the diffusion operator is of limited benefit if the agent-associated reactions (ODEs) are solved using a Forward Euler-based methodology (Table 2). Thus, improvements in one operator must also be thought of in the context of another operator, leading to a constant cycle of re-evaluation and implementation of numerical solvers.
Another critical aspect of solving hybrid multi-scale ABMs is the importance of correctly choosing algorithms with compatible time steps. The benefit of implementing a different numerical algorithm can be reduced if the algorithm cannot take sufficiently large time steps due to limitations of another numerical algorithm. For instance, in Case Study 2 using the RK4 algorithm to solve the agent-associated reactions could theoretically allow time steps upwards of 5–10 seconds. However, due to faster tissue diffusivity values of antibiotics (~10-fold higher than cytokines and chemokines) if we use the FTCS algorithm to solve extracellular molecule diffusion, it limits use of larger time steps in the RK4 algorithm (Table 3).
Lastly, it is important to understand the computational costs associated with expanding the size of a model. In our Case Study, transitioning from a 100×100 to a 200×200 simulation grid reduces the computational speed ~7-fold. We have also expanded the example hybrid multi-scale ABM to 3-dimensions (100×100 grid to 100×100×100 grid), which reduced the computational speed ~42-fold. Without a priori knowledge of the factors that contribute to computational costs, hybrid multi-scale ABMs may become too bulky or inefficient to provide novel insights into the complex biological systems they represent.
Discussion
Multi-scale models aim to replicate fundamental behaviors of biological systems, with constant exchange of information across scales, in order to better understand and predict system behavior as a whole. Coupling individual scale models, such as an agent-based model and differential equation models, to allow for the exchange of information, is typically accomplished by linking mathematical formulations across each scale. Thus, tools to link individual scale models and efficient methods of solving the subsequent linked models are important to advance the use of hybrid multi-scale ABMs in generating new biological knowledge from the wealth of available data.
Here, we describe the general framework for linking ABMs and differential equation models and solving the resulting hybrid multi-scale model, including layers, discretization, and operator splitting. The framework is extendable to different levels of model detail and adaptable to the focus and needs of the problem (e.g. inclusion of antibiotic treatment in an infection model, or moving from two to three dimensions). We review three numerical algorithms for solving extracellular molecule diffusion and two numerical algorithms for solving individual agent-associated reactions. Additionally, we demonstrate the merits of a tuneable resolution approach to reduce the complexity of a system and limit computational cost 49. We show how an appropriate choice of numerical algorithms and time steps can improve the computational tractability and efficiency of a hybrid multi-scale ABM. The most important factors in choosing these algorithms are: (1) familiarity with the underlying mathematics, (2) an understanding of relative process rates, and (3) computational resources such as time and power that are available. If unfamiliar with numerical methods, it is easier to implement an algorithm built around simple mathematical concepts, such as FTCS for solving extracellular molecule diffusion, than to start with a more mathematically sophisticated algorithm. As the user becomes comfortable with simpler implementations, more complex algorithms tend to build on simpler concepts in an incremental fashion. Second, it is necessary to have a basic understanding of relative rates (e.g. rates of diffusion compared to rates of agent-associated reactions) in order to determine whether solution of a hybrid multi-scale ABM will benefit from a particular numerical algorithm. For instance, a small molecule drug diffuses through tissue much faster than a extracellular protein molecule (such as a cytokine) so implementing a method that solves the agent-associated reactions with a time step larger than the time step required for solving diffusion of the drug through tissue has limited benefits as the diffusive rate restricts the overall time step (Table 3). Lastly, it is important to note that the extent of computational resources available such as power and computational time needed for each model, as new numerical methods can have diminishing returns. For instance, running a sensitivity analysis requires a large number of model simulations (e.g. thousands of simulations). Therefore, implementing a new numerical method for aspects of a model will have more benefits in this case as compared to single simulations.
At present, ABM platforms and languages such as NetLogo or SPARK do not accommodate the framework described in this manuscript to develop a hybrid multi-scale ABM. While in principle one can link those platforms with other platforms, e.g. MATLAB for solving ODEs, in practice we have found this difficult, if not impossible, for our models. This is largely due to communication between software packages slowing down the models to the point of uselessness. Platforms such as CHASTE are making advances towards re-usable and non-specific hybrid multi-scale ABMs (e.g. the code is able to model both cardiac tissue and crypt formation); however, there is no ‘one-size fits all’ platform that is flexible enough for a user to easily develop a hybrid multi-scale ABM 29,82. Therefore, a short-term solution may be stripping away the biological system/disease state-specific portions of models and publishing a baseline mathematical framework for a hybrid multi-scale ABM that can be modified by the user to suit a particular application. Although not ideal this would speed up development of hybrid multi-scale ABMs and also lower the barrier to creation of new models by inexperienced users. Issues of model modularity and reproducibility are of great interest to the multi-scale biological modeling community (see the Interagency Modeling and Analysis Group website at http://www.imagwiki.nibib.nih.gov) and may spur the development of more user-friendly packages 63.
Multi-scale models are becoming a more prevalent tool to understand systems-level biological phenomena. Modeling efforts are driving biological research from a descriptive field to a predictive field, especially in the context of pharmaceutical research. The ability to unify genomic, proteomic, metabolomics data using modeling constructs is an emerging technique to facilitate new drug-development and discovery along with new ways to re-purpose old drugs 12,47,53,58. Modeling approaches can predict new therapeutic targets by identifying key mechanisms of disease pathology, rapidly assess efficacy and toxicity of new drugs, or rapidly optimize drug concentrations and dosing schedules. We are only beginning to develop computational models capable of performing these studies. As more biological data are integrated into models and their complexity grows, efficient implementation of hybrid multi-scale ABMs becoming increasingly important. In this work, we demonstrate a framework and suggest tools that allow for efficient implementation of hybrid multi-scale ABMs to help guide the choice and development of both new model creation and existing platforms.
Acknowledgments
We thank Paul Wolberg and Joe Waliga for computational assistance. This research was supported in part through computational resources and services provided by Advanced Research Computing at the University of Michigan, Ann Arbor. This research was funded by the following NIH grants: R01 EB012579 (DEK and JJL) and R01 HL 110811 (DEK and JJL).
Footnotes
Conflicts of Interest
Nicholas Cilfone, Denise Kirschner, and Jennifer Linderman declare no conflicts of interests.
Ethical Standards
No human or animal studies were carried out by the authors for this article.
References
- 1.Adra S, Sun T, MacNeil S, Holcombe M, Smallwood R. Development of a three dimensional multiscale computational model of the human epidermis. PLoS One. 2010;5:e8511. doi: 10.1371/journal.pone.0008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alarcón T, Byrne HM, Maini PK. Towards whole-organ modelling of tumour growth. Prog Biophys Mol Biol. 2004;85:451–72. doi: 10.1016/j.pbiomolbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 3.An G, Mi Q, Dutta-Moscato J, Vodovotz Y. Agent-based models in translational systems biology. Wiley Interdiscip Rev Syst Biol Med. 2009;1:159–71. doi: 10.1002/wsbm.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson ARA, Chaplain MAJ, Rejniak KA, editors. Single-Cell-Based Models in Biology and Medicine. Basel: Birkhäuser Basel; 2007. [Google Scholar]
- 5.Angermann BR, et al. Computational modeling of cellular signaling processes embedded into dynamic spatial contexts. Nat Methods. 2012;9:283–9. doi: 10.1038/nmeth.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Athale Ca, Deisboeck TS. The effects of EGF-receptor density on multiscale tumor growth patterns. J Theor Biol. 2006;238:771–9. doi: 10.1016/j.jtbi.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 7.Athale C, Mansury Y, Deisboeck TS. Simulating the impact of a molecular “decision-process” on cellular phenotype and multicellular patterns in brain tumors. J Theor Biol. 2005;233:469–81. doi: 10.1016/j.jtbi.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Bailey AM, Lawrence MB, Shang H, Katz AJ, Peirce SM. Agent-based model of therapeutic adipose-derived stromal cell trafficking during ischemia predicts ability to roll on P-selectin. PLoS Comput Biol. 2009;5:e1000294. doi: 10.1371/journal.pcbi.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barakat HZ, Clark JA. On the Solution of the Diffusion Equations by Numerical Methods. J Heat Transfer. 1966;88:421. [Google Scholar]
- 10.Basak S, Behar M, Hoffmann A. Lessons from mathematically modeling the NF-κB pathway. Immunol Rev. 2012;246:221–38. doi: 10.1111/j.1600-065X.2011.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer AL, Beauchemin CaaA, Perelson AS. Agent-based modeling of host-pathogen systems: The successes and challenges. Inf Sci (Ny) Elsevier Inc. 2009;179:1379–1389. doi: 10.1016/j.ins.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg EL. Drug Discov Today. Vol. 19. Elsevier Ltd; 2013. Systems biology in drug discovery and development; pp. 113–125. [DOI] [PubMed] [Google Scholar]
- 13.Bird RB, Stewart WE, Lightfoot EN. Transport Phenomena. New York: John Wiley & Sons; 1994. [Google Scholar]
- 14.Braun Da, Fribourg M, Sealfon SC. Cytokine Response Is Determined by Duration of Receptor and Signal Transducers and Activators of Transcription 3 (STAT3) Activation. J Biol Chem. 2013;288:2986–93. doi: 10.1074/jbc.M112.386573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakrabarti A, Verbridge S, Stroock AD, Fischbach C, Varner JD. Multiscale models of breast cancer progression. Ann Biomed Eng. 2012;40:2488–500. doi: 10.1007/s10439-012-0655-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi T, Maurya MR, Tartakovsky DM, Subramaniam S. Stochastic operator-splitting method for reaction-diffusion systems. J Chem Phys. 2012;137:184102. doi: 10.1063/1.4764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christley S, An G. Agent-Based Modeling in Translational Systems Biology. In: Vodovotz Y, An G, editors. Complex Systems and Computational Biology Approaches to Acute Inflammation SE - 3. New York, NY: Springer New York; 2013. pp. 29–49. [Google Scholar]
- 18.Cilfone NA, Perry CR, Kirschner DE, Linderman JJ. Multi-scale modeling predicts a balance of tumor necrosis factor-α and interleukin-10 controls the granuloma environment during Mycobacterium tuberculosis infection. PLoS One. 2013;8:e68680. doi: 10.1371/journal.pone.0068680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa B. Spectral Methods for Partial Differential Equations. 2004;6 [Google Scholar]
- 20.Coveney PV, Fowler PW. Modelling biological complexity: a physical scientist’s perspective. J R Soc Interface. 2005;2:267–80. doi: 10.1098/rsif.2005.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Csomós P, Faragó I, Havasi Á. Weighted sequential splittings and their analysis. Comput Math with Appl. 2005;50:1017–1031. [Google Scholar]
- 22.Dada JO, Mendes P. Multi-scale modelling and simulation in systems biology. Integr Biol (Camb) 2011;3:86–96. doi: 10.1039/c0ib00075b. [DOI] [PubMed] [Google Scholar]
- 23.Daubechies I. The wavelet transform, time-frequency localization and signal analysis. IEEE Trans Inf Theory. 1990;36:961–1005. [Google Scholar]
- 24.Deisboeck TS, Wang Z, Macklin P, Cristini V. Multiscale cancer modeling. Annu Rev Biomed Eng. 2011;13:127–55. doi: 10.1146/annurev-bioeng-071910-124729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duhamel P, Vetterli M. Fast fourier transforms: A tutorial review and a state of the art. Signal Processing. 1990;19:259–299. [Google Scholar]
- 26.Fallahi-Sichani M, El-Kebir M, Marino S, Kirschner DE, Linderman JJ. Multiscale computational modeling reveals a critical role for TNF-α receptor 1 dynamics in tuberculosis granuloma formation. J Immunol. 2011;186:3472–83. doi: 10.4049/jimmunol.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fallahi-Sichani M, Flynn JL, Linderman JJ, Kirschner DE. Differential risk of tuberculosis reactivation among anti-TNF therapies is due to drug binding kinetics and permeability. J Immunol. 2012;188:3169–78. doi: 10.4049/jimmunol.1103298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fallahi-Sichani M, Kirschner DE, Linderman JJ. NF-κB Signaling Dynamics Play a Key Role in Infection Control in Tuberculosis. Front Physiol. 2012;3:170. doi: 10.3389/fphys.2012.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Figueredo GP, Joshi TV, Osborne JM, Byrne HM, Owen MR. On-lattice agent-based simulation of populations of cells within the open-source Chaste framework. Interface Focus. 2013;3:20120081. doi: 10.1098/rsfs.2012.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 31.Fornberg B. A practical guide to pseudospectral methods. Cambridge; New York: Cambridge University Press; 1996. p. x.p. 231. [Google Scholar]
- 32.Frieboes HB, et al. Computer simulation of glioma growth and morphology. Neuroimage. 2007;37(Suppl 1):S59–70. doi: 10.1016/j.neuroimage.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frigo M, Johnson SG. The Design and Implementation of FFTW3. Proc IEEE. 2005;93:216–231. [Google Scholar]
- 34.Geiser J, Tanoğlu G, Gücüyenen N. Comput Math with Appl. Vol. 62. Elsevier Ltd; 2011. Higher order operator splitting methods via Zassenhaus product formula: Theory and applications; pp. 1994–2015. [Google Scholar]
- 35.Gong C, Linderman JJ, Kirschner D. Harnessing the Heterogeneity of T Cell Differentiation Fate to Fine-Tune Generation of Effector and Memory T Cells. Front Immunol. 2014;5:1–15. doi: 10.3389/fimmu.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong C, Mattila JT, Miller M, Flynn JL, Linderman JJ, Kirschner D. J Theor Biol. 335C. Elsevier; 2013. Predicting lymph node output efficiency using systems biology; pp. 169–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gottlieb D, Shu CW. On the Gibbs Phenomenon and Its Resolution. SIAM Rev. 1997;39:644–668. [Google Scholar]
- 38.Guo Z, Sloot PMa, Tay JC. A hybrid agent-based approach for modeling microbiological systems. J Theor Biol. 2008;255:163–75. doi: 10.1016/j.jtbi.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 39.Hedengren JD, Edgar TF. Order reduction of large scale DAE models. Comput Chem Eng. 2005;29:2069–2077. [Google Scholar]
- 40.Hedengren JD, Edgar TF. In Situ Adaptive Tabulation for Real-Time Control. Ind Eng Chem Res. 2005;44:2716–2724. [Google Scholar]
- 41.Heidlauf T, Röhrle O. Modeling the Chemoelectromechanical Behavior of Skeletal Muscle Using the Parallel Open-Source Software Library OpenCMISS. Comput Math Methods Med. 2013;2013:517287. doi: 10.1155/2013/517287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holcombe M, et al. Modelling complex biological systems using an agent-based approach. Integr Biol (Camb) 2012;4:53–64. doi: 10.1039/c1ib00042j. [DOI] [PubMed] [Google Scholar]
- 43.Hou TY, Li R. Computing nearly singular solutions using pseudo-spectral methods. J Comput Phys. 2007;226:379–397. [Google Scholar]
- 44.Hunt CA, Kennedy RC, Kim SHJ, Ropella GEP. Agent-based modeling: a systematic assessment of use cases and requirements for enhancing pharmaceutical research and development productivity. Wiley Interdiscip Rev Syst Biol Med. 2013;5:461–80. doi: 10.1002/wsbm.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karlsen KH, Lie K, Natvig J, Nordhaug H, Dahle H. Operator Splitting Methods for Systems of Convection–Diffusion Equations: Nonlinear Error Mechanisms and Correction Strategies. J Comput Phys. 2001;173:636–663. [Google Scholar]
- 46.Kaul H, Cui Z, Ventikos Y. A multi-paradigm modeling framework to simulate dynamic reciprocity in a bioreactor. PLoS One. 2013;8:e59671. doi: 10.1371/journal.pone.0059671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim M, Gillies RJ, Rejniak Ka. Current Advances in Mathematical Modeling of Anti-Cancer Drug Penetration into Tumor Tissues. Front Oncol. 2013;3:278. doi: 10.3389/fonc.2013.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirschner DE, Chang ST, Riggs TW, Perry N, Linderman JJ. Toward a multiscale model of antigen presentation in immunity. Immunol Rev. 2007;216:93–118. doi: 10.1111/j.1600-065X.2007.00490.x. [DOI] [PubMed] [Google Scholar]
- 49.Kirschner DE, Hunt CA, Marino S, Fallahi-Sichani M, Linderman JJ. Tuneable resolution as a systems biology approach for multi-scale, multi-compartment computational models. Wiley Interdiscip Rev Syst Biol Med. 2014;6:289–309. doi: 10.1002/wsbm.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krinner A, Roeder I, Loeffler M, Scholz M. Merging concepts - coupling an agent-based model of hematopoietic stem cells with an ODE model of granulopoiesis. BMC Syst Biol. 2013;7:117. doi: 10.1186/1752-0509-7-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lauffenburger DA, Linderman JJ. Receptors: Models For Binding, Trafficking, and Signaling. New York: Oxford University Press; 1993. [Google Scholar]
- 52.LeVeque RJ. Society for Industrial and Applied Mathematics. SIAM; 3600 Market Street, Floor 6, Philadelphia, PA 19104: 2007. Finite Difference Methods for Ordinary and Partial Differential Equations: Steady-state and Time-Dependent Problems. [Google Scholar]
- 53.Linderman JJ, Kirschner DE. Drug Discov Today Dis Model. xxx. Elsevier Ltd; 2014. In silico models of M. tuberculosis infection provide a route to new therapies; pp. 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lucas TA. Operator Splitting for an Immunology Model Using Reaction-Diffusion Equations with Stochastic Source Terms. SIAM J Numer Anal. 2008;46:3113–3135. [Google Scholar]
- 55.Marino S, El-Kebir M, Kirschner D. A hybrid multi-compartment model of granuloma formation and T cell priming in tuberculosis. J Theor Biol Elsevier. 2011;280:50–62. doi: 10.1016/j.jtbi.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marino S, I, Hogue B, Ray CJ, Kirschner DE. A methodology for performing global uncertainty and sensitivity analysis in systems biology. J Theor Biol. 2008;254:178–96. doi: 10.1016/j.jtbi.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marino S, Linderman JJ, Kirschner DE. A multifaceted approach to modeling the immune response in tuberculosis. Wiley Interdiscip Rev Syst Biol Med. 2011;3:479–89. doi: 10.1002/wsbm.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Materi W, Wishart DS. Computational systems biology in drug discovery and development: methods and applications. Drug Discov Today. 2007;12:295–303. doi: 10.1016/j.drudis.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 59.Mitha F, Lucas Ta, Feng F, Kepler TB, Chan C. The Multiscale Systems Immunology project: software for cell-based immunological simulation. Source Code Biol Med. 2008;3:6. doi: 10.1186/1751-0473-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mugler DH, Scott RA. Fast fourier transform method for partial differential equations, case study: The 2-D diffusion equation. Comput Math with Appl. 1988;16:221–228. [Google Scholar]
- 61.Palsson S, et al. The development of a fully-integrated immune response model (FIRM) simulator of the immune response through integration of multiple subset models. BMC Syst Biol BMC Systems Biology. 2013;7:95. doi: 10.1186/1752-0509-7-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peaceman DW, Rachford HH., Jr The Numerical Solution of Parabolic and Elliptic Differential Equations. J Soc Ind Appl Math. 1955;3:28–41. [Google Scholar]
- 63.Petersen BK, Ropella GE, Hunt CA. Toward modular biological models: defining analog modules based on referent physiological mechanisms. BMC Syst Biol. 2014;8:95. doi: 10.1186/s12918-014-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pope SB. Computationally efficient implementation of combustion chemistry using in situ adaptive tabulation. Combust Theory Model. 1997;1:41–63. [Google Scholar]
- 65.Press WH. Numerical Recipes in C++: the art of scientific computing. 2. Cambridge, UK: Cambridge University Press; 2002. [Google Scholar]
- 66.Qutub Aa, Popel AS. Elongation, proliferation & migration differentiate endothelial cell phenotypes and determine capillary sprouting. BMC Syst Biol. 2009;3:13. doi: 10.1186/1752-0509-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qutub AA, Mac Gabhann F, Karagiannis ED, Vempati P, Popel AS. Multiscale models of angiogenesis. IEEE Eng Med Biol Mag. 2009;28:14–31. doi: 10.1109/MEMB.2009.931791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rao S, van der Schaft A, van Eunen K, Bakker BM, Jayawardhana B. A Model Reduction Method for Biochemical Reaction Networks. BMC Syst Biol. 2014;8:52. doi: 10.1186/1752-0509-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rapin N, Lund O, Bernaschi M, Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS One. 2010;5:e9862. doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ray JCJ, Flynn JL, Kirschner DE. Synergy between individual TNF-dependent functions determines granuloma performance for controlling Mycobacterium tuberculosis infection. J Immunol. 2009;182:3706–17. doi: 10.4049/jimmunol.0802297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riley KF, Hobson MP, Bence SJ. Mathematical Methods for Physics and Engineering: A Comprehensive Guide. Cambridge University Press; 2002. [Google Scholar]
- 72.Santoni D, Pedicini M, Castiglione F. Implementation of a regulatory gene network to simulate the TH1/2 differentiation in an agent-based model of hypersensitivity reactions. Bioinformatics. 2008;24:1374–80. doi: 10.1093/bioinformatics/btn135. [DOI] [PubMed] [Google Scholar]
- 73.Segovia-Juarez JL, Ganguli S, Kirschner D. Identifying control mechanisms of granuloma formation during M. tuberculosis infection using an agent-based model. J Theor Biol. 2004;231:357–76. doi: 10.1016/j.jtbi.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 74.Singer M, Pope S, Najm H. Modeling unsteady reacting flow with operator splitting and ISAT. Combust Flame. 2006;147:150–162. [Google Scholar]
- 75.Sloot PMa, Hoekstra AG. Multi-scale modelling in computational biomedicine. Brief Bioinform. 2010;11:142–52. doi: 10.1093/bib/bbp038. [DOI] [PubMed] [Google Scholar]
- 76.Southern J, et al. Multi-scale computational modelling in biology and physiology. Prog Biophys Mol Biol. 2008;96:60–89. doi: 10.1016/j.pbiomolbio.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stefanini MO, Wu FTH, Mac Gabhann F, Popel AS. The Presence of VEGF Receptors on the Luminal Surface of Endothelial Cells Affects VEGF Distribution and VEGF Signaling. PLoS Comput Biol. 2009;5 doi: 10.1371/journal.pcbi.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stern JR, Christley S, Zaborina O, Alverdy JC, An G. Integration of TGF-β- and EGFR-based signaling pathways using an agent-based model of epithelial restitution. Wound Repair Regen. 2012;20:862–71. doi: 10.1111/j.1524-475X.2012.00852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strang G. On the Construction and Comparison of Difference Schemes. SIAM J Numer Anal. 1968;5:506–517. [Google Scholar]
- 80.Sun T, Adra S, Smallwood R, Holcombe M, MacNeil S. Exploring hypotheses of the actions of TGF-beta1 in epidermal wound healing using a 3D computational multiscale model of the human epidermis. PLoS One. 2009;4:e8515. doi: 10.1371/journal.pone.0008515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sundnes J, Lines GT, Tveito A. An operator splitting method for solving the bidomain equations coupled to a volume conductor model for the torso. Math Biosci. 2005;194:233–48. doi: 10.1016/j.mbs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 82.Swat MH, Thomas GL, Belmonte JM, Shirinifard A, Hmeljak D, Glazier JA. Multi-scale modeling of tissues using CompuCell3D. Methods Cell Biol Elsevier Inc. 2012;110:325–66. doi: 10.1016/B978-0-12-388403-9.00013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–71. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trefethen LN. Unpublished Text. 1996. Finite Difference and Spectral Methods for Ordinary and Partial Differential Equations. [Google Scholar]
- 85.Walker DC, Southgate J. The virtual cell--a candidate co-ordinator for “middle-out” modelling of biological systems. Brief Bioinform. 2009;10:450–61. doi: 10.1093/bib/bbp010. [DOI] [PubMed] [Google Scholar]
- 86.Walpole J, Papin Ja, Peirce SM. Multiscale Computational Models of Complex Biological Systems. Annu Rev Biomed Eng. 2013:137–154. doi: 10.1146/annurev-bioeng-071811-150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang J, et al. Multi-scale agent-based modeling on melanoma and its related angiogenesis analysis. Theor Biol Med Model Theoretical Biology and Medical Modelling. 2013;10:41. doi: 10.1186/1742-4682-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Z, Bordas V, Sagotsky J, Deisboeck TS. Identifying therapeutic targets in a combined EGFR-TGFβR signalling cascade using a multiscale agent-based cancer model. Math Med Biol. 2012;29:95–108. doi: 10.1093/imammb/dqq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Z, Butner JD, Kerketta R, Cristini V, Deisboeck TS. Semin Cancer Biol. Elsevier Ltd; 2014. Simulating Cancer Growth with Multiscale Agent-Based Modeling; pp. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wise S, Kim J, Lowengrub J. Solving the regularized, strongly anisotropic Cahn–Hilliard equation by an adaptive nonlinear multigrid method. J Comput Phys. 2007;226:414–446. [Google Scholar]
- 91.Wise SM, Lowengrub JS, Cristini V. An Adaptive Multigrid Algorithm for Simulating Solid Tumor Growth Using Mixture Models. Math Comput Model Elsevier Ltd. 2011;53:1–20. doi: 10.1016/j.mcm.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wolff K, Barrett-Freeman C, Evans MR, Goryachev aB, Marenduzzo D. Modelling the effect of myosin X motors on filopodia growth. Phys Biol. 2014;11:016005. doi: 10.1088/1478-3975/11/1/016005. [DOI] [PubMed] [Google Scholar]
- 93.Yoshida H. Construction of higher order symplectic integrators. Phys Lett A. 1990;150:262–268. [Google Scholar]
- 94.Zhang L, et al. Developing a multiscale, multi-resolution agent-based brain tumor model by graphics processing units. Theor Biol Med Model. 2011:46. doi: 10.1186/1742-4682-8-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zingg DW, Chisholm TT. Runge–Kutta methods for linear ordinary differential equations. Appl Numer Math. 1999;31:227–238. [Google Scholar]



