Abstract
Efflux is by far the most common means of arsenic detoxification is by methylation catalyzed by a family of As(III) S-adenosylmethionine (SAM) methyltransferases (MTs) enzymes designated ArsM in microbes or AS3MT in higher eukaryotes. The protein sequence of more than 5000 AS3MT/ArsM orthologues have been deposited in the NCBI database, mostly in prokaryotic and eukaryotic microbes. As(III) SAM MTs are members of a large superfamily of MTs involved in numerous physiological functions. ArsMs detoxify arsenic by conversion of inorganic trivalent arsenic (As(III)) into mono-, di- and trimethylated species that may be more toxic and carcinogenic than inorganic arsenic. The pathway of methylation remains controversial. Several hypotheses will be examined in this review.
As a consequence of the environmental pervasiveness of arsenic, detoxifying systems are found in nearly every organism, from bacteria to humans (1). A common means to detoxify arsenic is by methylation catalyzed by As(III) S-adenosylmethionine methyltransferases enzymes (2, 3). In humans arsenic methylation paradoxically both detoxifies arsenic and simultaneously transforms it into carcinogenic species. Human AS3MT is a liver enzyme that is a member of a large superfamily of methyltransferases that are involved in many physiological functions (4). As(III) toxicity is due to its ability to react with protein sulfhydryl groups, and AS3MT detoxifies arsenic by conversion of into the methylated species methylarsenic (MAs) and dimethylarsenic (DMAs) (5). Biotransformation of small molecules into carcinogens is common (6), and arsenic biomethylation is associated with arsenic-related cancers by conversion of inorganic arsenic (As(III)) into carcinogenic trivalent MAs(III) and DMAs(III) (7–9).
The pathway of methylation remains controversial. One hypothesis proposed by Challenger (10, 11) is that the enzyme catalyzes a series of alternating oxidative methylations and reductions, using S-adenosylmethionine as the methyl donor to generate the pentavalent products methylarsenate (MAs(V)), dimethylarsenate (DMAs(V)) and a small amount of trimethylarsine oxide (TMAs(V)O). The trivalent species MAs(III), DMAs(III) and TMAs(III) are intermediates but not products. Most consistent with this hypothesis is that humans primarily excrete DMAs(V) and to a lesser extent MAs(V), but, until recently, little or no trivalent arsenicals were found in urine (12, 13). More recently, Hayakawa and coworkers (9) proposed an alternate pathway in which the preferred substrates of the methyltransferase are the glutathione (GSH) conjugates As(GS)3 and MAs(GS)2, and the products are the trivalent conjugates MAs(GS)2 and DMAs(GS). This pathway also involves a series of sequential oxidations and reductions, but SAM is reduced to S-adenosylhomocysteine (SAH), and GSH is oxidized to GSSG rather than changes in the oxidation state of arsenic, which remains trivalent throughout the catalytic cycle. The conjugates dissociate to unstable MAs(III) and DMAs(III), which rapidly oxidize nonenzymatically in air to MAs(V) and DMAs(V), the observed urinary species (13, 14). This review will examine these hypotheses in light of a new x-ray crystal structure (15) and biochemical analysis (16) of an As(III) SAM MT from the acidothermophilic alga Cyanidioschyzon merolae that grows in hot springs in Yellowstone National Park (2).
1) Arsenic binding proteins
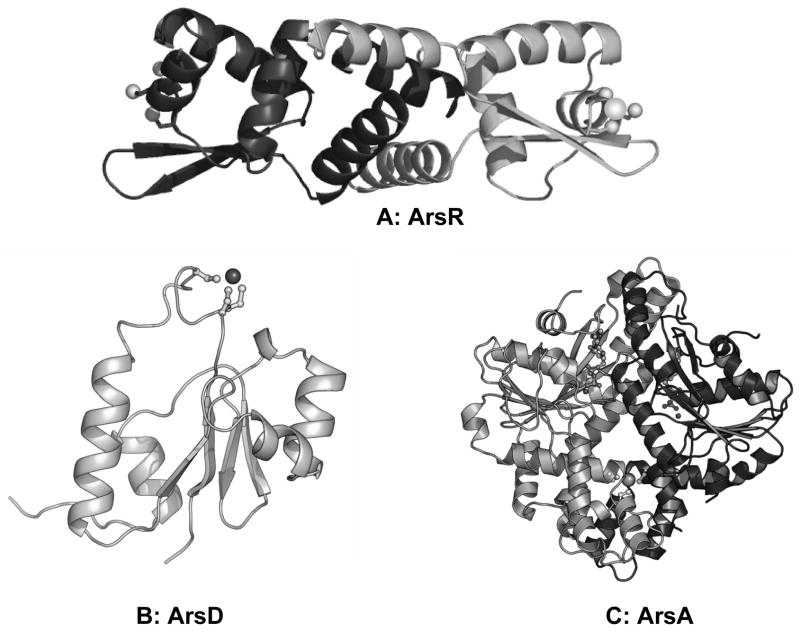
Arsenic is found in the organic and inorganic forms and it occurs in different valence states. The most common inorganic species are arsenate (As(V)) and arsenite (As(III)). In general, trivalent arsenite is more toxic than pentavalent arsenate and is primarily responsible for the biological effects of this metalloid. The detoxification mechanism of arsenic involves reduction, oxidation, methylation and demethylation (17). Reduction is the mechanism of arsenic reduced from pentavalent form to trivalent form, and oxidation is from trivalent to pentavalent form. Methylation is an alternate detoxification process in which inorganic arsenic is transformed into mono, di and trimethyl arsenicals, and demethylation is the reverse process of methylation. The major pentavalent products (DMAs(V)) and (TMAs(V)O) are approximately 100-fold and a 1,000-fold, respectively, less toxic than As(III) (18). The reduction of inorganic As(V) to As(III) is a prerequisite for its methylation and utilizes glutathione (GSH) and the ArsC arsenate reductase (19). The best characterized ars operon consists of three genes: arsR, arsB, and arsC (20). In addition to ArsC, ArsR is a trans-acting regulatory protein (21) and ArsB is an As(III) efflux protein (22). In addition to the three-gene chromosomal ars operon, some ars operons such as those carried by Escherichia coli plasmids R773 and R46 have five genes, arsRDABC, that encode two additional proteins, the ArsA ATPase, which is the catalytic subunit of the ArsAB efflux pump (23), and ArsD, an As(III) chaperone to the ArsAB pump (24). In each of these proteins, As(III) binds to two or three cysteine residues (25–27), except for ArsB, which has single cysteine residue that is not involved in As(III) binding (28). Escherichia coli plasmid R773 ArsR is a 117 residue homodimer with high affinity for the ars promoter (29) that binds As(III) at three cysteine residues, Cys32, Cys34, and Cys37 (30) (Fig. 1A). ArsD also has three conserved cysteine residues, Cys12, Cys13 and Cys18, that form an As(III) binding site (31) (Fig. 1B)
Fig. 1. Structures of As(III) binding proteins.
R773 ArsR binds As(III) to Cys32, Cys34 and Cys37. ArsD binds As(III) to Cys12, Cys13 and Cys18. ArsA binds As(III) to Cys113, Cys172, Cys444.
ArsA is an 583-amino acid ATPase with a high affinity metalloid binding domain and two nucleotide binding domains, A1 and A2, connected by a short linker (32). ArsD transfers As(III) to the ArsA metalloid binding site formed by cysteine residues Cys113, Cys172 and Cys422 (33) (Fig. 1C). Both three-gene and five-gene ars operons may be present in a single strain (for example in T. arsenitosidans 3As) (34). In addition ars opeons may include other genes such as the ArsH oxidoreductdase (35, 36), the ArsM As(III) SAM MT (3) or other genes of unknown function.
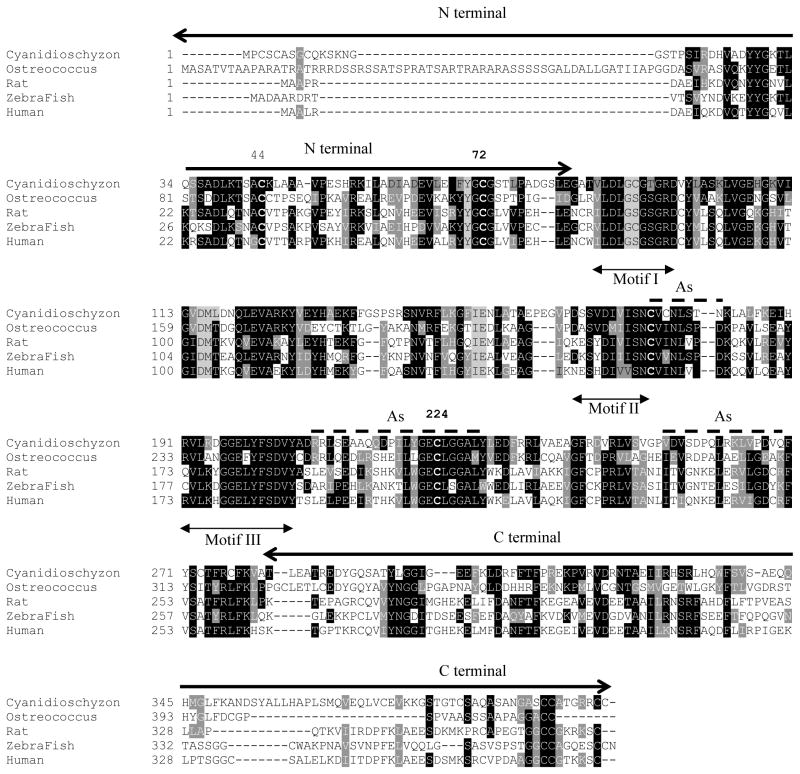
Biomethylation of arsenic is a common mechanism for its detoxification, and it has evolved in nearly all organisms, humans and most animal species (7, 37). As(III) SAM MTs share common sequence motifs that form the SAM binding site (Fig. 2). The recently solved structure of the C. merolae CmArsM demonstrates the As(III) and SAM binding sites and suggest a role for conserved cysteine residues Cys72, Cys174 and Cys224 in the arsenic methylation process (15).
Fig. 2. Structure-based alignment of CmArsM with five orthologous As(III) SAM MTs from an alga, rat, zebrafish, fungus and human.
Conversed cysteines are in bold, with numbering above. Black and grey shaded are conserved and similar residues. The dotted lines indicate the As(III) binding site, the double arrow are the three common SAM-binding motifs, and the thick arrows are N and C-terminal residues.
2) S-adenosylmethionine methyltransferases
S-adenosylmethionine (SAM or AdoMet) has an activated methyl group that is transferred by members of the MT superfamily to acceptor groups by SN2 displacement mechanisms, involving attack of a nucleophile on the methyl group of SAM with inversion of configuration and concomitant release of SAH. MTs are categorized based on the electron-rich, methyl accepting atom, usually O, N, C, S or As. SAM is involved in many essential chemical reactions and is the second most widely used and most multifunctional enzyme substrate after ATP (38). SAM plays a major role in many chemical reactions and serves as a target for various clinical and therapeutic studies (39) such as cancer (40, 41), Alzheimer’s disease (42) and Parkinson’s disease (43). SAM is used in the biosynthesis and modification of many types of biomolecules, including DNA, RNA, proteins, lipids and small molecules (44, 45). The highly favourable thermodynamics of SAM-dependent methyltransfer reactions enables SAM to be used in preference to other methyl donors such as folate. SAM-dependent methyltransferases are a well-studied diverse class of enzymes. To date, a text search of the keyword “methyltransferases” in the Protein Data Bank yields 825 unique structures. Of these, 147 structures have SAM bound, and 345 have the product SAH bound. SAM MTs are classified into nine classes (39). Among the available structures, the Rossmann fold is the best characterized type of fold that is composed of a basic seven-stranded β-sheet (46).
3) Structure of the CmArsM As(III) MT
CmArsM is a 400 amino acid residue orthologue of human AS3MTT that methylates As(III) to a final product of volatile TMAs(III) (2). A search of the NCBI site using CmArsM sequence yields 5480 unique sequences in 2934 species, of which 281 are in archaea, 4047 are in bacteria, 204 are in metazoa, 268 are in fungi, 99 are in plants and 581 others. CmArsM has been crystallized (47), and structures of CmArsM with or without bound SAM or As(III) have been reported (Fig. 3) (15). CmArsM adopts a compact globular structure with the typical Rossmann fold, the N-terminal β strand in the middle of sheet, and the strand topology is 6754123, with the 7th strand antiparallel to other sheets. All seven β-strands surrounded by helices forming a three-layer (αβα) sandwich. The SAM binding domain of CmArsM consists of 147 residues, and the secondary structure of this domain is highly conserved compared to other MTs. A search using the DALI server of this domain yielded the 1011 closest homologue structures. Out of 1011 molecules, 912 structures have Z-score 10 and above, and 877 structures can be superimposed with a root-mean-square (RMS) deviation of 3.0 Å, which suggests that the secondary structure of SAM binding is highly conserved. Analysis of multiple sequence alignment of the CmArsM with another alga, Ostreococcus, fish, fungal and mammalian (rat and human) demonstrates that they all have a high degree of conservation (Fig. 2). CmArsM belongs to the small molecule methyltranferase family (48). SAM-dependent methyltransferases of proteins and small molecules contained three common sequence motifs (49), and interactions between SAM and these conserved amino acids are critical for methyl group transfer to substrate (50). Motif I is the glycine-rich sequence of 87-(V/I)(L/V)DLG(C/S)G(T/S/G)G(R/I)D-98, the hall mark of SAM-binding motif hh(D/E)hGXGXG of SAM MTs, where h is a hydrophobic amino acid, and X is any amino acid. Motif II encompasses β-strand 4 in the structure of CmArsM, with sequences 164-(D/K/N)(S/A/E/K)S(V/Y/H)D(I/C)(V/I)(V/I)SN-173 and motif III corresponds to α5-β5 loop and β-strand 5 in the structure of CmArsM at residues 190 to 217. All three motifs are well conserved with the animal homologues such as sea urchin, sea squirt, mouse, rainbow trout, chicken, cow and chimpanzee (51).
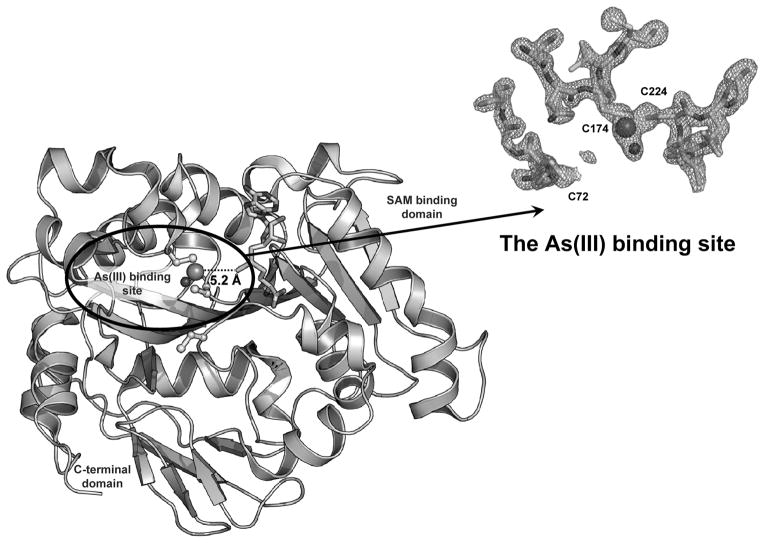
Fig. 3. Structure of CmArsM.
The crystal structures of CmArsM with bound As(III) and bound SAM were superimposed, yielding a model in which the methyl group of SAM is poised for transfer from SAM to As(III) a distance of 5.2 Å. Arrow: The electron density of the As(III) binding residues are fitted with stick models of Cys72, Cys174 and Cys224.
The As(III) binding domain of CmArsM has three modular components with a total of 48 residues. It is a novel and unique domain not found in non-As(III) MTs. The first module is between β4 and α5 with one 310 helix, the second module is between β5 and α8 with two short α helices. The third module is an insertion of 16 residues between β6 and β7 (15). In a comparison between CmArsM and orthologues, the first module adopts a similar secondary structure, and the sequences are highly conserved (Fig. 2) compare to other two modules. A CmArsM model with the As(III) and SAM bound structures superimposed reveals the active site in which the methyl group is transferred from SAM to inorganic As(III). In the crystal structure of CmArsM with bound As(III), the arsenic ion is bound to the thiolates of Cys174 and Cys224 at an average distance of 2.21 Å and forms a pyramidal site with a nonprotein ligand, chloride. Mutagenesis studies suggest that four conserved cysteine residues, Cys44, Cys72, Cys174, Cys224, are required for As(III) methylation, while only Cys174 and Cys224 are involved in MAs(III) methylation (16) (K. Marapakala and B.P. Rosen, unpublished results). This fact is further strengthened by comparing the location of reside Cys72 in the ligand-free, As(III)- and SAM-bound structures of CmArsM. In the SAM-bound structure, Cys72 moves closer to the As(III) binding site (6.58 Å) compare to ligand-free and As(III)-bound structures (8.23 Å). CmArsM has 17 cysteine residues, of which there are five in the N-terminal region and six in the C-terminal, with cysteine residues in the SAM (Cys92, Cys273 and Cys 277) and As(III) (Cys174, Cys176 and Cys224) binding domains (Fig. 2). Comparison of Cys72, Cys174 and Cys224 in CmArsR and the 100 closest homologues in the Blink database indicates that Cys72, Cys174 and Cys224 are conserved, suggesting conserved function. Cys44 and Cys92 are found in 83 and 72 respectively, of the 100 closest homologues, suggesting that they are not absolutely required for As(III) SAM MTs. Cys92 makes a hydrogen bond with the carboxyl group of the SAM cofactor, but this may not be necessary for catalysis. From mutagenic studies, however, Cys44 appears to be similar to Cys72 in methylation of As(III) but not MAs(III) (K. Marapakala and B.P. Rosen, unpublished results). The corresponding residue is missing in fungal sequences, but those have not been characterized, so it is not clear whether they are true orthologues. It is possible, for example, that the fungal enzymes methylate MAs(III) but not As(III), in which case the residues corresponding to Cys44 may not be required.
4) The catalytic mechanism of As(III) SAM MTs
As mentioned above, the pathway of methylation remains controversial. The hypothesis proposed by Challenger (10, 11) differs from that of Hayakawa and coworkers (9) in several major aspects including the nature of the substrates and the oxidation state of the products. To examine whether the substrate is As(OH)3 or As(GS)3, the fluorescence of engineered single-tryptophan derivates of CmArsM was used to analyze As(III) binding. Crystallographic data indicates that the Cys72 loop moves during the catalytic cycle, so Tyr70 in the Cys72 loop was changed to a tryptophan in an otherwise tryptophan-free derivative, creating a single tryptophan-derivate of CmArsMS3MT (16). This construct responded to As(III) binding with a large quenching of fluorescence. The rate of quenching was on the order of minutes, much slower than expected for a physiological reaction. In contrast, addition of pre-formed As(GS)3 evoked fluorescent quenching in milliseconds – several order of magnitude faster binding than As(OH)3, which is the solution form of As(III) at neutral pH (52). Similarly, binding of MAs(GS)2 was much faster than free MAs(OH)2. Other thiols such as 2-mercaptoethanol also increase the rate, but only GSH is present in millimolar levels in cell cytosol, and As(III) is nearly completely in complex with GSH (53). So it is likely that AS3MT recognizes as As(GS)3 preferentially over As(OH)3 in vivo. These results support Hayakawa’s hypothesis that GSH serves a chaperone-like function to bind intracellular As(III) and delivers it to As(III) SAM MTs. Similarly, As(GS)3 and MAs(GS)2 are substrates of the ABC efflux pumps MRP1 and MRP2, which excrete the conjugates into blood and bile (54, 55). Thus, As(GS)3 appears to be the common currency of intracellular arsenic.
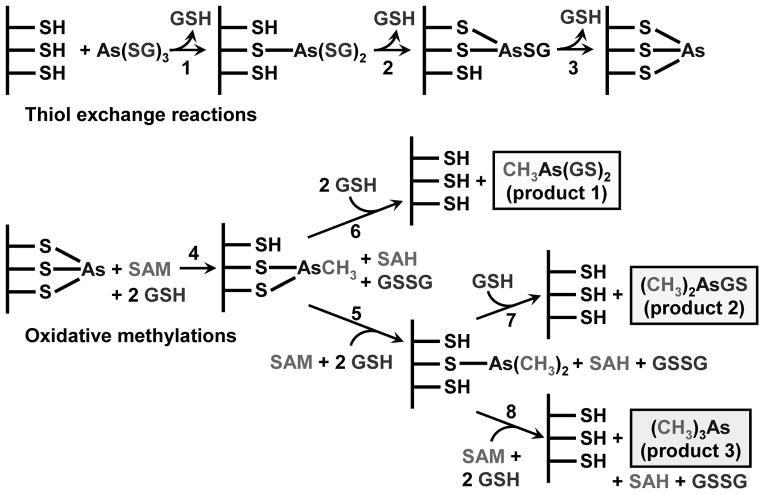
The other major difference between the Challenger and Hayakawa pathways is the oxidation state of the products. Challenger predicted that the products are pentavalent, while Hayakawa suggested that they are trivalent. Yet, to date, no study has demonstrated the presence of a pentavalent intermediate bound to an As(III) SAM MT, and the evidence that they directly generat As(V) metabolites is not strong. At long times of CmArsM catalysis, only DMAs(V) was observed, but DMAs(III) easily oxidizes in air (16). Importantly, the first methylation product of CmArsM is MAs(III), with no MAs(V) found, clearly in support of Hayakawa’s pathway. Another consideration is that no other SAM MT oxidizes its substrate (4). Lysine and arginine methyltransferases are good models for arsenic methyltransferases as they carry out three very similar methyl transfers without oxidation of the nitrogen (56). Nature is conservative, and there is no reason to consider that As(III) SAM MTs would use a different enzymatic mechanism from any other SAM MT. From these considerations, we have proposed a reaction scheme similar to Hayakawa’s in which the substrates and products are all trivalent glutathione conjugates (Fig. 4) (16).
Fig. 4. Proposed reaction scheme of As(III) S-adenosylmethionine methyltransferases.
As(GS)3 is proposed to be the actual substrate and binds to the active site cysteines by a series of thiol exchange reactions (reactions 1–3). The overall scheme involves two types of reactions, thiol exchange reactions (top) and oxidative methylations (bottom). After metalloid is bound, there is transfer of the methyl group from SAM, which becomes reduced to SAH (reaction 4). The enzyme-bound form of the methylated trivalent arsenic can dissociate from the enzyme in the reverse of the initial thiol exchange (reaction 5), producing MAs(GS)2, the first product. Alternatively, it can remain bound in the active site and undergo a second round of methylation (reaction 6). Exchange with GSH and dissociation (reaction 7) produces the second product, DMAs(GS)3. Alternatively, DMAs(III) can remain bound to the active site and undergo a third round of methylation (reaction 8), producing the final product, TMAs(III) gas. Exposure to oxygen results nonenzymatic oxidation to the frequently observed pentavalent species, which explains why human urine most commonly contains 60–80% DMAs(V) and only 10–30% MAs(V). This reaction scheme is based on data from (16).
Acknowledgments
This work was supported by National Institutes of Health Grant R37 GM55425.
Contributor Information
A. Abdul Ajees, Email: abdul.ajees@manipal.edu.
Barry P. Rosen, Email: brosen@fiu.edu.
References
- 1.Rensing C, Rosen BP. Heavy metals cycles (arsenic, mercury, selenium, others) In: Schaechter M, editor. Encyclopedia of Microbiology. Elsevier; Oxford, U.K: 2009. pp. 205–219. [Google Scholar]
- 2.Qin J, Lehr CR, Yuan C, Le XC, McDermott TR, Rosen BP. Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc Natl Acad Sci U S A. 2009;106:5213–5217. doi: 10.1073/pnas.0900238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci U S A. 2006;103:2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liscombe DK, Louie GV, Noel JP. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat Prod Rep. 2012;29:1238–1250. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- 5.Drobna Z, Xing W, Thomas DJ, Styblo M. shRNA silencing of AS3MT expression minimizes arsenic methylation capacity of HepG2 cells. Chemical research in toxicology. 2006;19:894–898. doi: 10.1021/tx060076u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guengerich FP. Metabolism of chemical carcinogens. Carcinogenesis. 2000;21:345–351. doi: 10.1093/carcin/21.3.345. [DOI] [PubMed] [Google Scholar]
- 7.Thomas DJ, Rosen BP. Arsenic methyltransferases. In: Kretsinger RH, Uversky VN, Permyakov EA, editors. Encyclopedia of Metalloproteins. Springer; New York: 2013. in press. [Google Scholar]
- 8.Styblo M, Drobna Z, Jaspers I, Lin S, Thomas DJ. The role of biomethylation in toxicity and carcinogenicity of arsenic: a research update. Environ Health Perspect. 2002;110(Suppl 5):767–771. doi: 10.1289/ehp.110-1241242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayakawa T, Kobayashi Y, Cui X, Hirano S. A new metabolic pathway of arsenite: arsenic-glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Arch Toxicol. 2005;79:183–191. doi: 10.1007/s00204-004-0620-x. [DOI] [PubMed] [Google Scholar]
- 10.Challenger F. Biological methylation. Sci Prog. 1947;35:396–416. [PubMed] [Google Scholar]
- 11.Challenger F. Biological methylation. Advances in enzymology and related subjects of biochemistry. 1951;12:429–491. doi: 10.1002/9780470122570.ch8. [DOI] [PubMed] [Google Scholar]
- 12.Vahter M. Methylation of inorganic arsenic in different mammalian species and population groups. Sci Prog. 1999;82 ( Pt 1):69–88. doi: 10.1177/003685049908200104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drobna Z, Del Razo LM, Garcia-Vargas GG, Sanchez-Pena LC, Barrera-Hernandez A, Styblo M, Loomis D. Environmental exposure to arsenic, AS3MT polymorphism and prevalence of diabetes in Mexico. J Expo Sci Environ Epidemiol. 2012;23:151–155. doi: 10.1038/jes.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le XC, Lu X, Ma M, Cullen WR, Aposhian HV, Zheng B. Speciation of key arsenic metabolic intermediates in human urine. Anal Chem. 2000;72:5172–5177. doi: 10.1021/ac000527u. [DOI] [PubMed] [Google Scholar]
- 15.Ajees AA, Marapakala K, Packianathan C, Sankaran B, Rosen BP. Structure of an As(III) S-adenosylmethionine methyltransferase: insights into the mechanism of arsenic biotransformation. Biochemistry. 2012;51:5476–5485. doi: 10.1021/bi3004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marapakala K, Qin J, Rosen BP. Identification of catalytic residues in the As(III) S-adenosylmethionine methyltransferase. Biochemistry. 2012;51:944–951. doi: 10.1021/bi201500c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu YG, Yoshinaga M, Zhao FJ, Rosen BP. Earth abides arsenic biotransformations. Annu Rev Earth and Planet Sci. 2014 doi: 10.1146/annurev-earth-060313-054942. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirano S, Kobayashi Y, Cui X, Kanno S, Hayakawa T, Shraim A. The accumulation and toxicity of methylated arsenicals in endothelial cells: important roles of thiol compounds. Toxicol Appl Pharmacol. 2004;198:458–467. doi: 10.1016/j.taap.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 19.Mukhopadhyay R, Rosen BP. Arsenate reductases in prokaryotes and eukaryotes. Environ Health Perspect. 2002;110(Suppl 5):745–748. doi: 10.1289/ehp.02110s5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlin A, Shi W, Dey S, Rosen BP. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol. 1995;177:981–986. doi: 10.1128/jb.177.4.981-986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J, Rosen BP. The ArsR protein is a trans-acting regulatory protein. Mol Microbiol. 1991;5:1331–1336. doi: 10.1111/j.1365-2958.1991.tb00779.x. [DOI] [PubMed] [Google Scholar]
- 22.Tisa LS, Rosen BP. Molecular characterization of an anion pump. The ArsB protein is the membrane anchor for the ArsA protein. J Biol Chem. 1990;265:190–194. [PubMed] [Google Scholar]
- 23.Dey S, Dou D, Tisa LS, Rosen BP. Interaction of the catalytic and the membrane subunits of an oxyanion- translocating ATPase. Archives of biochemistry and biophysics. 1994;311:418–424. doi: 10.1006/abbi.1994.1256. [DOI] [PubMed] [Google Scholar]
- 24.Lin YF, Walmsley AR, Rosen BP. An arsenic metallochaperone for an arsenic detoxification pump. Proc Natl Acad Sci U S A. 2006;103:15617–15622. doi: 10.1073/pnas.0603974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi W, Dong J, Scott RA, Ksenzenko MY, Rosen BP. The role of arsenic-thiol interactions in metalloregulation of the ars operon. J Biol Chem. 1996;271:9291–9297. doi: 10.1074/jbc.271.16.9291. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharjee H, Li J, Ksenzenko MY, Rosen BP. Role of cysteinyl residues in metalloactivation of the oxyanion-translocating ArsA ATPase. J Biol Chem. 1995;270:11245–11250. doi: 10.1074/jbc.270.19.11245. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Rawat S, Stemmler TL, Rosen BP. Arsenic binding and transfer by the ArsD As(III) metallochaperone. Biochemistry. 2010;49:3658–3666. doi: 10.1021/bi100026a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Dey S, Rosen BP. Soft metal thiol chemistry is not involved in the transport of arsenite by the Ars pump. J Bacteriol. 1996;178:911–913. doi: 10.1128/jb.178.3.911-913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Rosen BP. Metalloregulated expression of the ars operon. J Biol Chem. 1993;268:52–58. [PubMed] [Google Scholar]
- 30.Shi W, Wu J, Rosen BP. Identification of a putative metal binding site in a new family of metalloregulatory proteins. J Biol Chem. 1994;269:19826–19829. [PubMed] [Google Scholar]
- 31.Ye J, Ajees AA, Yang J, Rosen BP. The 1.4 Å crystal structure of the ArsD arsenic metallochaperone provides insights into its interaction with the ArsA ATPase. Biochemistry. 2010;49:5206–5212. doi: 10.1021/bi100571r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou T, Radaev S, Rosen BP, Gatti DL. Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. Embo J. 2000;19:1–8. doi: 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Salam AA, Rosen BP. Genetic mapping of the interface between the ArsD metallochaperone and the ArsA ATPase. Mol Microbiol. 2011;79:872–881. doi: 10.1111/j.1365-2958.2010.07494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arsene-Ploetze F, Koechler S, Marchal M, Coppee JY, Chandler M, Bonnefoy V, Brochier-Armanet C, Barakat M, Barbe V, Battaglia-Brunet F, Bruneel O, Bryan CG, Cleiss-Arnold J, Cruveiller S, Erhardt M, Heinrich-Salmeron A, Hommais F, Joulian C, Krin E, Lieutaud A, Lievremont D, Michel C, Muller D, Ortet P, Proux C, Siguier P, Roche D, Rouy Z, Salvignol G, Slyemi D, Talla E, Weiss S, Weissenbach J, Medigue C, Bertin PN. Structure, function, and evolution of the Thiomonas spp. genome. PLoS Genet. 2010;6:e1000859. doi: 10.1371/journal.pgen.1000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butcher BG, Deane SM, Rawlings DE. The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli. Appl Environ Microbiol. 2000;66:1826–1833. doi: 10.1128/aem.66.5.1826-1833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye J, Yang HC, Rosen BP, Bhattacharjee H. Crystal structure of the flavoprotein ArsH from Sinorhizobium meliloti. FEBS Lett. 2007;581:3996–4000. doi: 10.1016/j.febslet.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye J, Rensing C, Rosen BP, Zhu YG. Arsenic biomethylation by photosynthetic organisms. Trends in plant science. 2012;17:155–162. doi: 10.1016/j.tplants.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cantoni GL. Biological methylation: selected aspects. Annu Rev Biochem. 1975;44:435–451. doi: 10.1146/annurev.bi.44.070175.002251. [DOI] [PubMed] [Google Scholar]
- 39.Gana R, Rao S, Huang H, Wu C, Vasudevan S. Structural and functional studies of S-adenosyl-L-methionine binding proteins: a ligand-centric approach. BMC structural biology. 2013;13:6. doi: 10.1186/1472-6807-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaib H, Prebet T, Vey N, Collette Y. Histone methyltransferases: a new class of therapeutic targets in cancer treatment? Med Sci (Paris) 2011;27:725–732. doi: 10.1051/medsci/2011278014. [DOI] [PubMed] [Google Scholar]
- 41.Wagner JM, Hackanson B, Lubbert M, Jung M. Histone deacetylase (HDAC) inhibitors in recent clinical trials for cancer therapy. Clin Epigenetics. 2011;1:117–136. doi: 10.1007/s13148-010-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borroni B, Agosti C, Archetti S, Costanzi C, Bonomi S, Ghianda D, Lenzi GL, Caimi L, Di Luca M, Padovani A. Catechol-O-methyltransferase gene polymorphism is associated with risk of psychosis in Alzheimer Disease. Neurosci Lett. 2004;370:127–129. doi: 10.1016/j.neulet.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Zhu BT. CNS dopamine oxidation and catechol-O-methyltransferase: importance in the etiology, pharmacotherapy, and dietary prevention of Parkinson’s disease. Int J Mol Med. 2004;13:343–353. [PubMed] [Google Scholar]
- 44.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 45.Romano JD, Michaelis S. Topological and mutational analysis of Saccharomyces cerevisiae Ste14p, founding member of the isoprenylcysteine carboxyl methyltransferase family. Molecular biology of the cell. 2001;12:1957–1971. doi: 10.1091/mbc.12.7.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossmann MG, Argos P. The taxonomy of binding sites in proteins. Mol Cell Biochem. 1978;21:161–182. doi: 10.1007/BF00240135. [DOI] [PubMed] [Google Scholar]
- 47.Marapakala K, Ajees AA, Qin J, Sankaran B, Rosen BP. Crystallization and preliminary X-ray crystallographic analysis of the ArsM arsenic(III) S-adenosylmethionine methyltransferase. Acta crystallographica. 2010;66:1050–1052. doi: 10.1107/S1744309110027661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 49.Kagan RM, Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Archives of biochemistry and biophysics. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- 50.Kozbial PZ, Mushegian AR. Natural history of S-adenosylmethionine-binding proteins. BMC structural biology. 2005;5:19. doi: 10.1186/1472-6807-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas DJ, Li J, Waters SB, Xing W, Adair BM, Drobna Z, Devesa V, Styblo M. Arsenic (+3 oxidation state) methyltransferase and the methylation of arsenicals. Exp Biol Med (Maywood) 2007;232:3–13. [PMC free article] [PubMed] [Google Scholar]
- 52.Ramírez-Solis A, Mukopadhyay R, Rosen BP, Stemmler TL. Experimental and theoretical characterization of arsenite in water: insights into the coordination environment of As-O. Inorg Chem. 2004;43:2954–2959. doi: 10.1021/ic0351592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delnomdedieu M, Basti MM, Styblo M, Otvos JD, Thomas DJ. Complexation of arsenic species in rabbit erythrocytes. Chem Res Toxicol. 1994;7:621–627. doi: 10.1021/tx00041a006. [DOI] [PubMed] [Google Scholar]
- 54.Leslie EM. Arsenic-glutathione conjugate transport by the human multidrug resistance proteins (MRPs/ABCCs) J Inorg Biochem. 2011;108:141–149. doi: 10.1016/j.jinorgbio.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 55.Carew MW, Naranmandura H, Shukalek CB, Le XC, Leslie EM. Monomethylarsenic diglutathione transport by the human multidrug resistance protein 1 (MRP1/ABCC1) Drug Metab Dispos. 2011;39:2298–2304. doi: 10.1124/dmd.111.041673. [DOI] [PubMed] [Google Scholar]
- 56.Smith BC, Denu JM. Chemical mechanisms of histone lysine and arginine modifications. Biochimica et biophysica acta. 2009;1789:45–57. doi: 10.1016/j.bbagrm.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]