Abstract
In eukaryotes, the endoplasmic reticulum (ER) serves as the first membrane-enclosed organelle in the secretory pathway, with functions including protein folding, maturation and transport. Molecular chaperones, of the Hsp70 family of proteins, participate in assisting these processes and are essential to cellular function and survival. BiP is a resident Hsp70 chaperone in the ER of Saccharomyces cerevisiae. In this study the authors have created a partial differential equation model to examine how BiP interacts with the membrane-bound co-chaperone Sec63 in translocation, a process in which BiP assists in guiding a nascent protein into the ER lumen. It has been found that when Sec63 participates in translocation through localisation at the membrane, the spatial distribution of BiP is inhomogeneous, with more BiP at the surface. When translocation is inhibited through a disabling of Sec63’s membrane tether, the concentration of BiP throughout the ER becomes homogeneous. The computational simulations suggest that Sec63’s localisation and the resulting binding to BiP near the membrane surface of the ER enable a heterogeneous distribution of BiP within the ER, and may facilitate BiP’s role in translocation.
1 Introduction
Molecular chaperones participate in a wide range of processes essential to cellular function and survival. Found in all organisms, and ubiquitously distributed in the major compartments of eukaryotic cells, most are intricate players in the response to cellular stress [1]. Hsp70s assist in protein folding and maturation, assembly or disassembly of complexes, ribosomal RNA processing, translocation of newly synthesised proteins, suppression of aggregation and protein degradation (as reviewed in [2, 3]). The versatility among molecular chaperones is intriguing, for they have a single purpose: to bind protein substrates. The very high degree of conservation among Hsp70 proteins may favour a unique molecular mechanism common to all, whereas functional differences may depend on modulating co-chaperones such as Hsp40s and nucleotide exchange factors (NEFs) [4]. This phenomenon of a protein having multiple, sometimes competing functions, indicates a high level of systems control.
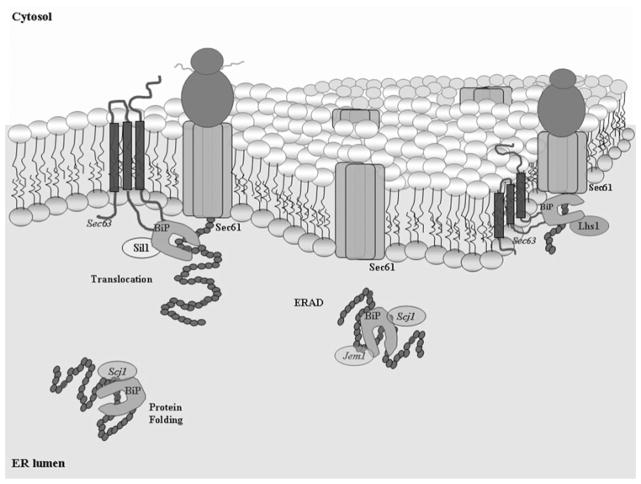
In the lumen of the endoplasmic reticulum (ER), a network of chaperones and cofactors ensures the proper folding of secretory proteins. One of the most abundant proteins of the ER is the Hsp70 molecular chaperone, BiP. Through biochemical and genetic experiments, BiP has been identified in critical cellular processes, including protein translocation of newly synthesised precursors across the ER membrane, folding and maturation, karyogamy and ER-associated degradation (ERAD) where unfolded or abnormally folded proteins are sent back to the cytosol for degradation [5–9]. Like other Hsp70 chaperones, BiP assists in folding of a protein by repeated adenosine triphosphate (ATP)-controlled cycles of binding and release. Co-chaperones, such as Hsp40s, stimulate the binding of molecular chaperones to the substrate and regulate chaperone activities [10]. In the ER of S. cerevisiae, co-chaperone Sec63 directly interacts with BiP, increasing its affinity for the nascent proteins proceeding through the translocation pore [11–13] (Fig. 1). Simultaneously within the ER lumenal environment, co-chaperones Scj1 and Jem1 associate with BiP during the processes of protein maturation and karyogamy, respectively [8, 9, 14]. BiP, Jem1 and Scj1 are all involved in the degradation of aberrant soluble proteins through ERAD [15]. As illustrated in Fig. 1, Sil1 and Lhs1 are the NEFs that play key roles in these processes by triggering substrate release [16, 17].
Fig. 1. Molecular chaperone BiP/Kar2 is required for efficient protein translocation into the ER lumen, protein folding and maturation and ERAD.
Interactions with selective co-chaperones include membrane protein Sec63 and freely diffusing Scj1 and Jem1. Intrinsic rates of peptide release are low; thus following ATP hydrolysis, nucleotide exchange is facilitated by Sil1 or Lhs1. In this illustration, Sec61 is a member of the ER membrane pore complex responsible for translocation and possibly the transport of aberrant proteins by ERAD
The regulation of Hsp70–Hsp40–NEF interactions is best understood for Escherichia coli homologues DnaK, DnaJ and GrpE. Mechanistic details have been experimentally explored and mathematically modelled [18–24]. Yet, E. coli is an organism that does not consist of membrane-bound compartments to perform distinct cellular functions. Additionally, many chaperone-mediated processes involve spatial aspects, such as the subcellular localisation of messenger RNA leading to translation of their encoded proteins [25]; subcompartments of the nucleus implicated in the processes of transcription and splicing [26, 27]; and the spatial localisation of BiP at the ER membrane maintaining the permeability barrier during protein translocation [28].
Yeast, S. cerevisiae, is a simple eukaryotic organism that compartmentalises selective processes and protein–protein interactions to specified organelles. Proteomic studies have verified the location of ER-resident proteins of S. cerevisiae and identified absolute levels of protein expression [29, 30]. These data suggest that the concentration of BiP exceeds the level of co-chaperones by at least an order of magnitude at normal growth conditions, and is significantly up-regulated during quality control mechanisms of the cell including the heat shock response and unfolded protein response (UPR) [31, 32]. It is also known that multiple BiPs can bind to substrates with varying affinities [33]. Experimentally, the co-chaperone Sec63 must be spatially localised at a suborganelle level – the ER membrane – in order for translocation to occur [34]. Given this evidence, we hypothesise that the spatial localisation of BiP and interactions with co-chaperones regulate its diversity and functionality in the ER of S. cerevisiae.
Thus, we describe our model development in Section 2, discuss the significance of our simulations in Section 3 and examine the sensitivity of our model to parameter perturbations in Section 4. In order to investigate the chaperone and co-chaperone interaction of BiP and Sec63, respectively, in ER translocation, we have introduced spatial components to our model. Subsequently, we have shown that Sec63s localisation and functional interaction with BiP in translocation provides an explanation for BiP’s heterogeneity in the ER.
2 Models
Modellers have attempted to discern the role of Hsp70 chaperones in assisting and accelerating translocation of proteins across membranes of organelles, specifically the ER and mitochondria of S. cerevisiae. Previous work has focused on transport mechanisms, including either the Brownian ratchet model [35], comparison to the power stroke model [36, 37] or a unifying mechanism of both termed entropic pulling [4]. We have examined the significance of spatial effects between BiP and the ER membrane co-chaperone Sec63 and implemented the reaction rates associated with previous models that evaluate a nascent protein as it transits through the translocation pore. Specifically, we are interested in how the BiP–Sec63 interaction enhances translocation of the nascent protein. Experiments exploring the BiP–Sec63 interaction in vitro suggest that Sec63 acts as an anchor to localise BiP within the proximity of the translocation channel, and accelerates the transit of a peptide through the membrane pore by regulating ATP hydrolysis of the chaperone [5, 34].
This work implements the experimental evidence [12, 16, 17, 38–42] with the added spatial component, including the ER membrane and lumenal regions. Developing spatially relevant models is important as in vivo experiments such as single particle tracking (SPT) and technology advancements in fluorescence imaging begin to capture spatial effects of protein localisation (reviewed in [43]). Estimates of species concentrations have been determined for S. cerevisiae [30] as well as binding affinities between BiP, Sec63 and synthetic peptides [38, 39]. When experimental data were not available, as in the case of the interactions between NEFs, Sil1 and Lhs1 and BiP, established estimates from the mammalian literature were used (supplementary material). However, a degree of uncertainty is inherent when evaluating kinetic rates as a result of in vitro experiments. We have incorporated these estimates into models which are used to elucidate in vivo mechanisms and varied the parameters by two orders of magnitude to test the sensitivity of the particular values. Owing to the high degree of homology between chaperones and co-chaperones among eukaryotes, we believe that this estimation procedure is an appropriate method.
Our goal is to use modelling to better understand the role of Sec63 in partitioning of BiP within the ER. To this end, we first constructed an ordinary differential equation (ODE) model to examine the reaction pathways and then extended this system to a partial differential equation (PDE) model in order to capture the spatiotemporal dynamics.
2.1 Model descriptions
2.1.1 Core ODE model
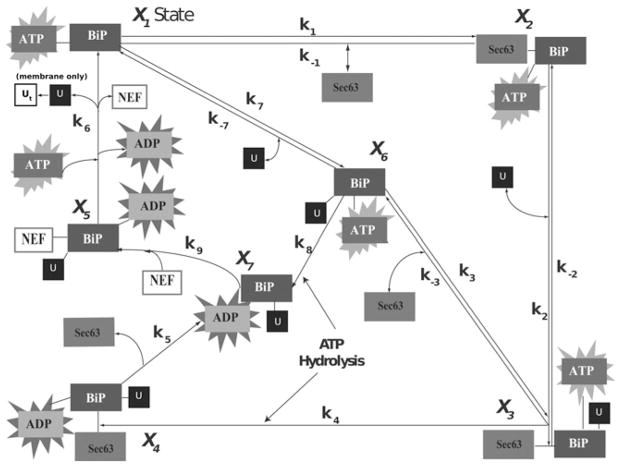
Our core model is described by a system of ODEs. It is a 7-state, 13-parameter model representing the interactions of BiP with the co-chaperone Sec63 and unfolded proteins. The schematic below, Fig. 2, depicts all plausible states of BiP binding to nascent proteins during protein translocation into the ER lumen. In general, molecular chaperones alternate between an ATP-bound state representing fast substrate binding/release rates and therefore a low affinity for proteins (upper right triangle, Fig. 2), and an ADP-bound state characteristic of slow association/dissociation kinetics and a high affinity for substrates (lower left triangle, Fig. 2) [44]. The states of the model are given in Table 1.
Fig. 2.
Schematic of our ODE model consisting of seven states that represent the interactions of BiP with co-chaperone Sec63, nascent and unfolded proteins (U), NEFs Sil1 and Lhs1 and required energy components ATP and ADP
Table 1.
List of state numbers and names for the core ODE model
| State number | State name |
|---|---|
| X1 | BiP–ATP |
| X2 | BiP–Sec63–ATP |
| X3 | BiP–Sec63–U–ATP |
| X4 | BiP–Sec63–U–ADP |
| X5 | BiP–U–NEF–ADP |
| X6 | BiP–U–ATP |
| X7 | BiP–U–ADP |
Under physiological conditions, BiP preferentially binds to ATP (X1) and associates with either unfolded proteins in the lumen or nascent proteins of the translocon (Fig. 1). BiP’s interaction with unfolded proteins can occur in the presence or absence of co-chaperone mediation (X1 → X6) [38, 40] where BiP preferentially binds to hydrophobic residues [33, 45]. ATP hydrolysis ensues (X6 → X7), trapping the peptide to form X7. Owing to low intrinsic rates of bound peptide released from the chaperone complex, NEFs further accelerate the cycle of protein folding (X7 → X5 → X1). The conventional mechanism of translocation proceeds with the binding of the co-chaperone Sec63 to BiP (X2) which synergistically stimulates peptide binding (X3) and ATP hydrolysis (X4). This coupling effect has been shown to mediate the molecular trapping of proteins that BiP would not bind on its own [38, 39]. However, it is also plausible that BiP could initially bind to the unfolded protein and then sequentially associate with its co-chaperone during translocation. This scenario is represented in states X1 → X6 → X3.
We have assumed that a single BiP is activated per Sec63 molecule located at the ER membrane. In vitro studies have indicated that the BiP–Sec63 interaction occurs transiently, and suggest that one Sec63 molecule could potentially activate at least ten BiP molecules [38]. On an average, E. coli experiments have determined that the incoming proteins present a new Hsp70 binding site for every 25 to 35 amino acids [46], which is consistent with S. cerevisiae literature that estimates a minimum of six to seven molecules of BiP bound to the endogenous protein, prepro α-factor [47]. For simplicity, our model assumes a 1:1 stoichiometry between BiP and unfolded proteins as well as between BiP and Sec63.
Simulations were realised until the values reached a steady-state level and then the final species concentrations were obtained for a range of initial conditions. We found that when the total amount of BiP is low, the Sec63-dependent pathway (Fig. 2, outer loop) accounted for most of the bound BiP. As the total amount of BiP in the system was increased, the Sec63-dependent pathway (Fig. 2, diagonal) dominated, and states such as BiP–U–ATP (X6) accounted for most of the BiP. The latter result confirmed an important point that BiP strongly interacts with unfolded protein, although it should be mentioned that the association rate to form X6 was chosen on the higher end of the range of experimental data [24]. This core ODE model served as a description of reaction kinetics between BiP, Sec63 and unfolded protein, and was a building block for constructing a spatially dependent model describing chaperone interactions in the ER. Details of the model are summarised in the Supplementary Material.
2.1.2 PDE model
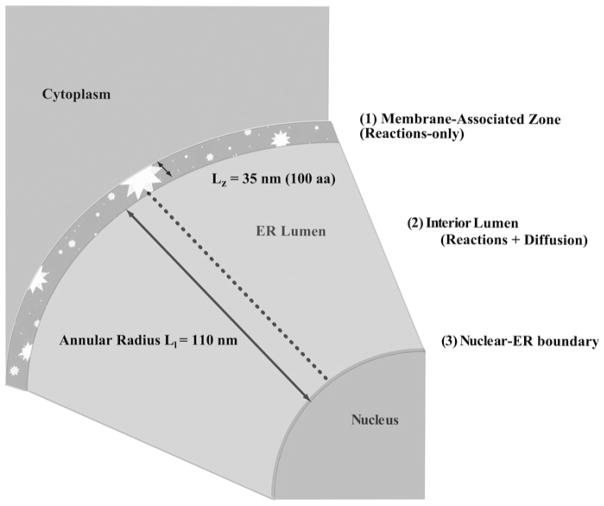
We constructed a PDE model, making use of the reaction kinetics from the ODE model, to determine the spatial distribution of BiP in the ER. The model incorporates: (i) chemical reactions representing transitions between states in the ODE system that take place at the inner membrane and (ii) diffusion into the lumen of the ER. This spatially dependent system of equations was approximated by the method of finite differences (Fig. 3). The irregular geometry of the ER was simplified to a sphere (surrounding a spherical nucleus) and assumed to be symmetric. With these assumptions, the (time-dependent) system can be modelled in one spatial dimension. We believe a PDE model is justified because BiP and (wild-type) Sec63 have a localised interaction at the membrane that precludes a well-mixed system.
Fig. 3. PDE model consists of a membrane-associated zone and lumen represented by reaction–diffusion equations.
Length of the membrane-associated zone, Lz, was taken to be 35 nm, while the annular radius of the lumen, Ll, is 110 nm ([51, 52]; Supplementary Material)
We define the membrane-associated zone as the membrane portion of the ER where protein–protein interactions take place between BiP and the other proteins in the system. Images attained by electron microscopy (EM) show that the membrane has a defined thickness [48], shown as 35 nm in Fig. 3. Sec63 spans the membrane [49] and associates with the Sec61-constituted pore channel [50]. We do, however, consider the entire region as well-mixed, represented by the boundary condition of the PDE.
The rate of change of the concentration of species k is the sum total of the concentrations of the free and bound species plus the diffusion in the interior.
| (1) |
Reactions and (one-dimensional) diffusion occurs in the lumen
| (2) |
with the nuclear-ER boundary condition given by
| (3) |
Here, Cz,k is the membrane-associated zone concentration and Cl,k represents the concentration in the lumen for diffusing species k. If this species exists only in the membrane-associated zone, then this term is absent. D is the diffusion coefficient (assumed to be the same for all ER lumenal species), and Rk is the reaction term for species k. Lz is the length of the membrane-associated zone (in one dimension), and Ll is the annular radius of the lumen. The variable x represents the distance from the membrane-associated zone for a particular point on the grid.
Discretising in space, we obtain
| (4) |
| (5) |
| (6) |
where represents the concentration of species k at grid point i in the lumen, and Δx is the spatial separation between two consecutive grid points. The boundary at x = 0 is the ER membrane, and the boundary at x = Ll is the nucleus. Given this formulation, the concentration of each species is tracked spatially and temporally.
Several assumptions were made to simplify the model and approximate the dynamics of the system. Reactions can occur in both the membrane-associated zone and the lumen. We define unfolded proteins as one species without distinguishing between proteins diffusing in the lumen from those transiting the translocation pore. Loss of proteins at the translocon (due to completed translocation) is offset by a production reaction (Ø → U ) that allows for replenishment of the U pool. The translocated U is designated as a non-reacting product Ut (Fig. 2). Thus, the total amount of unfolded protein in the model remains constant.
3 Simulation results
3.1 Model scenarios
Three scenarios were constructed to simulate different conditions in the ER. In each scenario, different species were allowed to diffuse in the lumen. In our model, we assume that if all the constituent species in a bound state can diffuse in a particular scenario, then the bound state can diffuse as well. The scenarios are:
The wild-type case that assumes that ER-resident chaperone BiP, NEFs Sil1 and Lhs1, soluble U and selective complexes diffuse freely throughout the lumen [16, 41, 53, 54]. Sec63 is an integral membrane protein and therefore remains embedded in the ER lipid bilayer [55].
In addition to the species outlined in Scenario 1, Sec63 and membrane-associated complexes are now allowed to diffuse into the lumen. Experimental data have shown that the presence of a soluble variant of Sec63 (63Jp) not localised to the ER membrane inhibits efficient protein translocation. These results suggest that 63Jp competes with the endogenous membrane-localised Sec63 by sequestering BiP from the vicinity of the translocon pore (Sec61 complex of Fig. 1) [34]. In this case, we can test whether the variant’s ability to diffuse into the lumen leads to a loss of BiP’s heterogeneous distribution in the ER.
When the BiP–Sec63 protein interactions are severely impaired, or in the absence of Sec63 within the system of scenario 1 (i.e. zero concentration), we have been able to mimic a situation in which ER protein translocation fails. Mutations in either Sec63 or BiP have been shown to inhibit translocation due to defects in Sec63 interacting with BiP (Sec63-1, [5, 56]) and, to varying degrees, BiP mutants display different translocation efficiencies into the lumen (kar2-113, kar2-159 and kar2-203; [57]).
It should be noted that only Scenario 1 represents physiological conditions. Scenarios 2 and 3 are special conditions that have been experimentally obtained using either protein variants or thermosensitive yeast strains. The species and states that diffuse in each scenario are given in Table 2.
Table 2.
Matrix describing which states and species diffuse in each scenario
| Diffusing species | BiP–ATP | BiP–Sec63–ATP | BiP–Sec63–U–ATP | BiP–Sec63–U–ADP | BiP–NEF–U–ADP | BiP–U–ATP | BiP–U–ADP | |
|---|---|---|---|---|---|---|---|---|
| 1 | BiP | |||||||
| U | ML | M | M | M | ML | ML | ML | |
| NEF | ||||||||
| 2 | BiP | |||||||
| Sec63 | ML | ML | ML | ML | ML | ML | ML | |
| NEF, U | ||||||||
| 3 | BiP | ML | ML | ML | ML | |||
| NEF, U | ||||||||
| Sec63 = 0 |
States that remain in the membrane-associated zone (M) are shaded in grey, while those states that can diffuse into the lumen as well (ML) are unshaded. Black represents states that are not present in the scenario
3.2 Ratio metric
From the ER PDE model, we determined the ratio of total BiP concentration localised to the membrane-associated zone compared to the total BiP concentration of the interior. This metric gives an indication of BiP’s spatial localisation within the ER subcompartments. The default scenario is to tether Sec63 to the membrane, while BiP, NEF and U are allowed to diffuse. From these scenarios, one can make predictions of the importance of these species on translocation. This is described by the equation
| (7) |
where the subscript z means the membrane-associated zone and l means the lumen. At steady-state, the concentrations of diffusing species in the membrane-associated zone and the lumen are the same, with the concentration of the non-diffusing species on the membrane-associated zone determining the divergence of the BiP ratio from unity.
3.3 Model results
Using the DASPK ODE/DAE solver [58], we ran simulations for each of the scenarios until steady-state (t = 105 s). The system starts from conditions where all proteins are free and present in the membrane-associated zone. Diffusion is fast, equalising gradients across the ER. Reactions then occur in the membrane-associated zone and in the lumen on a slower time scale. We analysed the concentration of each state in the membrane-associated zone and at interior grid points and then calculated the ratio of total BiP concentration in the membrane-associated zone compared to the total BiP concentration in the lumen. Simulations accounted for total Sec63 ranging from 0.55 to 1.37 mM (10 000–25 000 molecules) while BiP and NEF concentrations varied from 0.55 to 55 mM (104 – 106 molecules). These initial conditions were based on a range around populations given in the yeast genome database [30]. The amount of total unfolded protein (U ) was 5.5 mM or 105 molecules (see Section 4).
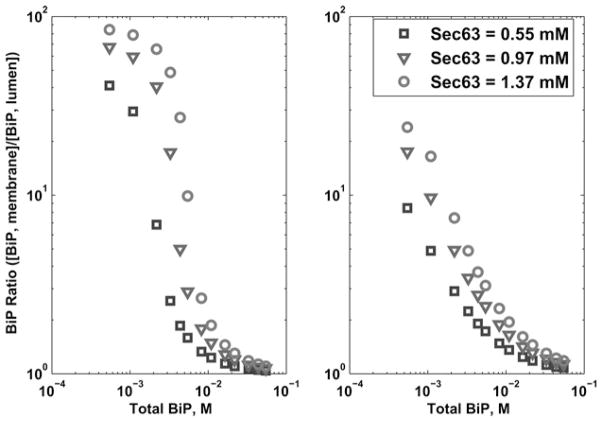
When Sec63 is appropriately integrated in the membrane, BiP preferentially remains in the membrane-associated zone (Fig. 4), resulting in an inhomogeneous distribution throughout the ER. This is expected from the model since BiP is reacting in the membrane-associated zone and forming non-diffusing species. When Sec63 is allowed to diffuse or is removed from the system entirely, BiP is uniformly distributed in the ER, resulting in a BiP ratio (7) of one for all total values of Sec63 and BiP. Thus, when translocation is inhibited, the concentration is homogeneous. When total BiP levels are low, most of it is involved in translocation, so the BiP ratio is high (left side of Fig. 4 graph). In the limit of high BiP levels, relatively less BiP is used in translocation, so its distribution tends towards homogeneity (right side of Fig. 4 graph). This calculation was initially performed with 11 spatial grid points and repeated for several grid sizes in order to increase the resolution of our results (e.g. up to 1000), yielding identical results for the ratios.
Fig. 4. Log–log plots for Scenario 1 (Sec63 is tethered to the membrane) detailing the BiP ratio (total BiP concentration on the membrane divided by the total BiP concentration in the lumen) for total BiP varying from 0.55 to 55 mM (104 to 106 molecules) for the modified ODE (left) and PDE (right) models.
Graphs show three sets of different total Sec63 concentrations (corresponding to 10 000, 17 700 and 25 000 molecules). When Sec63 is allowed to diffuse (Scenario 2) or is removed from the system (Scenario 3), the BiP ratio is uniformly one
We examined the results of an ODE model to describe the impact of the interaction between BiP and Sec63 on the chaperone distribution in the ER. In this model, we assumed that fast diffusion produced a well-mixed system for all species. We then scaled the concentrations of Sec63-derived states (i.e. X2, X3 and X4) with respect to the volume of the membrane-associated zone and the concentrations of the remaining states using the combined volume of the membrane zone and lumen. The calculation of the BiP ratio then proceeded as defined in the last section.
In comparison with the PDE model, we found that this modified ODE model resulted in a higher BiP ratio at low total BiP populations as shown in Fig. 4. This discrepancy occurs because in the ODE model, Sec63 has access to the entire BiP population for translocation. The PDE model, on the other hand, allows for the presence of BiP both in the membrane-associated zone and in the lumen. In this case, a percentage of BiP does not participate with Sec63 in translocation. Especially when BiP is scarce, this difference produces a higher BiP ratio in the ODE model against the PDE model. Given BiP’s known presence in both membrane-associated zone and in the lumen, we believe that a spatial model best describes the chaperone dynamics in the ER. Furthermore, the PDE model takes into account that the lumenal BiP is involved in other processes (e.g. protein folding and degradation).
The determination of an actual physiological value of the BiP ratio in Fig. 4 is complicated; many cellular factors affect it. For example, if BiP binds at higher stoichiometric ratios to Sec63 and U than 1:1, then there would be much more BiP found in the membrane-associated zone, and the BiP ratio would increase for the same initial conditions. BiP is involved in multiple non-translocation cellular functions [59], which may result in a smaller percentage available to translocation, and would describe the right side of Fig. 4. If either BiP or Sec63 is partially defective in their interaction with each other [39, 60], this scenario would lead to flattening of the curves towards homogeneity. Additionally, perturbations to the cellular environment, such as the induction of stress conditions in the ER, lead to membrane expansion [61] and a resulting increase in ER volume [62], yet simultaneously, the UPR increases the amount of BiP in the lumen to handle an unfolded protein load [32]. Thus, the BiP ratio would change dynamically in time. This project does not attempt a thorough investigation of protein folding in the ER nor perturbations to the system that, in turn, induce various quality control mechanisms; rather our model introduces the effect of dimensionality – specifically ER compartmentalisation – and shows that BiP’s interaction with Sec63 at the membrane brings about spatial localisation in the chaperone’s distribution within the ER, which will ultimately have implications in protein folding and other ER-related functions.
4 Sensitivity analysis
Although we found BiP’s concentration in the ER under wild-type conditions to be inhomogeneous (Scenario 1), we also wanted to examine how the degree of heterogeneity (as quantified in the BiP ratio) would change under variation in model parameters and initial conditions, to examine the influence of various model inputs on the system.
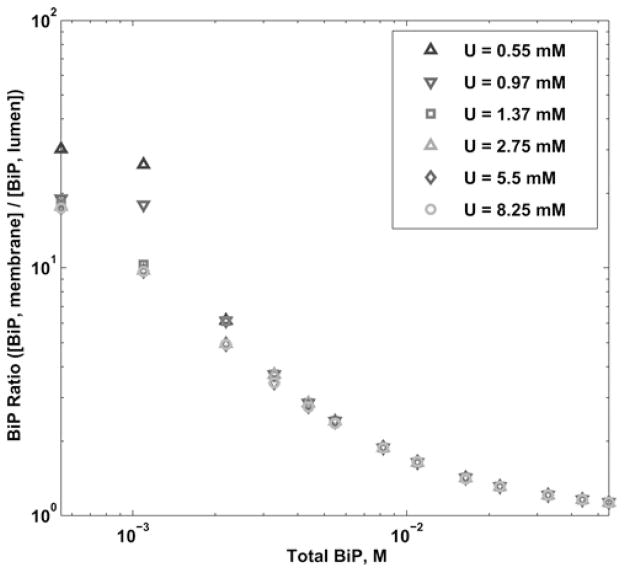
We first simulated the variation in total number of each protein species in the ER. The yeast genome database reports the total physiological population of BiP and Sec63 proteins at 337 000 and 17 700 molecules, respectively, but with those estimates there is a great deal of uncertainty. It is known that BiP levels in the yeast ER can fluctuate and are regulated by the UPR. Cellular conditions also dictate a large possible range in concentration of unfolded proteins. Thus, we simulated the model over a large range of values for all protein species, as summarised in Table 3. The resulting BiP ratio for variation in Sec63 is given in Fig. 4. We also examined a range of unfolded protein concentrations in our simulations from 0.55 to 8.25 mM (10 000–150 000 molecules). The ratio results for U are shown in Fig. 5. An estimate for U came from surface plasmon resonance experiments [39] using the peptide p5 at 2 mM or roughly 40 000 molecules in the ER. We found that this value of protein concentration resulted in less than a one per cent deviation in the BiP ratio from our standard conditions (at 5.5 mM or 100 000 molecules). Most of the U binds with free BiP, but the vast excess remains free and is not a factor of the BiP ratio calculation. Overall, our simulations with the aforementioned Sec63 and U concentrations have confirmed our previous result that when BiP levels are low, the BiP ratio is high and is affected by the amount of Sec63 in the system. As BiP levels are increased, the BiP ratio becomes insensitive to changes in all initial conditions.
Table 3.
Range of initial populations (and concentrations) for protein species
| Species | Initial population range | Initial concentration range, mM |
|---|---|---|
| BiP | 10 000–1 000 000 | 0.55–55 |
| Sec63 | 10 000–25 000 | 0.55–1.37 |
| NEF | 10 000–1 000 000 | 0.55–55 |
| U | 10 000–150 000 | 0.55–8.25 |
Fig. 5. Log–log plot of the BiP ratio in the wild-type scenario for a variation in total BiP and U, while fixing Sec63 = 0.97 mM.
Graph shows six sets of different total U population (from 0.55 to 8.25 mM)
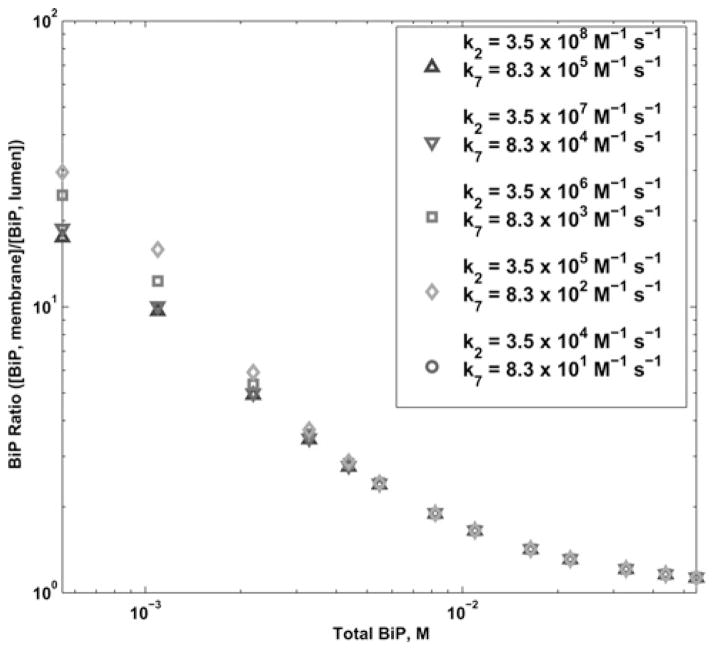
We next considered the binding rate between BiP and U as variable parameters (kinetic parameters k2 and k7, Fig. 6). BiP interacts with varying affinities to numerous types of unfolded proteins in the ER and these interactions are dependent on the exposed hydrophobic residues of the unfolded proteins [45]. Multiple BiPs can bind to a single translocating protein resulting in stoichiometric effects on the reaction, impacting the overall rate as well [35]. We varied the association rates over five orders of magnitude and recorded the BiP ratio for each simulation. As the association rate between BiP and U increases, there is less of these species in the free (as opposed to bound) state, but this does not affect the total amount of BiP bound to Sec63 at the membrane. Therefore we found that the BiP ratio was essentially the same for all kinetic values.
Fig. 6.
Log–log plot of the BiP ratio for variation in the Sec63-dependent (k2) and Sec63-independent BiP–U association rate (k7) from 3.5 × 104 to 3.5 × 108 M−1s−1 and 8.3 × 101 to 8.3 × 105 M−1s−1, respectively
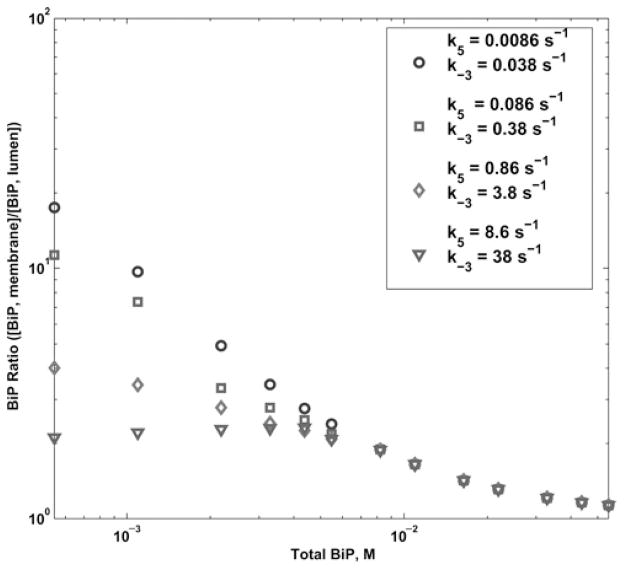
We examined the effect on the BiP ratio given a change in the Sec63 dissociation rate from the trimeric complex of BiP, Sec63 and U (Fig. 7). This is described by the kinetic rte constants k5 and k−3. The values from the literature are 0.0086 and 0.038 s−1 [38], respectively. Rate k5 is slower than the reactions upstream and downstream in the pathway (e.g. k4 = 0.016 s−1 and k6 = 0.267 s−1). Thus, a bottleneck occurs, resulting in a high concentration of the BiP–Sec63–U–ADP (X4) state. In vitro experiments [39, 63, 64] suggest that Sec63 may have a more transient interaction with BiP, which would allow for higher translocation efficiency. We increased the Sec63 dissociation rate to 10, 100 and 1000-fold, and found that the BiP ratio decreased at low levels of BiP and Sec63. We surmise this is due to translocating protein being shuttled out of the Sec63-dependent pathway at a faster rate, which in the wild-type scenario accounts for the excess BiP on the membrane.
Fig. 7.
Log–log plot of the BiP ratio for variation in Sec63 dissociation rate (k5, k−3) from 0.0086 to 8.6 s−1 and 0.038 to38 s−1, respectively
These results were also verified by local sensitivity analysis. Using SUNDIALS [65], we computed the sensitivities of the BiP ratio to perturbations of the reaction-rate parameters. If the BiP ratio is defined as a function G, by the chain rule we obtain the derived function
| (8) |
where C and p represent the states and parameters, respectively. The derivative ∂C/∂p is the standard sensitivity matrix while the second term ∂G/∂p equals zero because the BiP ratio has no explicit dependence on parameters. The BiP ratio sensitivities are then scaled by the BiP ratios and parameter values to allow for comparison. The results are summarised in Table 4.
Table 4.
Scaled sensitivities of the BiP ratio ranked by maximum value for 13 sets of BiP initial conditions in the range of 0.55 to 55 mM
| Parameter | Max, dG/dp | Min, dG/dp | Mean, dG/dp |
|---|---|---|---|
| k5 | 2.67 × 10−1 | 6.857 × 10−4 | 5.53 × 10−2 |
| k6 | 6.94 × 10−2 | 1.27 × 10−4 | 1.07 × 10−1 |
| k4 | 5.30 × 10−2 | 3.89 × 10−5 | 7.81 × 10−3 |
| k9 | 4.01 × 10−2 | 8.44 × 10−9 | 5.40 × 10−3 |
| k7 | 1.35 × 10−2 | 3.18 × 10−5 | 2.26 × 10−3 |
| k8 | 1.07 × 10−2 | 4.67 × 10−6 | 1.37 × 10−3 |
| k3 | 1.07 × 10−3 | 3.27 × 10−8 | 9.59 × 10−4 |
| k−2 | 5.54 × 10−3 | 3.28 × 10−6 | 6.27 × 10−4 |
| k−7 | 2.93 × 10−3 | 2.53 × 10−7 | 5.70 × 10−5 |
| k1 | 8.32 × 10−4 | 1.67 × 10−7 | 1.93 × 10−5 |
| k−3 | 4.09 × 10−4 | 1.61 × 10−8 | 3.43 × 10−6 |
| k2 | 4.85 × 10−5 | 6.85 × 10−8 | 2.68 × 10−6 |
| k−1 | 6.47 × 10−7 | 7.09 × 10−9 | 1.54 × 10−8 |
We took the minimum and maximum of 13 sets of initial conditions as defined in the last section as the range of our sensitivities. We then ranked the parameters by their maximum and calculated the mean of the ensemble. As Table 4 illustrates, the BiP ratio is robust to perturbations in parameters.
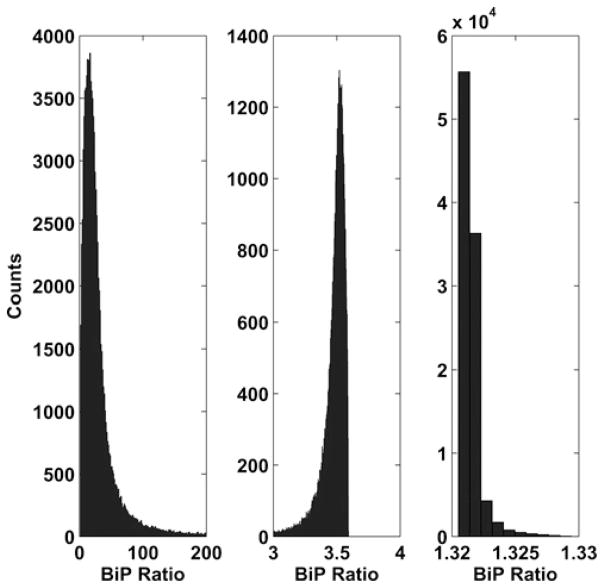
Finally, we examined the sensitivity of the BiP ratio by varying all the kinetic parameters simultaneously. We used uniform random numbers to select each parameter value in the range [0.1pi, 10pi], where pi is the nominal value of the parameter. From this method, we generated 105 sets of parameters to serve as the input for the simulations. We then simulated the model, increasing the total BiP concentration from 0.55 to 55 mM, and recording the resulting distributions of the BiP ratio in a histogram as presented in Fig. 8. Other metrics were considered [66], but we wanted to examine the global sensitivity of the BiP ratio and to vary more than one parameter at a time.
Fig. 8.
Histograms of the BiP ratio for 104, 105 and 106 BiP molecules (0.55, 5.5 and 55 mM, respectively) while varying all kinetic parameters simultaneously
We found that at the low end of the BiP concentration range (0.55 mM), the BiP ratio has a wide spread of values for the simulations. When BiP is scarce, changes in kinetic parameters have a significant effect on whether the BiP is bound to Sec63 at the membrane or diffuses freely in the lumen. Despite this sensitivity, most simulations yielded a highly heterogeneous BiP distribution (BiP ratio = 3.5–163.7, 95% confidence interval for total BiP = 0.55 mM), confirming that most of the BiP participates in translocation. The tails of the BiP ratio distributions are due to the combination of parameters all being near their maximum values. As the total BiP in the ER increases, more of the BiP is not interacting with Sec63 (but rather freely diffusing in the lumen); thus the reaction parameters have much less effect. Further studies concluded that no one parameter dominates the variation of the BiP ratio. The qualitative behaviour of the model was reproduced in all 105 simulations.
In summary, the BiP distribution is heterogeneous with a high concentration of BiP near the membrane for the wild-type condition where Sec63 is tethered to the membrane. In contrast, for situations where Sec63 is allowed to freely diffuse into the lumen, the BiP concentration is homogeneous. Is Sec63 recruiting BiP to the membrane? In the model there is no driving force for BiP diffusion towards the membrane. However, BiP has a relatively fast diffusion rate, which means that a BiP molecule readily traverses both the lumen and the membrane-associated zone. When it arrives at the membrane-associated zone, it has a high probability of binding to Sec63, and this binding results in a preferential localisation at the membrane. This supports the model that the J-domain co-chaperones act to modulate BiP localisation in the ER.
5 Conclusion
We have developed a spatial PDE model describing the chaperone activity in the ER of S. cerevisiae through reaction–diffusion equations. From the simulations, we found that the concentration of BiP in the system was inhomogeneous, with the concentration being greater on the membrane, particularly as BiP levels in the ER decrease. Our simulations showed, however, that when Sec63 was untethered and allowed to diffuse freely into the lumen, the BiP distribution was homogeneous. This is consistent with the absence of BiP (translocation failure) at the membrane observed experimentally in this situation. Sec63’s localisation and functional interaction with BiP in translocation provides an explanation for BiP’s heterogeneity in the ER.
Supplementary Material
Acknowledgments
6 Acknowledgments
The authors gratefully acknowledge funding from NIH Grant GM07529 and NSF IGERT Grant DGE02-21715. We would also like to thank David Raden (U. Delaware) and Theresa Yuraszeck (UC Santa Barbara) for their helpful discussions.
References
- 1.Georgopoulos C, Welch W. Role of the major heat shock proteins as molecular chaperones. Annu Rev Cell Biol. 1993;9:601–634. doi: 10.1146/annurev.cb.09.110193.003125. [DOI] [PubMed] [Google Scholar]
- 2.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanisms. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck T, Wright C, Brodsky J. The activities and function of molecular chaperones in the endoplasmic reticulum. Semin Cell Dev Biol. 2007;18:751–761. doi: 10.1016/j.semcdb.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de los Rios P, Ben-Zvi A, Slutsky O, Azem A, Goloubinoff P. Hsp70 chaperones accelerate protein translocation and the unfolding of stale protein aggregates by entropic pulling. Proc Natl Acad Sci. 2006;103:6166–6171. doi: 10.1073/pnas.0510496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodsky JL, Schekman R. A Sec63p–BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123(6 Pt 1):1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Latterich M, Schekman R. The Karyogamy gene KAR2 and novel proteins are required for ER-membrane fusion. Cell. 1994;78(1):87–98. doi: 10.1016/0092-8674(94)90575-4. [DOI] [PubMed] [Google Scholar]
- 7.McCracken AA, Brodsky JL. Evolving questions and paradigm shifts in endoplasmic reticulum-associated degradation (ERAD) BioEssays. 2003;25(9):868–877. doi: 10.1002/bies.10320. [DOI] [PubMed] [Google Scholar]
- 8.Nishikawa S, Endo T. The yeast Jem1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J Biol Chem. 1997;272(20):12889–12892. doi: 10.1074/jbc.272.20.12889. [DOI] [PubMed] [Google Scholar]
- 9.Schlendstedt G, Harris S, Risse B, Lill R, Silver P. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/Kar2 via a conserved domain that specifies interactions with Hsp70s. J Cell Biol. 1995;129:979–988. doi: 10.1083/jcb.129.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hennessy F, Nicoll W, Zimmermann R, Cheetham M, Blatch G. Not all J domains are created equal: implications for the specificity of Hsp40–Hsp70 interactions. Prot Sci. 2005;14:1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scidmore MA, Okomura HH, Rose M. Genetic interactions between KAR2 and SEC63, encoding eukaryote homologues of DnaK and DnaJ in the endoplasmic reticulum. Mol Biol Cell. 1993;4:1145–1159. doi: 10.1091/mbc.4.11.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyman SK, Schekman R. Interaction between BiP and Sec63 is required for the completion of protein translocation into the ER of Saccharomyces cerevisiae. J Cell Biol. 1995;131(5):1163–1171. doi: 10.1083/jcb.131.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young B, Craven R, Reid P, Willer M, Stirling C. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 2001;20:262–271. doi: 10.1093/emboj/20.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silberstein S, Schlenstedt G, Silver P, Gilmore R. A role of the DnaJ homologue Scj1 in protein folding in the yeast endoplasmic reticulum. J Cell Biol. 1998;143:921–933. doi: 10.1083/jcb.143.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153(5):1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyson J, Stirling C. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 2000;19:6440–6452. doi: 10.1093/emboj/19.23.6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steel G, Fullerton D, Tyson J, Stirling C. Coordinated activation of Hsp70 Chaperones. Science. 2004;303:98–101. doi: 10.1126/science.1092287. [DOI] [PubMed] [Google Scholar]
- 18.Gisler SM, Pierpaoli EV, Christen P. Catapult mechanism renders the Chaperone action of Hsp70 unidirectional. J Mol Biol. 1998;9:833–840. doi: 10.1006/jmbi.1998.1815. [DOI] [PubMed] [Google Scholar]
- 19.Mayer MP, Schröder H, Rüdiger S, Paal K, Bukau B. Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J Mol Biol. 1999;289:1131–1134. doi: 10.1006/jmbi.1999.2844. [DOI] [PubMed] [Google Scholar]
- 20.Mayer MP, Schröder H, Paal K, Bukau B. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat Struct Biol. 2000;7:586–593. doi: 10.1038/76819. [DOI] [PubMed] [Google Scholar]
- 21.Schmid D, Bacci A, Gehring H, Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 22.Laufen T, Mayer MP, Beisel C, Klostermeier D, Reinstein J, Bukau B. Mechanism of regulation of Hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci USA. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chesnokova LS, Slepenkov SV, Protasevich I, Sehorn MG, Brouillette CG, Witt SN. Deletion of DnaK’s Lid strengthens binding to the nucleotide exchange factor, GrpE: a kinetic and thermodynamic analysis. Biochemistry. 2003;42:9028–9046. doi: 10.1021/bi0346493. [DOI] [PubMed] [Google Scholar]
- 24.Hu B, Mayer MP, Tomita M. Modeling Hsp70-mediated protein folding. Biophys J. 2006;91(2):496–507. doi: 10.1529/biophysj.106.083394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holt CE, Bullock S. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326(5957):1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 27.Misteli T, Spector DL. The cellular organization of gene expression. Curr Opin Cell Biol. 1998;10:322–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- 28.Haigh N, Johnson A. A new role for BiP: closing the aqueous translocon pore during protein integration into the ER membrane. J Cell Biol. 2002;156:261–270. doi: 10.1083/jcb.200110074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huh W-K, Falvo JV, Gerke LC, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 30.Ghaemmaghami S, Huh W-K, Bower K, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 31.Mager WH, Herreira PM. Stress response of yeast. Biochem J. 1993;290(Pt. 1):1–13. doi: 10.1042/bj2900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Travers K, Patil CK, Wodicka L, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 33.Flynn G, Pohl J, Flocco M, Rothman J. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 34.Corsi AK, Schekman R. The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J Cell Biol. 1997;137(7):1483–1493. doi: 10.1083/jcb.137.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liebermeister W, Rapoport TA, Heinrich R. Ratcheting in posttranslational protein translocation: a mathematical model. J Mol Biol. 2001;305(3):643–656. doi: 10.1006/jmbi.2000.4302. [DOI] [PubMed] [Google Scholar]
- 36.Elston T. The Brownian ratchet and power stroke models for posttranslational protein translocation into the endoplasmic reticulum. Biophys J. 2002;82(3):1239–1253. doi: 10.1016/S0006-3495(02)75480-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chauwin J, Oster G, Glick B. Strong precursor-pore interactions constrain models for mitochondrial protein import. Biophys J. 1998;74:1732–1743. doi: 10.1016/S0006-3495(98)77884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misselwitz B, Staeck O, Rapaport TA. J-Proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol Cell. 1998;2(5):593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 39.Misselwitz B, Staeck O, Matlack KES, Rapaport TA. Interaction of BiP with the J-domain of the Sec63p component of the endoplasmic reticulum protein translocation complex. J Biol Chem. 1999;274(29):20110–20115. doi: 10.1074/jbc.274.29.20110. [DOI] [PubMed] [Google Scholar]
- 40.Mayer M, Reinstein J, Buchner J. Modulation of the ATPase cycle of BiP by peptides and proteins. J Mol Biol. 2003;330:137–144. doi: 10.1016/s0022-2836(03)00556-4. [DOI] [PubMed] [Google Scholar]
- 41.Kabani M, Beckerich J, Gaillardin C. Sls1p Stimulates Sec63p-mediated activated of Kar2p in a conformation-dependent manner in the yeast endoplasmic reticulum. Mol Cell Biol. 2000;20:6923–6935. doi: 10.1128/mcb.20.18.6923-6934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Keyzer J, Steel G, Hale S, Humphries D, Stirling C. Nucleotide binding by Lhs1p is essential for its nucleotide exchange activity and for function in vivo. J Biol Chem. 2009;284:31564–31571. doi: 10.1074/jbc.M109.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levi V, Gratton E. Exploring dynamics in living cells by tracking single particles. Cell Biochem Biophys. 2007;48:1–15. doi: 10.1007/s12013-007-0010-0. [DOI] [PubMed] [Google Scholar]
- 44.Liberek K, Marszalek J, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blond-Elguindi S, Dower W, Lipshutz R, Sprang S, Sambrook J, Geiting M-JH. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75(4):717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 46.Rudiger S, Buchberger A, Bukau B. Interaction of Hsp70 chaperones with substrates. Nat Struct Biol. 1997;4:342–349. doi: 10.1038/nsb0597-342. [DOI] [PubMed] [Google Scholar]
- 47.Matlack K, Misselwitz B, Plath K, Rapoport T. BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell. 1994;97:553–564. doi: 10.1016/s0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
- 48.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4(12):2311–2324. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Craig EA, Huang P, Aron R, Andrew A. The diverse roles of J-proteins: the obligate Hsp70 Co-chaperone. Rev Physiol Biochem Pharmacol. 2006;156:1–21. doi: 10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- 50.Römisch K. Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane. J Cell Sci. 1999;112:4185–4191. doi: 10.1242/jcs.112.23.4185. [DOI] [PubMed] [Google Scholar]
- 51.Johnson A, van Waes MA. The translocon: a dynamic gateway at the ER membrane. Ann Rev Cell Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 52.Beck M, Förster F, Ecke M, et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306(5700):1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- 53.Rose MD, Misra LM, Vogel JP. Kar2, a karyogamy gene, is the yeast homologue of the mammalian BiP/Grp78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 54.Craven RA, Egerton M, Stirling CJ. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 1996;15(11):2640–2650. [PMC free article] [PubMed] [Google Scholar]
- 55.Feldheim D, Rothblatt J, Schekman R. Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol Cell Biol. 1992;12:3288–3296. doi: 10.1128/mcb.12.7.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum of yeast. J Cell Biol. 1989;109:2641–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brodsky JL, Goeckeler J, Schekman R. BiP and Sec63p are required for both co- and post-translational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brenan KE, Campbell SL, Petzold LR. SIAM Classics in Applied Mathematics Series. 2 New York: 1996. The numerical solution of initial value problems and differential-algebraic equations. [Google Scholar]
- 59.Fewell SW, Travers K, Weissman JS, Brodsky JL. The action of molecular chaperones in the early secretory pathway. Ann Rev Gene. 2001;35(1):149–191. doi: 10.1146/annurev.genet.35.102401.090313. [DOI] [PubMed] [Google Scholar]
- 60.Awad W, Estrada I, Shen Y, Hendershot L. BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP. Proc Natl Acad Sci USA. 2008;105:1164–1169. doi: 10.1073/pnas.0702132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187(4):525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Despa F. Dilation of the endoplasmic reticulum in beta cells due to molecular crowding: kinetic simulations of extension limits and consequences of proinsulin synthesis. Biophys Chem. 2009;140(1–3):115–121. doi: 10.1016/j.bpc.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 63.Pierpaoli EV, Sandmeier E, Schonfeld H-J, Christen P. Control of the DnaK chaperone cycle by substoichiometric concentrations of the co-chaperones DnaJ and GrpE. J Biol Chem. 1997;273(12):6643–6649. doi: 10.1074/jbc.273.12.6643. [DOI] [PubMed] [Google Scholar]
- 64.Suh WC, Lu CZ, Gross CA. Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with the cochaperone DnaJ. J Biol Chem. 1999;274:30534–30539. doi: 10.1074/jbc.274.43.30534. [DOI] [PubMed] [Google Scholar]
- 65.Hindmarsh AC, Brown PN, Lee SL, Serban R, Woodward CS. SUNDIALS: suite of nonlinear and differential/algebraic solvers. ACM Trans Math Soft. 2005;31(3):363–396. [Google Scholar]
- 66.Stelling J, Gilles ED, Francis JD. Robustness properties of circadian clock architectures. Proc Natl Acad Sci USA. 2004;101(36):13210–13215. doi: 10.1073/pnas.0401463101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.