Abstract
Cladoniamides are indolotryptoline natural products that derive from indolocarbazole precursors. Here, we present a microbial platform to artificially redirect the cladoniamide pathway to generate unnatural bisindoles for drug discovery. Specifically, we target glycosyltransferase, halogenase, and oxidoreductase genes from the phylogenetically-related indolocarbazole rebeccamycin and staurosporine pathways. We generate a series of novel compounds, reveal details about the substrate specificities of a number of enzymes, and set the stage for future efforts to develop new catalysts and compounds by engineering of bisindole genes. The strategy for structural diversification we use here could furthermore be applied to other natural product families with known biosynthetic genes.
Keywords: Combinatorial engineering, bisindole biosynthesis, glycosyltransferase, halogenase, flavin-dependent monooxygenase, synthetic pathways
Natural products are an important source of available chemotherapeutics.1 However, development of new natural product-based drugs has been on the decline over the last decades, for reasons ranging from decreased investment to the structural complexity of natural products and high rates of re-discovery.2 With the progress in understanding the biosynthetic pathways of these small molecules and the advances in genetics and genomics, new techniques, such as genome and metagenome mining, have been developed to search new natural products from both culturable and unculturable microbes.3,4 Furthermore, approaches like mutational and combinatorial biosynthesis have been applied to explore new chemical space to prevent the lapse in the discovery pipelines.5
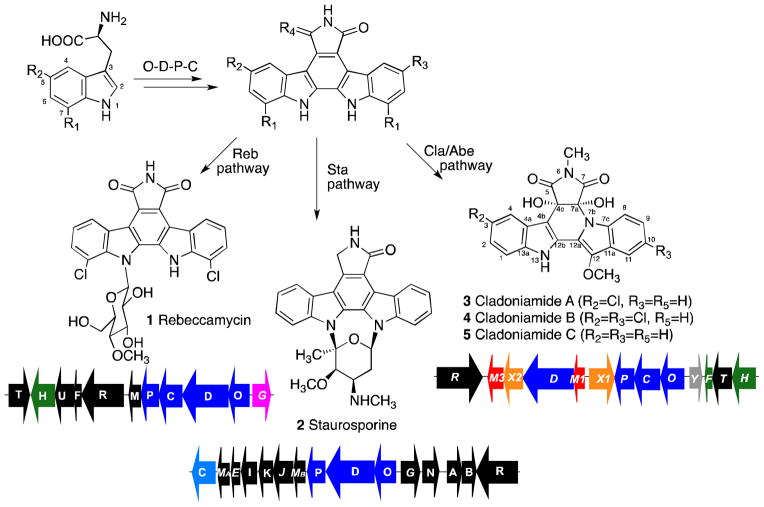
Among microbial natural products, bisindoles that derive from the oxidative dimerization of tryptophan possess diversified biological activities. Well-known examples of such bisindoles include rebeccamycin (1), a DNA-topoisomerase I inhibitor, and staurosporine (2), a polykinase inhibitor, analogues of which progressed into Phase II/III clinical trials against various cancers. Biosynthetic work has established a core set of enzymes, including a chromopyrrolic acid synthase (StaD/RebD), as critical for formation of the indolocarbazole scaffold embedded in molecules such as staurosporine and rebeccamycin.6 Recently, by exploiting degenerate primers specific for chromopyrrolic synthase genes, the Brady, Zhang, and Zhu groups isolated a number of new bisindole genes. These efforts have led to the discovery of new indolocarbazoles, such as erdasporines7 and streptocarbazoles,8 along with non-indolocarbazoles such as spiroindimicins9 and methyarcyriarubin,10 along with the indolotrypolines BE-5401711 and borregomycin A.12 Structurally, BE-54017 and borregomycin A are closely related to the indolotryptoline cladoniamides (3-9), which were reported by the Andersen and Davies laboratories from a Streptomyces strain and proposed to derive from indolocarbazoles.13
Indolocarbazoles themselves derive from a four-enzyme pathway from L-tryptophan. This pathway involves (1) production of an indolepyruvate (IPA) imine from L-tryptophan (or a chlorinated derivative) by an amino acid oxidase (StaO/RebO);14 (2) radical coupling of the IPA enamine by chromopyrrolic acid synthase (StaD/RebD) to give an indolepyruvate enamine dimer, that spontaneously converts to chromopyrrolic acid in oxygen;15 (3) a cytochrome P450 (StaP/RebP) catalyzed aryl-aryl coupling, giving an unstable intermediate;16 and (4) interception of a StaP-derived product by a flavin-dependent enzyme (StaC/RebC),17 ultimately generating the desired indolocarbazole aglycone (Scheme 1, Figure S1). In the case of rebeccamycin and staurosporine, the indolocarbazole scaffolds are then tailored through additional chemistry, including glycosyltransfer and methyltransfer reactions.6 A combination of genetic, biochemical, and heterologous biotransformation studies have now revealed that indolotryptolines also derive, as hypothesized,13 from the enzymatic processing of indolocarbazole precursors.11,18 Indeed, we recently reported the full series of chemical transformations that occur in cladoniamide biosynthesis to convert an indolocarbazole to an indolotryptoline.18 These transformations consist of (5) cis diol formation across the C4c-C7a double bond by ClaX1; (6) N-methylation of the succinimide ring by ClaM1; (7) formation of the indolotryptoline scaffold by ClaX2; and (8) installation of O-methylation by ClaM3 (Figure S1).
Scheme 1.
Biosynthesis of bisindoles. Two molecules of L-tryptophan are converted to an indolocarbazole by the action of four enzymes (RebODPC/StaODPC/ClaODPC). Pathway-specific enzymes then generate the final rebeccamycin (1), staurosporine (2), and cladoniamide (3-5) structures. All gene clusters are shown with the O-D-P-C genes in blue. Genes investigated in the combinatorial engineering study are chlorinase gene rebH (in green), glucosyltransferase gene rebG (in pink), and oxidoreductase gene staC (light blue). The unique cladoniamide genes are shown in red (methyltransferase genes), orange (flavoenzyme genes claX1 and claX2), green (halogenase and associated reductase gene), gray (unknown, claY), and black (regulator and transporter genes). For the rebeccamycin pathway: R1=Cl, R2=R3=H, R4=O; for the staurosporine pathway: R1=R2=R3=H, R4=2H; for the cladoniamide pathway: R1=R5=H, R2=R3=H/Cl, R4=O.
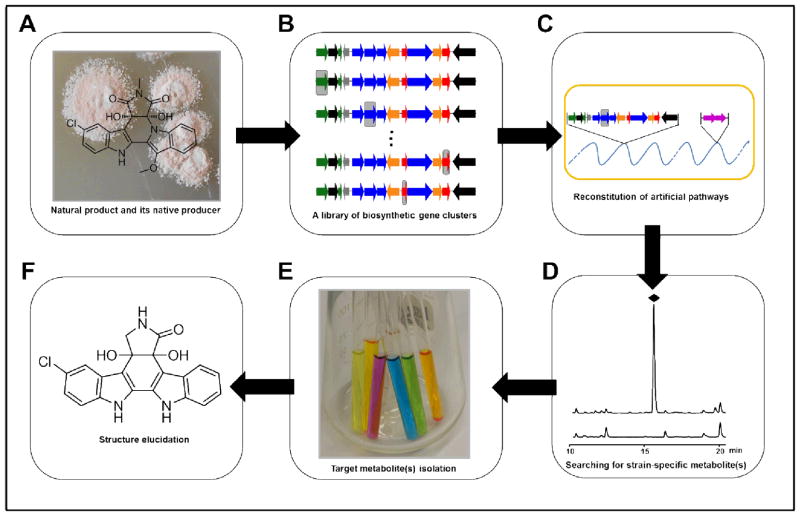
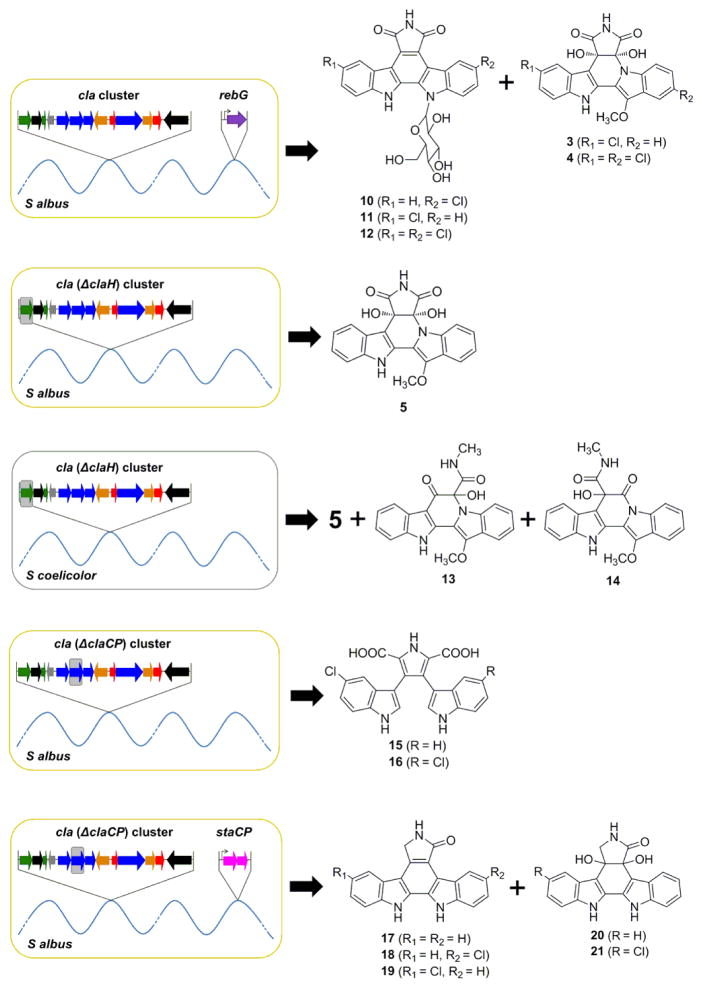
Previous studies on the indolocarbazole 1 and 2 gene clusters not only revealed their biosynthetic pathways, but also provided a genetic toolbox for combinatorial biosynthesis for production of indolocarbazole compounds.19 The recent isolation and characterization of a number of new bisindole gene cluster potentially enriches this molecular toolbox. With the biosynthetic gene cluster for cladoniamides (cla cluster) in hand, we sought to utilize indolocarbazole genes to artificially redirect the cladoniamide biosynthetic route for synthesis of novel bis-indole compounds and to test the substrate specificity of Cla enzymes. The microbial platform we designed for this purpose consists of (i) a “base” biosynthetic gene cluster (cla cluster), (ii) selected genes taken from phylogenetic-related clusters, (iii) genetically-, genomically-, and metabolically-friendly heterologous hosts (Scheme 2).
Scheme 2.
A microbial platform for combinatorial construction of new bisindole compounds. (A) Natural product and its native producer. (B) A library of biosynthetic clusters arise from isolation and modification of the natural biosynthetic gene cluster. Gray boxes indicate inactivated genes. (C) Reconstitution of the artificial pathways in optimized heterologous hosts. (D) Metabolic analysis for strain-specific metabolite(s). (E) Large-scale fermentation for target compound(s) purification. (F) Structure elucidation.
First, we generated a small library of engineered cla clusters, each of which carries an inactive cla gene constructed by target gene-inactivation. Introduction of these clusters into heterologous hosts are expected to generate biosynthetic intermediates or shunt products from the cladoniamide pathway. Indeed, our recent studies have shown that introduction of cla (ΔclaM1) or cla (ΔclaM3) clusters into S. albus or S. coelicolor resulted in the detection of a series of bis-indole compounds.18,20 Next, we reconstituted artificial bis-indole pathways by co-introducing intact or engineered cla clusters and genes from indolocarbazole gene clusters into S. albus or S. coelicolor. Specifically, we chose three genes from indolocarbazole systems to introduce into the cladoniamide production system. First, we are interested in the possibility of generating glycosylated cladoniamides, and we reasoned that the glucosyltransferase RebG, from the rebeccamycin pathway, is the characterized enzyme most likely to accept an indolotryptoline as a substrate. Prior studies had shown some substrate flexibility for this enzyme, including the ability to glycosylate a minimal carbazole ring, as well as a brominated indolocarbazole.21 We sought to probe whether RebG was able to glycosylate cladoniamide, or any of the intermediates along the ClaX1-ClaM1-ClaX2-ClaM3 cascade. Second, we targeted RebH, the 7-L-tryptophan halogenase from the rebeccamycin pathway.22 We reasoned that if we eliminated the putative 5-L-tryptophan halogenase gene claH from the cladoniamide cluster, introduction of rebH might give access to cladoniamides with altered halogenation patterns. Third, we targeted StaC, the flavin-dependent monooxygenase from the staurosporine pathway. This enzyme controls the oxidative outcome in the last step of staurosporine aglycone formation, giving a pyrrolinone ring, in contrast to the maleimide ring observed in rebeccamycin and cladoniamide.23,24 Through elimination of claC and introduction of staC, we anticipated that we could generate cladoniamides and/or cladoniamide intermediates with a pyrrolinone ring.
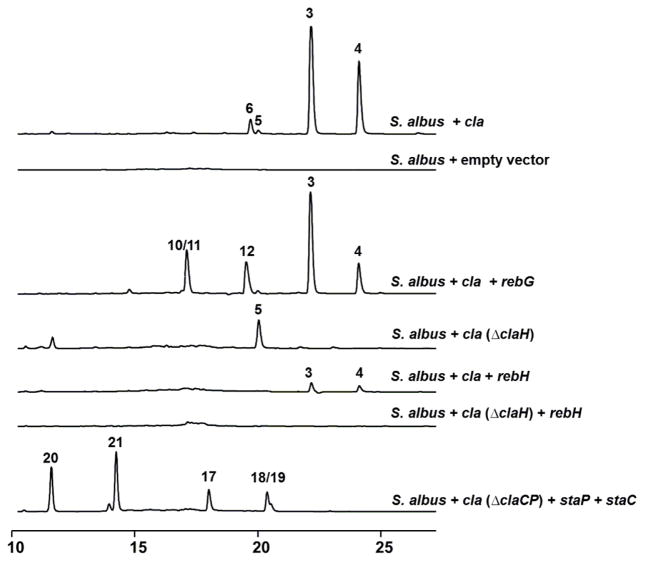
We first introduced rebG into the S. albus strain carrying the complete cla cluster. HPLC analysis of the ethyl acetate extract of S. albus + cla + rebG showed four major peaks (Figure 1), two (tR: 22.0 and 24.1 min) of which were identified to be cladoniamide A (3) and B (4) by comparison with controls (S. albus + empty vector and S. albus + cla, the latter accumulates cladoniamides A, B, C and D (3-6)20). Another two earlier-eluted peaks (tR: 17.1 and 19.5 min) displayed UV-Vis spectra characteristic of the indolocarbazole chromophore and were characterized as a mixture of glucosyl-3-chloroarcyriaflavin A (10) and glucosyl-9-chloroarcyriaflavin A (11) and pure glucosyl-3,9-chloroarcyriaflavin A (12) by MS and NMR analysis (Tables S3–S5 and Figures S2–S3). These three molecules are new analogues of demethylrebeccamycin. These results suggest that RebG favors indolocarbazole substrates and cannot readily glycosylate either cladoniamide A or B, but RebG is able to accept alternately chlorinated indolocarbazoles.
Figure 1.
Analytical HPLC profiles of various genetic mutants constructed for the combinatorial study (detection wavelength = 348 nm).
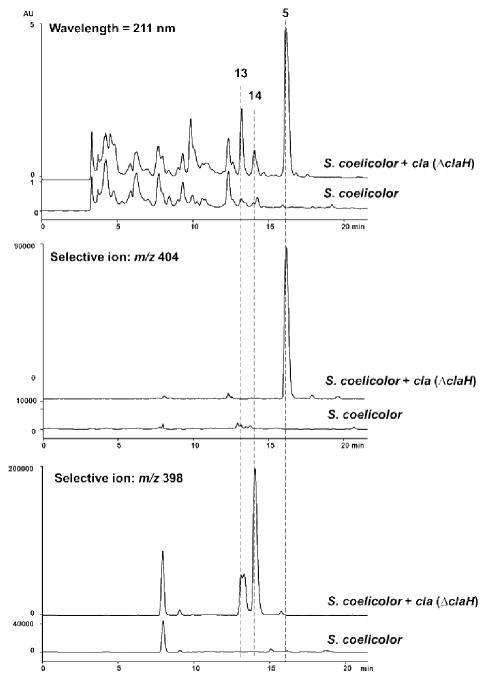
Next, to probe whether we could modify the natural cla pathway to obtain new bisindoles with chlorine atom(s) at alternative positions, the cla (ΔclaH) cluster was transferred into both S. albus and S. coelicolor strains. From the S. albus + cla (ΔclaH) heterologous production system, cladoniamide C (5) was identified to be the major product (Figure 1), while from the S. coelicolor + cla (ΔclaH) system, along with the production of cladoniamide C (5), two new metabolites (13 and 14) were also produced (Figure 2). They were identified as new, non-chloro derivatives of cladoniamides D/E and F/G by UV/Vis and LC-MS analysis (Figure 2, Table S3, and Figure S4). Next, rebH was introduced into S. albus + cla (ΔclaH), but no cladoniamides or new products were detected. We also co-expressed rebH with the intact cla cluster in S. albus, but this strain produced a very low amount of cladoniamides A and B (Figure 1). These results suggest that early Cla enzymes might not be able to accept 7-chloro-L-tryptophan, and the addition of RebH could hijack L-tryptophan from the cla pathway.
Figure 2.
Production of non-chlorinated cladoniamides C (5), H (13) and I (14) from engineered strain S. coelicolor YD17. S. coelicolor YD17 produces 5 as a major clone-specific metabolite along with cladoniamide H (13) and I (14) (detection wavelength = 211 nm). Selective ion monitoring at m/z = 404 reveals 5 as clone-specific metabolite. Selective ion monitoring at m/z = 398 reveals 13 and 14 as clone-specific metabolites.
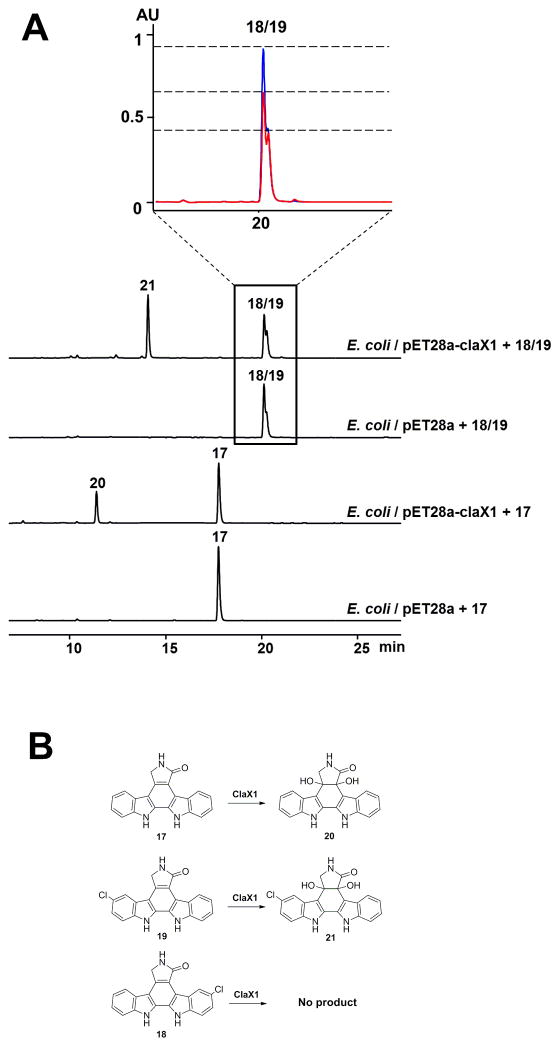
Finally, StaC has been shown to be a branch point where it diverges the sta pathway from reb/cla pathways. We sought to probe the possibility of artificially redirecting the product of StaC (K252c, 17) back to cla pathway to generate bisindoles with staurosporine-like pyrrolinone rings. To achieve this, we first constructed an S. albus strain harboring the cla (ΔclaC) cluster, in which claC was in-frame deleted. This strain unexpectedly accumulated 9-chloro-chromopyrrolic acid (15) and 9,9′-dichloro-chromopyrrolic acid (16) (Figures S5–S6), instead of producing the chlorinated derivatives of arcyriaflavin A, K252c, and 7-hydroxy-K252c,25 suggesting a polar effect on the downstream claP gene. We thus introduced both staP and staC to the S. albus + cla (ΔclaCP) system, in order to construct a pathway that would provide staurosporine aglycone (K252c, 17) or its chlorinated derivatives for the downstream Cla enzymes. Extracts of S. albus + cla (ΔclaCP) + staPC had five major clone-specific metabolites compared to control strains (Figure 1). The compound 17 (tR: 18.0 min) showed identical UV-vis spectrum as K252c, and it was characterized as K252c by comparing its LC-MS and 1H NMR data (Table S3 and Figure S7) with the reported data from K252c.26 The HPLC peaks for compounds 18 and 19 are largely overlapped, and both of them display similar UV-Vis spectra as K252c (17) (Figure S4). They were collected together by semi-prep HPLC and analyzed by ESI-MS, revealing a single MS signal (m/z 346, [M+H]+) with the signature of a chlorine atom. They were identified to be 3-chloro and 9-chloro derivatives of K252c based on the 1H NMR data (Figure S7). Compounds 20(tR: 11.6 min) and 21 (tR: 14.2 min) were identified as 4c/7a diol derivatives of K252c and Cl-K252c based on their MS, 1D and 2D NMR data (Table S6–S7, and Figure S8–S9). Interestingly, in contrast to Cl-K252c, which we isolated as a mixture of 3-chloro and 9-chloro K252c (18/19), the 4c/7a diol derivative of Cl-K252c (21) was purified as single component. The structure of 21 implies that ClaX1 can accept only one of the two Cl-K252c molecules as a substrate, and the accumulation of 21 furthermore suggests that the downstream enzyme ClaM1 cannot utilize compound 21 as a substrate (Figure S1). To further solidify the above conclusions, E. coli-based biotransformation assays were performed. Specifically, the mixture of 3-chloro and 9-chloro derivatives of K252c (18/19) was fed to the culture of E. coli strain expressing ClaX1, and metabolic profiling by HPLC revealed the production of 21. Furthermore, only one of the two Cl-K252c’s was consumed (Figure 3). Similarly, 20 was produced when K252c (17) was fed to this strain. Both 20 and 21 are new compounds.
Figure 3.
In vivo biotransformation of 17 and 18/19 with claX1. (A) HPLC traces for in vivo biotransformation assay. Insert, only the peak at the left was consumed in the in vivo biotransformation assay of E. coli/pET28a-claX1 + 18/19, which is assumed to be 19, based on the corresponding production of 21. Blue, E. coli/pET28a + 18/19. Red, E. coli/pET28a-claX1 + 18/19. Detection wavelength: 348 nm. (B) Scheme for conversion by ClaX1.
We tested the biological activities of selected new compounds generated from our combinatorial platform, and we observed IC50 values of 1.42 μM for 10/11, 3.24 μM for 12, and 4.42 μM for 18/19 against colon cancer cell line HCT-116.
Microbial natural products and their biosynthetic gene clusters are shaped by long evolutionary processes, involving joining of subclusters and gene mutation, acquisition, loss, duplication, and functional divergence.27,28 Understanding the biosynthetic pathways of natural products that putatively share the same ancestor can give insights into the naturally occurring modes of structural diversification, and facilitate efficient manipulation of these pathways for the generation of unnatural products. Previous studies by Brady and our group established that the rare indolotryptoline scaffolds derive from more common indolocarbazole structures. In this study, we introduce rebG, the glucosyltransferase from the rebeccamycin pathway into a cla-containing strain. Our results show that RebG can intercept the chlorinated arcyriaflavin A intermediates from the cladoniamide pathway to generate new glycosylated indolocarbazoles (Scheme 3). However, the enzyme appears unable to react with cladoniamide itself, or any intermediate beyond the ClaX1 substrate, likely reflecting its need for a substrate with an intact carbazole.21 In a next set of experiments, we show that elimination of the chlorinase gene claH can give the full suite of non-chlorinated cladoniamides. However, halogenation at the 7 position of L-tryptophan, through introduction of 7-L-tryptophan chlorinase rebH, apparently hijacks tryptophan precursors away from the cladoniamide pathway, suggesting that early cladoniamide enzymes are unable to accept 7-chloro-L-tryptophan precursors (Scheme 3). Finally, we interrogate introduction of a pyrrolinone-containing indolocarbazole into the cladoniamide pathway through deletion of claC and introduction of staPC. We find that ClaX1 can accept an indolocarbazole with a pyrrolinone ring to generate 20 or 21, however, downstream enzyme(s) are unable to accept substrates with the pyrrolinone ring. Interestingly, whereas StaP/StaC appear to be able to process a substrate with chlorine at either the 3 or 9 position, ClaX1 is highly specific to the positioning of chlorine on the ring, tolerating chlorination at the 3 position but not the 9 position (Scheme 3). Overall, our results suggest that unique substrate specificities have been molded into cladoniamide and indolocarbazole enzymes during their distinct evolutionary histories, making them best suited to react with native substrates.
Scheme 3.
Combinatorial biosynthesis of bis-indole metabolites.
The platform developed for this study gave access to a number of new compounds, including glucosyl-3,9-dichloroarcyriaflavin A (12) and the 4c/7a diol of Cl-K252c (21). Additionally, this platform should be highly suitable for precursor-directed biosynthesis. For instance, feeding of synthetic indolocarbazole precursors to the S. albus + cla (ΔclaCP) system should allow us to generate new compounds and enable us to further assess substrate tolerance of the Cla enzymes late in the pathway. Finally, this combinatorial work contributes to knowledge about bisindole enzyme substrate specificities, laying the groundwork for future efforts to engineer bisindole enzymes for higher substrate tolerance, to ultimately enable the production of an even larger suite of bisindole molecules.
Methods
Combinatorial engineering
For construction of a library of engineered cla clusters, vector pYD1 carrying the whole biosynthetic gene cluster was subjected to a series of targeted gene-inactivation protocols by using REDIRECT technique,29 resulting in modified cla clusters, including cla (ΔclaH) and cla (ΔclaC).
For the construction of strain Streptomyces + cla cluster + rebG, the coding region of rebG was amplified from the genomic DNA of Lechevalieria aerocolonigenes DSM 44217 using Phusion DNA polymerase. NdeI and XbaI double digested PCR products were inserted into the corresponding sites of pYLD20,30 resulting in pYLDrebG. After confirmation by DNA sequencing, the plasmids were introduced into Streptomyces strains carrying cla cluster by conjugation.31 Thiostrepton-, apramycin- and kanamycin-resistant clones were confirmed for site-specific integration by PCR with the primer pairs for rebG (Table S2).
For the construction of strain Streptomyces + cla (ΔclaH) cluster, Streptomyces + cla (ΔclaH) cluster + rebH, and Streptomyces + cla cluster + rebH, the vector carrying the cla (ΔclaH) cluster was passaged through E. coli ET12567/pUZ8002 before being introduced via conjugation into S. albus J107432 and S. coelicolor M114633 to afford S. albus YD22 and S. coelicolor YD17, respectively. Clones were confirmed for integration by PCR with the primer pairs for claH (Table S2). The co-expression of rebH with the intact or engineered cla cluster was performed similarly as described for the co-expression of rebG.
For the construction of strains Streptomyces + cla (ΔclaCP) cluster and Streptomyces + cla (ΔclaCP) cluster + staPC, the vector carrying the cla (ΔclaC), in which claP was also inactivated due to a polar effect, was introduced into S. albus J1074 to generate S. albus YD23. The coding regions of staP and staC genes were amplified from pET vectors (generously provided by the Walsh and Drennan laboratories) using Phusion DNA polymerase. NdeI and XbaI double digested PCR products of staC and XbaI-digested PCR products of staP were inserted into the corresponding sites of pYLD20, resulting in pYLDstaCP. After confirmation by DNA sequencing, the plasmid was introduced into S. albus YD23 by conjugation. Thiostrepton-, apramycin- and kanamycin-resistant clones were confirmed for site-specific integration by PCR with the primer pairs for staP and staC (Table S2).
Production and analysis of metabolites from engineered Streptomyces strains
Cultivation of Streptomyces mutants was performed as described previously.20 At the end of fermentation, the cultures were extracted twice with 1 volume of ethyl acetate, and combined extracts were dried under vacuum and then dissolved in methanol.
Analysis of bisindole compounds production was performed with an Agilent 1260 high-pressure liquid chromatography (HPLC) apparatus with a diode array UV detector, fitted with an Phenomenex Luna C18(2) column (5 μm, 4.6 mm ID × 250 mm). Elution was performed at 1 mL/min with a mobile-phase mixture consisting of a linear gradient of water and acetonitrile (85:15 to 20:80, 0 to 25 min; 0:100, 25 to 30 min), both of which contain 0.05% (v/v) trifluoroacetic acid. LC-MS was performed under the same conditions (except that the flow rate was set at 0.8 mL/min) on an Agilent 6120 Quadrupole LC/MS system operated in both positive and negative ion electrospray modes.
Isolation, purification, and structural elucidation of bisindoles from Streptomyces mutants
Ethyl acetate extracts from Streptomyces cultures (3~6 L) were dried under vacuum and then dissolved in methanol. The resulting crude extracts were fractionated on reversed-phase C8 flash column with MeOH/water gradient elution, and Sephadex LH-20 with MeOH elution. Metabolites of interest, as tracked by analytical HPLC as described above, were purified from these fractions by reversed-phase semi-preparative HPLC (Phenomenex Luna C18(2), 5 μm, 10 mm ID × 250 mm) (Figure S10). Compounds 10/11 (yield: 5.5 mg/L) were purified using 30:70 acetonitrile:water. Compound 12 (yield: 5 mg/L) was purified using 37:63 acetonitrile:water. Compound 15 (yield: 6.8 mg/L) was purified using 54:46 acetonitrile:water. Compound 16 (yield: 8 mg/L) was purified using 58:42 acetonitrile:water. Compound 17 (yield: 2.4 mg/L) was purified using 60:40 acetonitrile:water. Compound 18/19 (yield: 2 mg/L) were purified using 70:30 acetonitrile:water. Compound 20 (yield: 4.5 mg/L) was purified using 45:55 acetonitrile:water. Compound 21 (yield: 5 mg/L) was purified using 50:50 acetonitrile:water.
The 1H- and 13C-NMR spectra were recorded on a Bruker AV-600 MHz spectrometer with a 5 mm CPTCI cryoprobe. The DMSO-d6 signals (2.50 ppm for 1H and 39.5 ppm for 13C) were used as references. High-resolution mass spectrometry data were acquired using a Micromass LCT time-of-flight (TOF) mass spectrometer equipped with an electrospray ion source. Structure elucidation of 10, 11, 12, 20, 21 is described in Supplementary Methods.
Supplementary Material
Acknowledgments
We are grateful to Jessie Chen, Elena Polishchuk, and Xiaoyan Deng for assistance with the cytotoxicity work. Financial support was provided by the Canadian Institutes of Health Research and Genome British Columbia. Yi-Ling Du is supported by a Michael Smith Foundation for Health Research Fellowship.
Footnotes
Supporting tables, figures, methods, and references. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 3.Winter JM, Behnken S, Hertweck C. Genomics-inspired discovery of natural products. Curr Opin Chem Biol. 2011;15:22–31. doi: 10.1016/j.cbpa.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Donia MS, Ruffner DE, Cao S, Schmidt EW. Accessing the hidden majority of marine natural products through metagenomics. ChemBioChem. 2011;12:1230–1236. doi: 10.1002/cbic.201000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirschning A, Hahn F. Merging chemical synthesis and biosynthesis: a new chapter in the total synthesis of natural products and natural product libraries. Angew Chem Int Ed. 2012;51:4012–4022. doi: 10.1002/anie.201107386. [DOI] [PubMed] [Google Scholar]
- 6.Sánchez C, Méndez C, Salas JA. Indolocarbazole natural products: occurrence, biosynthesis, and biological activity. Nat Prod Rep. 2006;23:1007–1045. doi: 10.1039/b601930g. [DOI] [PubMed] [Google Scholar]
- 7.Chang FY, Ternei MA, Calle PY, Brady SF. Discovery and synthetic refactoring of tryptophan dimer gene clusters from the environment. J Am Chem Soc. 2013;135:17906–17912. doi: 10.1021/ja408683p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu P, Yang C, Wang Y, Liu P, Ma Y, Xu L, Su M, Hong K, Zhu W. Streptocarbazoles A and B, two novel indolocarbazoles from the marine-derived actinomycete strain Streptomyces sp. FMA. Org Lett. 2012;14:2422–2425. doi: 10.1021/ol3008638. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Liu Z, Li S, Yang T, Zhang Q, Ma L, Tian X, Zhang H, Huang C, Zhang S, Ju J, Shen Y, Zhang C. Spiroindimicins A-D: new bisindole alkaloids from a deep-sea-derived actinomycete. Org Lett. 2012;14:3364–3367. doi: 10.1021/ol301343n. [DOI] [PubMed] [Google Scholar]
- 10.Chang FY, Brady SF. Characterization of an environmental DNA-derived gene cluster that encodes the bisindolylmaleimide methylarcyriarubin. ChemBioChem. 2014;15:815–821. doi: 10.1002/cbic.201300756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang FY, Brady SF. Cloning and characterization of an environmental DNA-derived gene cluster that encodes the biosynthesis of the antitumor substance BE-54017. J Am Chem Soc. 2011;133:9996–9999. doi: 10.1021/ja2022653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang FY, Brady SF. Discovery of indolotryptoline antiproliferative agents by homology-guided metagenomic screening. Proc Natl Acad Sci. 2013;110:2478–2483. doi: 10.1073/pnas.1218073110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams DE, Davies J, Patrick BO, Bottriell H, Tarling T, Roberge M, Andersen RJ. Cladoniamides A-G, tryptophan-derived alkaloids produced in culture by Streptomyces uncialis. Org Lett. 2008;10:3501–3504. doi: 10.1021/ol801274c. [DOI] [PubMed] [Google Scholar]
- 14.Nishizawa T, Aldrich CC, Sherman DH. Molecular analysis of the rebeccamycin L-amino acid oxidase from Lechevalieria aerocolonigenes ATCC 39243. J Bacteriol. 2005;187:2084–2092. doi: 10.1128/JB.187.6.2084-2092.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asamizu S, Hirano S, Onaka H, Koshino H, Shiro Y, Nagano S. Coupling reaction of indolepyruvic acid by StaD and its product: implications for biosynthesis of indolocarbazole and violacein. ChemBioChem. 2012;13:2495–2500. doi: 10.1002/cbic.201200535. [DOI] [PubMed] [Google Scholar]
- 16.Howard-Jones AR, Walsh CT. Nonenzymatic oxidative steps accompanying action of the cytochrome P450 enzymes StaP and RebP in the biosynthesis of staurosporine and rebeccamycin. J Am Chem Soc. 2007;129:11016–11017. doi: 10.1021/ja0743801. [DOI] [PubMed] [Google Scholar]
- 17.Goldman PJ, Ryan KS, Hamill MJ, Howard-Jones AR, Walsh CT, Elliott SJ, Drennan CL. An unusual role for a mobile flavin in StaC-like indolocarbazole biosynthetic enzymes. Chem Biol. 2012;19:855–865. doi: 10.1016/j.chembiol.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du Y-L, Williams DE, Patrick BO, Andersen RJ, Ryan KS. Reconstruction of cladoniamide biosynthesis reveals non-enzymatic routes to bisindole diversity. ACS Chem Biol. doi: 10.1021/cb500728h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sánchez C, Zhu L, Braña AF, Salas AP, Rohr J, Méndez C, Salas JA. Combinatorial biosynthesis of antitumor indolocarbazole compounds. Proc Natl Acad Sci U S A. 2005;102:461–466. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du YL, Ding T, Ryan KS. Biosynthetic O-methylation protects cladoniamides from self-destruction. Org Lett. 2013;15:2538–2541. doi: 10.1021/ol401036f. [DOI] [PubMed] [Google Scholar]
- 21.Zhang C, Albermann C, Fu X, Peters NR, Chisholm JD, Zhang G, Gilbert EJ, Wang PG, Van Vranken DL, Thorson JS. RebG- and RebM-catalyzed indolocarbazole diversification. ChemBioChem. 2006;7:795–804. doi: 10.1002/cbic.200500504. [DOI] [PubMed] [Google Scholar]
- 22.Yeh E, Garneau S, Walsh CT. Robust in vitro activity of RebF and RebH, a two-component reductase/halogenase, generating 7-chlorotryptophan during rebeccamycin biosynthesis. Proc Natl Acad Sci U S A. 2005;102:3960–3965. doi: 10.1073/pnas.0500755102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groom K, Bhattacharya A, Zechel DL. Rebeccamycin and staurosporine biosynthesis: insight into the mechanisms of the flavin-dependent monooxygenases RebC and StaC. ChemBioChem. 2011;12:396–400. doi: 10.1002/cbic.201000580. [DOI] [PubMed] [Google Scholar]
- 24.Asamizu S, Shiro Y, Igarashi Y, Nagano S, Onaka H. Characterization and functional modification of StaC and RebC, which are involved in the pyrrole oxidation of indolocarbazole biosynthesis. Biosci Biotechnol Biochem. 2011;75:2184–2193. doi: 10.1271/bbb.110474. [DOI] [PubMed] [Google Scholar]
- 25.Howard-Jones AR, Walsh CT. Staurosporine and rebeccamycin aglycones are assembled by the oxidative action of StaP, StaC, and RebC on chromopyrrolic acid. J Am Chem Soc. 2006;128:12289–12298. doi: 10.1021/ja063898m. [DOI] [PubMed] [Google Scholar]
- 26.Liu R, Zhu T, Li D, Gu J, Xia W, Fang Y, Liu H, Zhu W, Gu Q. Two indolocarbazole alkaloids with apoptosis activity from a marine-derived actinomycete Z2039-2. Arch Pharm Res. 2007;30:270–274. doi: 10.1007/BF02977605. [DOI] [PubMed] [Google Scholar]
- 27.Osbourn A. Secondary metabolic gene clusters: evolutionary toolkits for chemical innovation. Trends Genet. 2010;26:449–457. doi: 10.1016/j.tig.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Fischbach MA, Walsh CT, Clardy J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc Natl Acad Sci U S A. 2008;105:4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du YL, Dalisay DS, Andersen RJ, Ryan KS. N-carbamoylation of 2,4-diaminobutyrate reroutes the outcome in padanamide biosynthesis. Chem Biol. 2013;20:1002–1011. doi: 10.1016/j.chembiol.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. The John Innes Foundation; Colney, Norwich: 2000. [Google Scholar]
- 32.Chater KF, Wilde LC. Streptomyces albus G mutants defective in the SalGI restriction-modification system. J Gen Microbiol. 1980;116:323–334. doi: 10.1099/00221287-116-2-323. [DOI] [PubMed] [Google Scholar]
- 33.Gomez-Escribano JP, Bibb MJ. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol. 2011;4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.