Abstract
Gene expression is epigenetically regulated through DNA methylation and covalent chromatin modifications, such as acetylation, phosphorylation, ubiquitination, sumoylation, and methylation of histones. Histone methylation state is dynamically regulated by different groups of histone methyltransferases and demethylases. The trimethylation of histone 3 (H3K4) at lysine 4 is usually associated with the activation of gene expression, whereas trimethylation of histone 3 at lysine 27 (H3K27) is associated with the repression of gene expression. The polycomb repressive complex contains the H3K27 methyltransferase Ezh2 and controls dimethylation and trimethylation of H3K27 (H3K27me2/3). The Jumonji domain containing-3 (Jmjd3, KDM6B) and ubiquitously transcribed X-chromosome tetratricopeptide repeat protein (UTX, KDM6A) have been identified as H3K27 demethylases that catalyze the demethylation of H3K27me2/3. The role and mechanisms of both JMJD3 and UTX have been extensively studied for their involvement in development, cell plasticity, immune system, neurodegenerative disease, and cancer. In this review, we will focus on recent progresses made on understanding JMJD3 in the regulation of gene expression in development and diseases. This article is part of a Directed Issue entitled: Epigenetics dynamics in development and disease.
Keywords: Jumonji, Jmjd3, Utx, Histone demethylation, H3K27
1. Introduction
The most common modifications on histones are acetylation and methylation (Cloos et al., 2008), which cause steric effects on other DNA or histone modifiers, leading to either gene expression or repression. Methylation of histones occurs on arginine or lysine residues, which can be mono-, di-, or tri-methylated, differentially affecting other histone modifications, and hence, chromatin structure and gene expression. Histones are methylated by the enzymes, histone methyltransferases, and histone methylation can be antagonized either indirectly or directly. Peptidylarginine deiminase 4 (PADI4) is the first class of such enzymes to be identified that can indirectly reverse arginine methylation (Cuthbert et al., 2004; Wang et al., 2004). The second class identified, Lysine specific demethylase 1 (LSD1), directly reverses the methylation of the mono- and di-methyl groups on lysines 4 and 9 on histone 3 (H3K4 and H3K9) (Metzger et al., 2005; Shi et al., 2004). Conversely, enzymes of the third and largest class contain a Jumonji C (JmjC) domain and can remove all three mono-, di-, or tri-methyl groups (Klose et al., 2006; Tsukada et al., 2006). Jumonji domain-containing protein D3 (JMJD3), also called lysine-specific demethylase 6B (KDM6B) is a member of this family of JmjC histone demethylases, and along with ubiquitously transcribed X-chromosome tetratricopeptide repeat protein (UTX), specifically demethylates H3K27 (Swigut and Wysocka, 2007). JMJD3 and UTX specifically decrease H3K27 dimethylation (H3K27me2) and H3K27 trimethylation (H3K27me3) (Hong et al., 2007; Xiang et al., 2007). When H3K27 is trimethylated, it is typically associated with inactive gene promoters; whereas when H3K27 is monomethylated, it is associated with active gene promoters (Barski et al., 2007). Although JMJD3 and UTX play important roles in the epigenetic regulation of gene expression, altering cellular memory and reprogramming the fate of cells, this review will primarily focus on the role of JMJD3. Specifically, JMJD3 is involved in several cellular processes, such as differentiation, proliferation, senescence, and apoptosis. The regulation of JMJD3 is highly gene- and context-specific and is involved in several tissue responses, such as vertebrate development, cancer, inflammatory diseases, and neurodegenerative diseases.
1.1. JMJD3 and development
Embryonic stem cells (ESCs) repress developmental genes by utilizing H3K27 trimethylation, but ESC deficiency in JMJD3 does not seem to affect stem cell maintenance and self-renewal capacity (Mansour et al., 2012; Ohtani et al., 2013). During differentiation, H3K27 methylation is removed in a tissue- and cell-specific manner, and the demethylases, JMJD3 and UTX, are directly involved in embryogenesis into the three germ layers, endoderm, mesoderm, and ectoderm, of a developing vertebrate.
1.1.1. Endoderm
JMJD3 and UTX drive the formation of the germ layer, endoderm, which gives rise to the gastrointestinal tract, respiratory tract, endocrine glands, and the auditory and urinary systems. Endoderm commitment is controlled by the WNT signaling pathway and the transforming growth factor-beta (TGF-β) superfamily member, NODAL/Activin A, which utilizes the transcription factors, SMAD2/3 (Mfopou et al., 2010; Schier, 2009). Upon treatment with NODAL/Activin A, JMJD3 recruitment to SMAD2/3 genes is associated with decreased enrichment of H3K27me3 marks at SMAD2/3 genes (Kim et al., 2011) (Fig. 1A). Knockdown of JMJD3 and UTX in human ESCs, but not mouse ESCs, inhibits endoderm formation, which can be rescued by continuous activation of WNT signaling (Jiang et al., 2013), suggesting that JMJD3 and UTX are essential, at least for human, endodermal development. JMJD3 also binds to other factors essential for endoderm development. Embyronic T-box transcription factors, Eomesodermin (Eomes) and Tbx3, are essential for endoderm formation (Fig. 1B). Eomes is normally maintained in a transcriptionally poised state in ES cells. During early endoderm differentiation, TBX3 associates with JMJD3 at the enhancer region of Eomes, bringing its enhancer in close proximity to its promoter to drive its own expression. This DNA looping of Eomes allows for a self-activating loop, thereby, maintaining endoderm fate (Kartikasari et al., 2013) and preventing abnormal development (Fig. 1B). Endodermal differentiation into lung tissue is also defective in mice with global knockout of Jmjd3 (Li et al., 2014a). Mice are born smaller with thickened alveolar cell walls and inadequate air space, and expression of markers of lung differentiation is decreased. Perinatal death ensues within 30 min after birth due to respiratory failure (Li et al., 2014a; Satoh et al., 2010). However, time of death and the precise phenotype of Jmjd3 knockout mice are dependent on the gene dosage and the gene deletion strategy used to generate the mice (Fig. 2). Jmjd3 is important for lung development in a stage-dependent manner (Fig. 3A). Jmjd3 regulates lung development via regulating SP-B expression with transcription factor Nkx2.1 and epigenetic factor Brg1 (Fig. 3B). In Jmjd3 knockout mice, in which exons 4–5 have been deleted, the mice die before embryonic day 6.5 (E6.5) (Ohtani et al., 2013). However, deletion of exons 14–21, including the JmjC domain, lead to a frameshift, and the mice die perinatally (Satoh et al., 2010). Whereas knockout of Jmjd3 in mice generated by a gene trap strategy, inserting a neo-cassette between exons 1 and 2, show postnatal lethality with normal lung development (Burgold et al., 2012). Although whether JMJD3 is absolutely essential for lung development remains contradictory, it is clear that JMJD3 is a key regulator in early endoderm specification and endoderm differentiation into lung tissue. However, whether JMJD3 mediates late endoderm commitment or differentiation into the gastrointestinal tract, endocrine glands, or auditory or urinary systems has not been investigated.
Fig. 1.
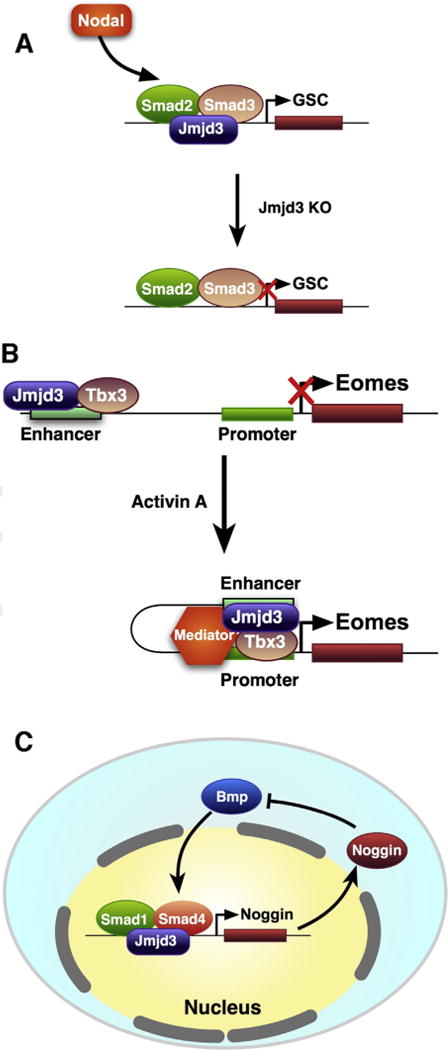
Role of Jmjd3 in development. (A) Negative feedback of Noggin to regulate Jmjd3-mediated activation of Noggin via inhibition of BMP activity. Upon treatment with Nodal, JMJD3 recruitment to SMAD2/3 genes is associated with decreased enrichment of H3K27me3 marks at SMAD2/3 genes (Kim et al., 2011), leading to the expression of the homeobox protein goosecoid (GSC) initiation of endoderm development. (B) During early endoderm differentiation, TBX3 associates with JMJD3 at the enhancer region of Eomes, bringing its enhancer in close proximity to its promoter to drive its own expression. This DNA looping of Eomes allows for a self-activating loop, thereby, maintaining endoderm fate (Kartikasari et al., 2013) and preventing abnormal development. (C) Negative feedback of Noggin to BMP activity via Jmjd3 regulation.
Fig. 2.
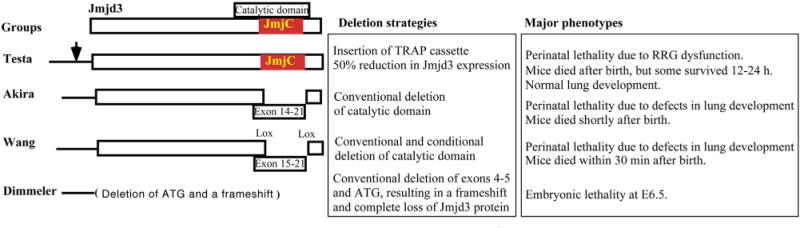
Deletion strategies of JMJD3 and their associated mouse phenotypes.
Fig. 3.
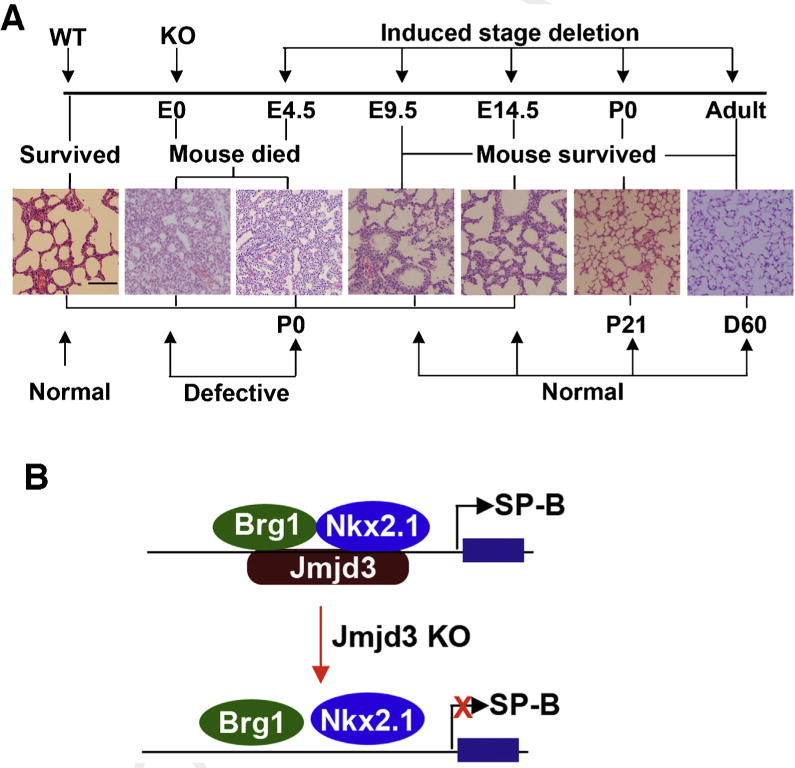
Stage-specific role of JMJD33 in lung development and function. (A) JMJD3 regulates lung development in a stage-dependent manner. H&E staining showing the stage-dependent effects of Jmjd3 deletion on lung architecture and its correlation with embryo viability. Bar = 200 μm, (B). A proposed model explaining how JMJD3 specifically upregulates SP-B expression by interacting with Nkx2.1 and Brg1 at the SP-B promoter.
1.1.2. Mesoderm
JMJD3 and UTX are also involved in the formation of the germ layer, mesoderm, but JMJD3 can partially compensate for the loss of UTX during ESC differentiation into mesoderm (Morales Torres et al., 2013). Mesodermal development leads to the formation of muscle tissue, spleen, cartilage, bone, skin, kidneys, gonads, heart, blood vessels, and blood cells. Jmjd3 deficiency in ESCs significantly increases repressive H3K27me3 marks on the promoter of the mesodermal regulator, Brachyury and decreases its expression, leading to impaired recruitment of β-catenin (Ohtani et al., 2013), which is a prerequisite for WNT-induced mesoderm differentiation. During late mesoderm differentiation, JMJD3 is also essential for normal organ development. Jmjd3 knockout in ESCs reduces endothelial cell differentiation as well as cardiac progenitor cell differentiation (Ohtani et al., 2013). In addition, Jmjd3 knockout mice at E17.5 demonstrate impaired spleen development, with smaller size and hyperemic areas (Li, 2014a). Furthermore, JMJD3 affects bone formation. Endochondral bone formation and ossification begins with multipotent mesenchymal stem cell (MSC) differentiation into chondrocytes. This cartilage maturation during endochondral bone formation is regulated by JMJD3 and its association with the transcription factor, RUNX2, promoting proliferation and hypertrophy of chondrocytes (Zhang et al., 2015). JMJD3 can also direct MSCs to differentiate preferentially into one lineage over another lineage. Specifically, depletion of Jmjd3 leads to a decrease in osteogenic differentiation, while increasing adipogenic differentiation (Gomez-Sanchez et al., 2013). Studies implicating the involvement of microRNAs in MSC differentiation (Bengestrate et al., 2011; Inose et al., 2009; Li et al., 2008; Yang et al., 2011) has led to the discovery that the specific microRNA, MIR146A, interacts with the 3′ UTR of JMJD3, inhibiting its function and decreasing osteogenesis (Huszar and Payne, 2014). H3K27me3 marks are notably increased in bone marrow MSCs in a mouse model of osteoporosis, suggesting that JMJD3 may contribute to the development of osteoporosis (Burgold et al., 2012). Similarly, MSCs, isolated from dental tissue, have decreased odontogenic differentiation capacity upon Jmjd3 knockdown (Xu et al., 2013). JMJD3 is recruited to bone morphogenetic protein (BMP) 2 promoters with subsequent removal of gene silencing H3K27me3 marks on odontogenic master transcription genes (Xu et al., 2013). All of these findings provide evidence that JMJD3 is involved in early mesoderm specification and differentiation, and it is essential for cardiovascular development and bone formation.
1.1.3. Ectoderm
Formation of the germ layer, ectoderm, is also regulated by JMJD3 and partially by UTX (Morales Torres et al., 2013). The ectoderm can differentiate into the nervous system, including the spine, peripheral nerves, and brain. Key regulators in neurogenesis, Pax6, Sox1, and Nestin, harbor bivalent marks, H3K4me3/H3K27me3, which are dynamically regulated during differentiation, and Jmjd3 can directly regulate expression of these key regulators in neurogenesis (Burgold et al., 2008). Jmjd3 is also a key regulator in Shh-dependent neural tube development (Shi et al., 2014). In fact, Jmjd3 is essential for ESC commitment into neural lineages. In the developing spinal cord, trimethylation of H3K27 also regulates BMP activity, which, in turn, leads to JMJD3 interaction with the transcription factors, SMAD1/4, to activate the notochord-derived BMP antagonist, Noggin (Akizu et al., 2010) (Fig. 1C). This negative feedback loop ensures rigorous and anatomically defined spinal cord formation. Aberrant neural development effects of Jmjd3 inactivation can also be observed after birth. Inactivation of Jmjd3 in mice generated by a gene trap strategy leads to disruption of the neuronal network of the pacemaker of the respiratory rhythm generator (Burgold et al., 2012). This causes respiratory failure in mice, further emphasizing the importance of Jmjd3 in normal vertebrate development. In the retina of the eye, H3K27 methylation and demethylation play important roles in retinal neuron proliferation and maturation, where expression of Bhlhb4 in retinal cells is associated with decreased H3K27me3 marks (Iida et al., 2014a). In a similar study, it was shown that during late retinal development, deficiency in Jmjd3 leads to decreased expression of Bhlhb4, which is associated with lower levels of H3K27me3 marks at the Bhlhb4 gene (Iida et al., 2014b). These studies suggest that Jmjd3 expression is required for correct maturation of retinal cells. Ectoderm can also give rise to hair, nails, sweat glands, outer layer of skin, and epithelia of various organs. However, no specific role for JMJD3 in the development of these tissues has been investigated. Overall, the important role of JMJD3 in the formation of all three germ layers suggests that JMJD3 is fundamental in cell fate and plasticity.
1.2. JMJD3 and cell plasticity
Not only does JMJD3 play a role in cellular differentiation in the developing embryo, it regulates cellular processes in differentiated tissues. Specifically, Jmjd3 is intimately involved in tissue repair. Following bone injury, osteoclasts are responsible for bone resorption, and abnormal osteoclast differentiation can lead to osteoporosis. RANKL stimulation of the osteoclast cell surface receptor, RANK, leads to osteoclast differentiation. This is accompanied by JMJD3-mediated demethylation of H3K27me3 at the Nfatc1 gene, a gene responsible for bone mass (Yasui et al., 2011). These studies suggest that JMJD3 mediates osteoclast differentiation after bone injury and may limit the onset of osteoporosis. JMJD3 has also been implicated in skin repair (Shaw and Martin, 2009). Whereas Polycomb genes, which are involved in histone methylation, are significantly downregulated during murine skin repair, the demethylases, Jmjd3 and Utx, are markedly upregulated, leading to less Polycomb-mediated silencing of the wound repair genes, Myc and Egfr (Shaw and Martin, 2009). Whether JMJD3 is directly involved in other types of tissue repair is not yet known, but JMJD3 is likely involved in the cellular plasticity involved in tissue repair. In fact, JMJD3 may play a direct role in the reprogramming of adult cells into a pluripotent state. The ability to revert differentiated cells back into a pluripotent, embryonic stem cell-like state has been a breakthrough for patient-specific disease modeling and drug testing. This cellular reprogramming is induced by the exogenous addition of the transcription factors, Sox2, Oct4, c-Myc, and Klf4. OCT4 is critical for generating induced pluripotent stem cells (iPSCs) and maintaining pluripotency (Lowry et al., 2008; Pesce and Scholer, 2001). Activation of OCT4 occurs in parallel with the recruitment of JMJD3 to chromatin, suggesting that JMJD3 is involved in cellular reprogramming (Apostolou and Hochedlinger, 2013). Our own research has shown that JMJD3 is a potent negative regulator of cellular reprogramming (Zhao et al., 2013) (Fig. 4). Ablation of Jmjd3 in mouse embryonic fibroblasts increases iPSC formation, whereas, ectopic Jmjd3 expression inhibits reprogramming by both histone demethylase-dependent and -independent mechanisms (Zhao et al., 2013). Further understanding of the role of JMJD3 in cellular reprogramming and tissue repair may lead to therapeutic test-beds of iPSCs and enhance wound repair.
Fig. 4.
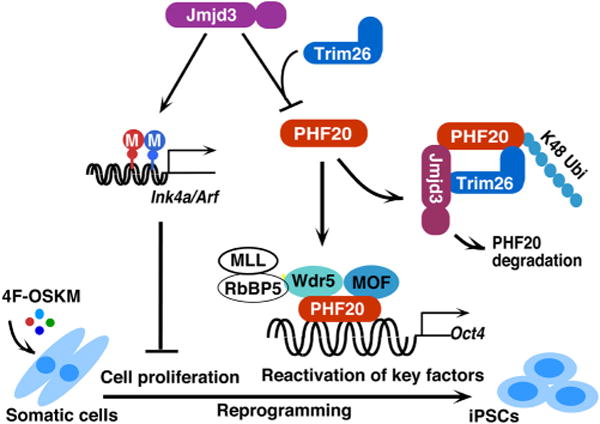
A working model to illustrate how JMJD3 negatively regulates cellular reprogramming through demethylase-dependent and independent pathways. JMJD3 upregulates Ink4a/Arf and p21 by removal of H3K27 methylation through its H3K27me2/3 demethylase activity. Increased amounts of Ink4a and Arf induce cell senescence or apoptosis and reduce cell proliferation, which leads to decrease in efficiency and kinetics of reprogramming. Importantly, JMJD3 protein also targets PHF20 for ubiquitination and degradation by recruiting an E3 ligase Trim26 in an H3K27 demethylase activity-independent manner. PHF20 is required for the reactivation of key core reprogramming factors such as Oct4 through interaction with WDR5. Thus, downregulation of PHF20 protein by JMJD3 leads to the inhibition of reprogramming efficiency.
JMJD3 also modulates blood cell growth. Deletion of JMJD3 in stem cells limits endothelial cell differentiation (Ohtani et al., 2013). In endothelial progenitor cells, CD34+ stem cells, and mesioangioblasts, in which histone methylation and acetylation are inhibited, endothelial nitric oxide synthase (eNOS) gene expression is increased concomitantly with reduction of repressive H3K27me3 marks and increased expression of JMJD3 (Ohtani et al., 2011). In addition, inhibition of JMJD3 can lead to an increase in cellular apoptosis and cellular senescence. Therefore, JMJD3 is involved, not only in cellular plasticity, but also in other cellular processes. JMJD3 may serve to prevent abnormal tissue growth associated with several pathologies by augmenting cell death and by controlling unlimited cell growth.
JMJD3 is also involved in cellular aging. As cells age, they cease to divide, and this aging process can contribute to the loss of the ability of adult stem cells to self-renew (Agger et al., 2009). When MSCs undergo cellular senescence, histone methyltransferases are downregulated and JMJD3 is upregulated, which is balanced by changes in histone acetylation status (Jung et al., 2010). Gene expression of two key regulators of cellular senescence, the tumor suppressor proteins, p16INKA and p14ARF, is silenced by H3K27me3 marks on the INK4a/ARF-gene locus in growing cells (Agherbi et al., 2009). Conversely, oncogenic-RAS-induced senescence leads to JMJD3 expression, which contributes to the activation of p16INK4A (Martinelli et al., 2011). Cellular senescence is also induced by the loss of the tumor repressor gene, B cell translocation gene 3 (BTG3) through increased JMJD3 expression and subsequent p16INK4A activation (Lin et al., 2012). In mouse fibroblasts, this senescence can be overcome by inhibition of JMJD3, which leads to their immortalization (Agherbi et al., 2009; Martinelli, 2011). Although it may seem counterintuitive that JMJD3 regulates the differentiation of some cells, while regulating the senescence of other cells, cell fate decisions are likely dependent a multitude of intracellular and extracellular signals that regulate the function of JMJD3. Cellular senescence can also occur within the nervous system. Schwann cells, which support neurons, are the glia cells of the peripheral nervous system. Tumorigenic stimuli or injury can lead to uncontrolled proliferation of Schwann cells, generating neurofibromas and schwannomas. Activation of JMJD3 leads to the removal of H3K27 marks from the Ink4a/Arf-locus and switches Schwann cells from a proliferative state to a senescent state to prevent overproliferation after nerve injury and during regeneration (Gomez-Sanchez et al., 2013). In this setting, JMJD3 plays a beneficial role in limiting tissue transformation and progression a pathological disease state. In primary human fibroblasts, senescence-associated heterochromatin foci (SAHF) are formed to stabilize the senescence state of cells, and the non-histone protein retinoblastoma (RB) protein, which is essential for SAHF, is demethylated at its lysine 810 amino acid residue (Zhao et al., 2015). This demethylation is associated with increased senescence and SAHF formation (Zhao et al., 2015), and this senescence of fibroblasts can promote epithelial cell growth and tumorigenesis (Krtolica et al., 2001). Whether the functional role of JMJD3 in cellular senescence is beneficial or detrimental may depend on the cell and tissue type, in which JMJD3 is expressed and activated.
1.3. JMJD3 in neurodegenerative diseases
The causes of peripheral neuropathies are very heterogeneous and can be caused by a genetic predisposition, inflammation-mediated damage, or physical injury. Schwann cells myelinate axons in the peripheral nervous system and play a role in inherited demyelinating diseases. These cells are crucial for repair of the spinal cord and peripheral nerves, and JMJD3 limits Schwann cell overproliferation after nerve injury and during regeneration, thus protecting against neurofibroma (Gomez-Sanchez et al., 2013). Not only can JMJD3 affect the peripheral nervous system, it can also affect the central nervous system. In Parkinson’s disease, an association exists in the polarization of microglia cells between a pro-inflammatory microglia M1 phenotype and an anti-inflammatory microglia M2 phenotype, and suppression of JMJD3 augments the M1 pro-inflammatory response by inhibiting M2 microglia polarization and leading to neuronal cell death (Tang et al., 2014). This was associated with increased H3K27 marks in the midbrain of aged mice, suggesting that JMJD3 plays a role in microglia polarization and may alter immune pathogenesis of Parkinson’s disease (Tang et al., 2014). Furthermore, in a subtype of neurons that is associated with Parkinson’s disease, the mid-brain type dopaminergic (DA) neurons, vitamin C enhances their differ entiation (He et al., 2014). Vitamin C decreases H3K27me3 marks on gene promoters in cells of the DA phenotype (He et al., 2014), suggesting that JMJD3 mediates their differentiation. The involvement of JMJD3 was confirmed using Jmjd3 knockdown or Jmjd3 inhibition experiments, which demonstrated that H3K27me3 changes are essential for vitamin C-mediated mid-brain type DA neuron differentiation (He et al., 2014). In addition to Parkinson’s disease, JMJD3 has been implicated in Alzheimer’s disease. Alzheimer’s disease is thought to be associated with the accumulation of amyloid-β in neuronal cells and activation of tumor suppressor genes. A member of the p53 tumor suppressor family, p63, was shown to have dual functions. The full-length form of the tumor suppressor p63, including its transactivation domain (TAp63γ), antagonizes neuronal apoptosis in response to amyloid-β stimulation (Fonseca et al., 2012b) as well as in neurogenesis (Fonseca et al., 2012a). TAp63γ interacts with JMJD3 to regulate neural-specific gene expression, which alters p63 methylation state, levels, and nuclear accumulation during neurogenesis (Fonseca et al., 2012a), suggesting a protective role for JMJD3 in Alzheimer’s disease. Most neurodegenerative diseases are associated with neural cell death, and dead or injured neurons are not easily replaced. Neurogenesis may be key in circumventing neurodegenerative disease progression. Neurogenesis in the adult subventricular zone requires JMJD3, and JMJD3 interacts with the promoters and the enhancers of neuronal genes (Park et al., 2014). It is clear that JMJD3 is involved in differentiation of cells of the nervous system, but whether stimulating or antagonizing JMJD3 will truly prove beneficial is not yet known. Any therapeutic effects on neurodegenerative diseases may be context-dependent.
1.4. JMJD3 in cancer
The histone demethylase gene, UTX, was the first mutated histone demethylase gene to be associated with cancer (van Haaften et al., 2009), suggesting that cancer cell growth is regulated by H3K27 methylation levels at site-specific promoters or enhancers. In fact, the tumor suppressor protein p53 physically interacts with JMJD3, and these two proteins have overlapping promoter and enhancer binding sites (Williams et al., 2014). JMJD3 is also associated with several types of syndromes that can give rise to cancer. Myelodysplastic syndromes (MDS) are associated with ineffective hematopoiesis and abnormal blood cell differentiation into one or more blood cell lineages and can lead to acute lymphoblastic leukemia. Peripheral blood CD34+ stem cells in patients with lower-risk MDS have increased expression of JMJD3 and an increased ability to form erythroid colonies upon inhibition of JMJD3 (Wei et al., 2013a, 2013b). This suggests that JMJD3 is intimately involved in hematopoietic lineage determination, and its inhibition may become a therapeutic option for anemia, which manifests in patients with MDS. MDS may transform into diseases such as acute lymphoblastic leukemia. Patients with T-cell acute lymphoblastic leukemia (T-ALL), harboring excess T lymphocytes, have poor prognosis with a 25% relapse rate. Whereas UTX functions as a tumor suppressor in T-ALL, JMJD3 demethylates H3K27 on oncogenes, allowing for initiation and maintenance of T-ALL (Ntziachristos et al., 2014). JMJD3 is also involved in other blood cancers. In Hodgkin’s Lymphoma, during the differentiation of B cells into centroblasts, JMJD3 expression increases (Anderton et al., 2011). Furthermore, known gene targets of JMJD3 are upregulated in Hodgkin’s Lymphoma and ablation of JMJD3 restores H3K27me3 enrichment at these gene targets (Anderton et al., 2011).
The importance of JMJD3 in cancer is not limited to cancers of the blood, but is evident in solid tumors as well. In breast cancer cells, JMJD3 is recruited to estrogen receptor enhancers, leading to activation of the anti-apoptotic protein, BCL2. This epigenetic state affects ligand dependency during estrogen receptor-mediated gene expression (Svotelis et al., 2011). Moreover, transfection of the lung cancer cell line, A549, with JMJD3 leads to increased cell proliferation and decreased expression of the cellular senescence marker, p21 (Tian et al., 2012), suggesting that increased endogenous JMJD3 expression may contribute to lung cancer development. Histone modifications such as acetylation and methylation have been shown to predict the risk of developing prostate cancer (Seligson et al., 2005). In fact, an H3K27 methyltransferase is associated with prostate cancer progression (Varambally et al., 2002), and JMJD3 expression is upregulated in prostate cancer, with a further increase during metastasis (Xiang, 2007). These findings suggest that histone methylation status at H3K27 is highly associated with prostate cancer. JMJD3 is also upregulated in renal cell carcinoma (Shen et al., 2012), and carcinogens have been shown to increase JMJD3 expression and dimethylation of H3K27 in kidney cancer cells (Guo et al., 2014). Conversely, loss of JMJD3 augments the aggressiveness of pancreatic cancer cells and increases liver metastasis (Yamamoto et al., 2014). Furthermore, JMJD3 expression is markedly increased in pancreatic precancerous lesions, but its decrease is associated with malignant grade progression. Similarly, JMJD3 expression levels are lower in colorectal carcinoma tissue compared to normal tissue (Tokunaga et al., 2014), which is associated with increased expression of cell cycle related genes and protection against apoptosis. Analysis of glioma patient databases of gene expression revealed high expression levels of JMJD3 in patients with high-grade glioma (Perrigue et al., 2015). Moreover, JMJD3 expression is induced upon glioblastoma stem cell differentiation through modulation of the INK4a/ARF-locus and through chromatin-independent, p53 protein stabilization (Ene et al., 2012).
Epithelial-mesenchymal transition (EMT) plays a crucial role in malignant transformation, tumor progression, and metastasis. Cells lose their polarity and adhesion properties and gain migratory and invasive properties. In mammary epithelial cells, JMJD3 allows for TGF-β-induced EMT through expression of the EMT-specific gene, SNAI1, leading to breast cancer invasion (Ramadoss et al., 2012). This suggests that JMJD3 is key regulator in cancer metastasis. Colon cancer cells also have the ability to undergo EMT, and vitamin D leads to EMT gene expression, including ZEB1, ZEB2, and SNAI1, through upregulation of JMJD3 (Pereira et al., 2012). Regulation of JMJD3 by vitamin D ensures precise gene activation by removing repressive marks in key genes associated with colon cancer cells (Pereira et al., 2011).
1.5. JMJD3 in the immune system
JMJD3 can enhance both pro-inflammatory and anti-inflammatory responses (as reviewed in Salminen et al., 2014) involved in infection and wound healing. In cytokine-stimulated human leukemia monocyte (THP-1) cells, gene expression and proteomic profiling revealed that JMJD3 knockdown decreases expression of key inflammatory markers, heat shock protein beta-1 (HspB1), and increases tripartite motif protein (TRIM5), glutathione peroxidase (Gpx), glia maturation factor-gamma (GMFG), caspase recruitment domain family member 14 (CARMA2, and dUTP pyrophosphatase) (Das et al., 2013, 2010). Changes in these pro- and anti-inflammatory markers are involved in the NF-κB, chemokine, and CD40 signaling pathways, and knockdown of Jmjd3 inhibits several NF-κB-regulated inflammatory genes (Das et al., 2010). The bacterial toxin, lipopolysaccharide (LPS), which signals through NF-κB, decreases JMJD3 expression in human gingival tissue-derived keratinocytes (de Camargo Pereira et al., 2013), suggesting that endogenous JMJD3 may limit further pro-inflammatory cytokine gene expression.
Macrophages can adopt different phenotypes and are classified as M1 phenotype or M2 phenotype. Classically activated M1 macrophages are typically responsible for phagocytosing endogenous apoptotic cells and foreign bodies, such as bacteria, parasites, and viruses, as well as presenting antigens to induce an adaptive immune response. M2 macrophages promote tissue remodeling and wound healing by downregulating the pro-inflammatory response induced by M1 macrophages. Macrophages, both M1 and M2, are an integral part of both the acute and chronic phases of inflammation and have been implicated in atherosclerosis, where they engulf excess cholesterol and become foam cells. Serum amyloid A proteins (SAAs) are also associated with atherosclerosis and are primarily produced by the liver. Upon release into the circulation, SAAs associate with high-density lipoprotein (HDL) particles for cholesterol transport to the liver. SAAs can induce macrophage pro-inflammatory cytokine production and foam cell formation. SAA-stimulation of macrophages leads to increased Jmjd3 expression, which is associated with reduced H3K27me3 marks, allowing for gene expression (Yan et al., 2014). Furthermore, Jmjd3 silencing and depletion inhibits macrophage pro-inflammatory cytokine secretion, and Jmjd3 is essential for SAA-induced foam cell formation (Yan et al., 2014), suggesting a possible therapeutic role in atherosclerosis. In wound healing, specifically in patients with Type 2 Diabetes (T2D), macrophages of the pro-inflammatory M1 phenotype mediate chronic inflammation. Differentiation of bone marrow stem/progenitor cells into macrophages is associated with an enrichment of H3K27me3 at the interleukin (IL)-12 gene promoter, which silences IL-12 gene expression (Gallagher et al., 2014). Under diabetic conditions, JMJD3 augments IL-12 expression, which can be regulated by Jmjd3 inhibition, suggesting that Jmjd3 inhibition could become a treatment option for diabetic wounds. Altogether, JMJD3 is intimately involved in modulating sterile inflammation, including atherosclerosis and wound healing, and the clinical benefits of targeting JMJD3 during these processes may depend on the disease state.
JMJD3 also plays a role in response to bacteria, parasites, or viruses. Upon macrophage administration of LPS, JMJD3 is recruited to the transcription start sites at over 70% of LPS-inducible genes (De Santa et al., 2009, 2007). However, deletion of Jmjd3 did affect the expression of most target genes except for a few hundred inflammatory genes, and most target genes had undetectable H3K27me3 marks, suggesting H3K27 demethylation-independent JMJD3-mediated gene activation (De Santa et al., 2009). JMJD3 also plays a role in the resistance of the macrophage cell line, RAW 264.7 cells, to the anthrax lethal toxin, Bacillus anthracis (Das et al., 2010), suggesting that the ability of JMJD3 to promote a pro-inflammatory response may prove beneficial in certain settings. During latency, the genome of the herpes simplex virus 1 (HSV-1) is associated with H3K27me3 marks, and inhibitors of histone demethylases, JMJD3 and UTX, limit the ability of HSV-1 to become reactivated in sensory neurons (Messer et al., 2015). JMJD3 is also induced by the EBV oncogene in the Epstein–Barr virus-associated malignancy, Hodgkin’s Lymphoma [63]. JMJD3 may preferentially regulate a particular subset of macrophages depending on the inciting infection. Macrophages have the ability to polarize between a pro-inflammatory M1 phenotype and an anti-inflammatory M2 phenotype and Jmjd3 plays a role in this polarization and activation (as reviewed in (Van den Bossche et al., 2014)). In one study, depletion of Jmjd3 indicated that Jmjd3 is not essential for M1 responses, but is imperative for M2 polarization in response to parasitic helminth infections (Satoh et al., 2010). In another study, M2 macrophages treated with IL-4 had decreased H3K27 methylation at M2 marker gene promoters along with increased Jmjd3 expression, which is mediated by STAT6 signaling (Ishii et al., 2009). Although compensatory mechanisms may exist for the loss of Jmjd3, JMJD3 may actually fine-tune cell-specific pro-inflammatory and anti-inflammatory immune responses, which may also be mediated by H3K27-independent mechanisms.
JMJD3 is also important in autoimmune diseases. Specifically, Jmjd3-deficient mice are resistant to experimental autoimmune encephalomyelitis (EAE) (Liu et al., 2015). In systemic lupus erythemastosus, which is characterized by overactive T cells and overstimulated B cells, CD4+ T cells have decreased hematopoietic progenitor kinase 1 (HPK1) expression. The promoter of this negative regulator of T cell-mediated immune responses harbors increased H3K27me3 marks with decreased JMJD3 binding (Zhang et al., 2011). Knockdown of Jmjd3 decreases this binding and increases H3K27me3 enrichment, suggesting that Jmjd3 expression in CD4+ T cells contributes to T cell activation and B-cell stimulation (Zhang et al., 2011). In a recent study, JMJD3 has been shown to play a role in systemic sclerosis, whereby JMJD3 overexpression is associated with decreased H3K27me3 marks in CD4+ T cells (Wang et al., 2015). JMJD3 is critically important in CD4+ T cell differentiation. Naïve CD4+ T cell activation leads to an immediate increase in Jmjd3 expression (Liu et al., 2015), and the absence of Jmjd3 affects CD4+ T cell differentiation. However, the mechanisms of how Jmjd3 affects this differentiation are controversial. The T-box transcription factor T-bet, which is required for T helper (Th)1 cell differentiation and interferon (IFN)-gamma production, has the ability to recruit JMJD3 to its target genes (Miller and Weinmann, 2010). Deletion of Jmjd3, specifically in T cells, inhibits the differentiation of naïve T cells into Th1 helper T cell or regulatory T cells, but promotes the differentiation into Th2 helper T cells and Th17 cells (Li et al., 2014b; Liu et al., 2015). Furthermore, adoptive transfer of these Jmjd3-deficient T cells limits colitis in a mouse model of disease (Li et al., 2014b). Conversely, another study showed deletion of Jmjd3 in CD4± T cells impaired Th17 cell differentiation (Liu et al., 2015). Furthermore, this study showed that ectopic expression of Jmjd3 induced differentiation of Jmjd3-deficient CD4± T cells into Th17 cells, which was associated with the demethylation of H3K27 on Th17 cytokine genes, Il17, Il17f, and Il22 as well as Rorc, which encodes for Th17 transcription factor Rorγt (Liu et al., 2015). The reasons for the discrepancies in the two studies are not clear. In one study, in which Jmjd3 deletion impaired Th17 differentiation, Jmjd3 was deleted by targeting exons 14–20, and in the other study, in which Jmjd3 deletion promoted Th17 differentiation, Jmjd3 was deleted by targeting exons 15–21. These discrepancies are in agreement with the different phenotypes in Jmjd3 knockout mice that are dependent on gene dosage and the gene deletion strategy (Burgold et al., 2012; Ohtani et al., 2013; Satoh et al., 2010). Furthermore, slight differences in the purity of naïve CD4± T cells as well as tissue type derivation of the cells may have resulted in different results. In the study by Liu et al., Jmjd3 deletion did not change the ability of naïve T cells to differentiate into Th1 cells. However, in the study by Li et al., Jmjd3 deletion inhibited the differentiation of naïve T cells into Th1 cells. In addition, Jmjd3 ablation in macrophages markedly affects the expression of two genes, which mediate recruitment of T cells and Th1 cell polarization (De Santa, 2009). The role of Jmjd3 may be highly dependent on the specific cell type. Overall, JMJD3 is involved in modulating immune cells in response to several conditions: to sterile inflammatory mediators, to injury, to infectious agents and to self (autoimmunity). Therefore, inhibition of JMJD3 may have different affects, depending on cell type and under different conditions.
1.6. JMJD3 and histone demethylase independent function
JMJD3 can regulate gene expression independent of its demethylase activity (Miller et al., 2010; Zhao et al., 2013). In quiescent, differentiated cells, broad changes in gene expression are not as imperative as in cells undergoing differentiation. Demethylase proteins, such as UTX and JMJD3, which antagonize H3K27me3-mediated gene repression, are still expressed in terminally differentiated cells. This is perplexing since incorrect timing of gene activation could be detrimental to a cell. In the transformed murine thymoma cell line, EL4, the epigenetic profile is already established and set, but interaction between JMJD3 and the T-bet, the lineage-defining T-box factor for T helper 1 (Th1) cell development, still exists (Miller et al., 2008). JMJD3 mediates an interaction between a BRG-1-containing SWI/SNF remodeling complex, which is required for promoter remodeling and optimal gene expression (Miller et al., 2010). Therefore, the demethylase-independent activity of JMJD3 is still related to alterations in chromatin structure, although not directly. We also demonstrated that JMJD3 functions in a histone demethylase-independent manner (Zhao et al., 2013). Depletion of Jmjd3 increases somatic reprogramming into a pluripotent state in a histone demethylase-independent manner. JMJD3 targets PHD finger protein 20 (PHF20) for ubiquitination and subsequent degradation through Trim26, an E3 ligase (Zhao et al., 2013). PHF20 is required for maintenance of pluripotency and cellular reprogramming. Therefore, JMJD3 may limit iPSC formation in a histone demethylase-independent manner. However, JMJD3 can also modulate iPSC formation in a histone demethylase-dependent manner by targeting dimethylation at the INK4a/ARF locus (Fig. 2). Besides reverting cells back into a pluripotent state, Jmjd3 can mediate or inhibit stem cell differentiation into terminally differentiated cells in a histone demethylase-independent manner (Fig. 4). During glioblastomagenesis, stem cell differentiation occurs in a chromatin-independent manner through stabilization of the p53 protein (Ene et al., 2012). During glioblastoma formation cellular terminal differentiation may become blocked and lead to malignant transformation. Dysfunction of JMJD3 may contribute to glioblastoma formation by blocking this cellular terminal differentiation (Ene et al., 2012). JMJD3 can also demethylate non-histone proteins. During cellular senescence, JMJD3 can demethylate the non-histone retinoblastoma (RB) protein at its lysine 810 residue (Zhao et al., 2015). The histone demethylase activity of JMJD3 may also be regulated by its intracellular localization (Kamikawa and Donohoe, 2014). JMJD3 contains two classical nuclear localization sequences at its N-terminus (Kamikawa and Donohoe, 2014). Forced nuclear localization of JMJD3 leads to markedly enhanced H3K27me3 demethylation, whereas cytosolic localization may led to dimethylation of non-histone proteins (Zhao et al., 2015). During SAHF formation, JMJD3 is colocalized with the Golgi protein, GM130 (Zhao et al., 2015) and may demethylate non-histone proteins undergoing posttranslational modifications within the Golgi. Both JMJD3 demethylase-dependent and -independent pathways may mediate efficient cellular reprogramming and differentiation, but the independent pathway may play a major role in terminally differentiated cells and cancer.
1.7. Future prospects
Targeting JMJD3 for the development of therapeutics to treat a number of diseases may prove feasible and efficacious. Using a structure-function approach and subsequent screening of a GlaxoSmithKline corporate compound collection (approximately two million compounds), Kruidenier L et al. identified GSK-J1 as a specific JMJD3 catalytic site inhibitor (Kruidenier et al., 2012). This small molecule inhibitor was further modified by addition of cell-penetrating ethyl esters, yielding GSK-J4, which inhibited LPS-induced cytokine expression in human macrophages from healthy volunteers. This molecule was further shown to inhibit the cell proliferation of all human T-ALL cell lines tested, and GSK-J4 did not affect the growth of myeloid leukemia cells, stromal cells, or hematopoietic progenitor cells (Ntziachristos et al., 2014). GSK-J4 has been used to treat cells isolated from pediatric brainstem glioma (Hashizume et al., 2014) patients with a lysine to methionine (K27M) substitution on histones H3.1 and H3.3 (Wu et al., 2014). Preclinical studies treating K27M-mutant brainstem glioma cell lines using GSK-J4 demonstrated marked inhibition of cell growth (Wu et al., 2014). Treating cells with a JMJD3 inhibitor did not increase the levels of H3K27me3 marks on the K27M-mutant, but rather indirectly promoted the methylation of the non-mutant protein allele (Wu et al., 2014). Furthermore, Rotili et al. developed pan-histone demethylase inhibitors, which can target all Jumonji C demethylases, including JMJD3, and these inhibitors have the capacity to induce cell cycle arrest and increase apoptosis in human prostate LNCaP cells and colon HCT116 cancer cells (Rotili et al., 2014).
Alternative to using small molecule inhibitors, a new technology, (clustered regularly interspaced short palindromic repeats)/Cas9-based gene editing (Hsu Patrick et al., 2014; Qi et al., 2013), may be used to delete JMJD3 for therapeutic applications. This gene editing can use RNA-guided DNA recognition to precisely edit the gene of interest (Qi), with minimal off-target effects. This technology can also be used as a screening approach to identify downstream effectors of JMJD3 and to identify which genes are essential for mediating JMJD3 responses.
Inhibition of JMJD3 may provide benefit in certain circumstances, but inhibition may actually produce deleterious responses. If ablation of JMJD3 increases pancreatic cancer cell metastasis (Yamamoto et al., 2014), inhibiting JMJD3 using GSK-J4 or gene editing may be detrimental. In addition, low levels of JMJD3 expression may be associated with colorectal cancer (Tokunaga et al., 2014), and thus, inhibition of JMJD3 in certain cancers may not provide benefit. Alternatively, complete JMJD3 ablation may have more profound effects on cells and tissues compared with a small molecule inhibitor. However, because JMJD3 is involved in a plethora of cellular processes in diverse cell types, globally administered JMJD3 inhibitors may produce off-target and side effects. Therefore, designing cell-specific agonists or antagonist would help control for any non-specific effects. Whether JMJD3 should be activated or inhibited for therapeutic efficacy may be highly dependent on the tissue or disease being targeted. The use of JMJD3 inhibitors may prove particularly effective in treating diseases involving gene mutations, gene dosage defects, or gene imprinting, where increasing expression of the non-mutant, inactivated, or imprinted allele may alter disease state. Conversely, in diseases, such as some types of cancer, activating JMJD3 may be a more effective treatment by suppressing cancer stem cell proliferation, preventing malignant transformation, and promoting tumor suppression.
The future identification of non-histone proteins, which are demethylated or directed for ubiquitination and subsequent degradation, will provide evidence of other key molecules that are downstream and dependent on JMJD3. Also, further investigations are needed to characterize the intracellular localization of JMJD3 and to identify alternative functions of JMJD3 within these intracellular compartments. The mechanisms of how JMJD3 RNA and protein are degraded have not been determined, but may help regulate JMJD3 levels and downstream histone demethylase-dependent and -independent pathways.
Acknowledgments
This work is supported, in part, by grants from the NIH (R01CA101795, R01CA090327) and Cancer Prevention and Research Institution of Texas (CPRIT).
Footnotes
This article is part of a Directed Issue entitled: Epigenetics dynamics in development and disease.
References
- Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–6. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agherbi H, Gaussmann-Wenger A, Verthuy C, Chasson L, Serrano M, Djabali M. Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence. PLoS ONE. 2009;4:e5622. doi: 10.1371/journal.pone.0005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akizu N, Estaras C, Guerrero L, Marti E, Martinez-Balbas MA. H3K27me3 regulates BMP activity in developing spinal cord. Development (Cambridge, England) 2010;137:2915–25. doi: 10.1242/dev.049395. [DOI] [PubMed] [Google Scholar]
- Anderton JA, Bose S, Vockerodt M, Vrzalikova K, Wei W, Kuo M, et al. The H3K27me3 demethylase, KDM6B, is induced by Epstein-Barr virus and overexpressed in Hodgkin’s lymphoma. Oncogene. 2011;30:2037–43. doi: 10.1038/onc.2010.579. [DOI] [PubMed] [Google Scholar]
- Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–71. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bengestrate L, Virtue S, Campbell M, Vidal-Puig A, Hadaschik D, Hahn P, et al. Genome-wide profiling of microRNAs in adipose mesenchymal stem cell differentiation and mouse models of obesity. PLoS ONE. 2011;6:e21305. doi: 10.1371/journal.pone.0021305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, et al. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE. 2008;3:e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgold T, Voituron N, Caganova M, Tripathi PP, Menuet C, Tusi BK, et al. The H3K27 demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival. Cell Rep. 2012;2:1244–58. doi: 10.1016/j.celrep.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–40. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–53. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Das ND, Jung KH, Chai YG. The role of NF-kappaB and H3K27me3 demethylase, Jmjd3, on the anthrax lethal toxin tolerance of RAW 264.7 cells. PLoS ONE. 2010;5:e9913. doi: 10.1371/journal.pone.0009913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Das ND, Jung KH, Park JH, Lee HT, Han D, et al. Proteomic changes induced by histone demethylase JMJD3 in TNF alpha-treated human monocytic (THP-1) cells. Mol Immunol. 2013;56:113–22. doi: 10.1016/j.molimm.2013.04.013. [DOI] [PubMed] [Google Scholar]
- de Camargo Pereira G, Guimaraes GN, Planello AC, Santamaria MP, de Souza AP, Line SR, et al. Porphyromonas gingivalis LPS stimulation downregulates DNMT1, DNMT3a, and JMJD3 gene expression levels in human HaCaT keratinocytes. Clin Oral Invest. 2013;17:1279–85. doi: 10.1007/s00784-012-0816-z. [DOI] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–94. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009;28:3341–52. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene CI, Edwards L, Riddick G, Baysan M, Woolard K, Kotliarova S, et al. Histone demethylase Jumonji D3 (JMJD3) as a tumor suppressor by regulating p53 protein nuclear stabilization. PLoS ONE. 2012;7:e51407. doi: 10.1371/journal.pone.0051407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca MB, Nunes AF, Morgado AL, Sola S, Rodrigues CM. TAp63gamma demethylation regulates protein stability and cellular distribution during neural stem cell differentiation. PLoS ONE. 2012a;7:e52417. doi: 10.1371/journal.pone.0052417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca MB, Nunes AF, Rodrigues CM. c-Jun regulates the stability of anti-apoptotic DeltaNp63 in amyloid-beta-induced apoptosis. J Alzheimer’s Dis: JAD. 2012b;28:685–94. doi: 10.3233/JAD-2011-111547. [DOI] [PubMed] [Google Scholar]
- Gallagher KA, Joshi A, Carson WF, Schaller M, Allen R, Mukerjee S, et al. Epigenetic changes in bone marrow progenitor cells influence the inflammatory phenotype and alter wound healing in type 2 diabetes. Diabetes. 2014 doi: 10.2337/db14-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Sanchez JA, Gomis-Coloma C, Morenilla-Palao C, Peiro G, Serra E, Serrano M, et al. Epigenetic induction of the Ink4a/Arf locus prevents Schwann cell overproliferation during nerve regeneration and after tumorigenic challenge. Brain: J Neurol. 2013;136:2262–78. doi: 10.1093/brain/awt130. [DOI] [PubMed] [Google Scholar]
- Guo X, Zhang Y, Zhang Q, Fa P, Gui Y, Gao G, et al. The regulatory role of nickel on H3K27 demethylase JMJD3 in kidney cancer cells. Toxicol Ind Health. 2014 doi: 10.1177/0748233714552687. [DOI] [PubMed] [Google Scholar]
- Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20:1394–6. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XB, Kim M, Kim SY, Yi SH, Rhee YH, Kim T, et al. Vitamin C facilitates dopamine neuron differentiation in fetal midbrain through TET1- and JMJD3-dependent epigenetic control manner. Stem Cells (Dayton, Ohio) 2014 doi: 10.1002/stem.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A. 2007;104:18439–44. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Patrick D, Lander Eric S, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huszar JM, Payne CJ. MIR146A inhibits JMJD3 expression and osteogenic differentiation in human mesenchymal stem cells. FEBS Lett. 2014;588:1850–6. doi: 10.1016/j.febslet.2014.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida A, Iwagawa T, Baba Y, Satoh S, Mochizuki Y, Nakauchi H, et al. Roles of histone H3K27 trimethylase Ezh2 in retinal proliferation and differentiation. Dev Neurobiol. 2014a doi: 10.1002/dneu.22261. [DOI] [PubMed] [Google Scholar]
- Iida A, Iwagawa T, Kuribayashi H, Satoh S, Mochizuki Y, Baba Y, et al. Histone demethylase Jmjd3 is required for the development of subsets of retinal bipolar cells. Proc Natl Acad Sci U S A. 2014b;111:3751–6. doi: 10.1073/pnas.1311480111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inose H, Ochi H, Kimura A, Fujita K, Xu R, Sato S, et al. A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci U S A. 2009;106:20794–9. doi: 10.1073/pnas.0909311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114:3244–54. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang J, Zhang Y. Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Res. 2013;23:122–30. doi: 10.1038/cr.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JW, Lee S, Seo MS, Park SB, Kurtz A, Kang SK, et al. Histone deacetylase controls adult stem cell aging by balancing the expression of polycomb genes and jumonji domain containing 3. Cell Mol Life Sci: CMLS. 2010;67:1165–76. doi: 10.1007/s00018-009-0242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikawa YF, Donohoe ME. The localization of histone H3K27me3 demethylase Jmjd3 is dynamically regulated. Epigenetics: Off J DNA Methylation Soc. 2014;9:834–41. doi: 10.4161/epi.28524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartikasari AE, Zhou JX, Kanji MS, Chan DN, Sinha A, Grapin-Botton A, et al. The histone demethylase Jmjd3 sequentially associates with the transcription factors Tbx3 and Eomes to drive endoderm differentiation. EMBO J. 2013;32:1393–408. doi: 10.1038/emboj.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Yoon S-J, Chuong E, Oyolu C, Wills AE, Gupta R, et al. Chromatin and transcriptional signatures for nodal signaling during endoderm formation in hESCs. Dev Biol. 2011;357:492–504. doi: 10.1016/j.ydbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–27. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A. 2001;98:12072–7. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruidenier L, Chung C-w, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–8. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A. 2008;105:13906–11. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang HY, Chepelev I, Zhu Q, Wei G, Zhao K, et al. Stage-dependent and locus-specific role of histone demethylase Jumonji D3 (JMJD3) in the embryonic stages of lung development. PLoS Genet. 2014a;10:e1004524. doi: 10.1371/journal.pgen.1004524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zou J, Wang M, Ding X, Chepelev I, Zhou X, et al. Critical role of histone demethylase Jmjd3 in the regulation of CD4+ T-cell differentiation. Nat Commun. 2014b;5:5780. doi: 10.1038/ncomms6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY, Cheng YC, Yang HC, Lin WC, Wang CC, Lai PL, et al. Loss of the candidate tumor suppressor BTG3 triggers acute cellular senescence via the ERK-JMJD3-p16(INK4a) signaling axis. Oncogene. 2012;31:3287–97. doi: 10.1038/onc.2011.491. [DOI] [PubMed] [Google Scholar]
- Liu Z, Cao W, Xu L, Chen X, Zhan Y, Yang Q, et al. The histone H3 lysine-27 demethylase Jmjd3 plays a critical role in specific regulation of Th17 cell differentiation. J Mol Cell Biol. 2015 doi: 10.1093/jmcb/mjv022. [DOI] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–8. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–13. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- Martinelli P, Bonetti P, Sironi C, Pruneri G, Fumagalli C, Raviele PR, et al. The lymphoma-associated NPM-ALK oncogene elicits a p16INK4a/pRb-dependent tumor-suppressive pathway. Blood. 2011;117:6617–26. doi: 10.1182/blood-2010-08-301135. [DOI] [PubMed] [Google Scholar]
- Messer HGP, Jacobs D, Dhummakupt A, Bloom DC. Inhibition of H3K27me3-specific histone demethylases JMJD3 and UTX blocks reactivation of herpes simplex virus 1 in trigeminal ganglion neurons. J Virol. 2015;89:3417–20. doi: 10.1128/JVI.03052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Mfopou JK, Chen B, Sui L, Sermon K, Bouwens L. Recent advances and prospects in the differentiation of pancreatic cells from human embryonic stem cells. Diabetes. 2010;59:2094–101. doi: 10.2337/db10-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Weinmann AS. Molecular mechanisms by which T-bet regulates T-helper cell commitment. Immun Rev. 2010;238:233–46. doi: 10.1111/j.1600-065X.2010.00952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–93. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales Torres C, Laugesen A, Helin K. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLOS ONE. 2013;8:e60020. doi: 10.1371/journal.pone.0060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–7. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K, Vlachojannis GJ, Koyanagi M, Boeckel JN, Urbich C, Farcas R, et al. Epigenetic regulation of endothelial lineage committed genes in pro-angiogenic hematopoietic and endothelial progenitor cells. Circ Res. 2011;109:1219–29. doi: 10.1161/CIRCRESAHA.111.247304. [DOI] [PubMed] [Google Scholar]
- Ohtani K, Zhao C, Dobreva G, Manavski Y, Kluge B, Braun T, et al. Jmjd3 controls mesodermal and cardiovascular differentiation of embryonic stem cells. Circ Res. 2013;113:856–62. doi: 10.1161/CIRCRESAHA.113.302035. [DOI] [PubMed] [Google Scholar]
- Park DH, Hong SJ, Salinas RD, Liu SJ, Sun SW, Sgualdino J, et al. Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 2014;8:1290–9. doi: 10.1016/j.celrep.2014.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F, Barbachano A, Silva J, Bonilla F, Campbell MJ, Munoz A, et al. KDM6B/JMJD3 histone demethylase is induced by vitamin D and modulates its effects in colon cancer cells. Hum Mol Genet. 2011;20:4655–65. doi: 10.1093/hmg/ddr399. [DOI] [PubMed] [Google Scholar]
- Pereira F, Barbachano A, Singh PK, Campbell MJ, Munoz A, Larriba MJ. Vitamin D has wide regulatory effects on histone demethylase genes. Cell Cycle (Georgetown, Tex) 2012;11:1081–9. doi: 10.4161/cc.11.6.19508. [DOI] [PubMed] [Google Scholar]
- Perrigue PM, Silva ME, Warden CD, Feng NL, Reid MA, Mota DJ, et al. The histone demethylase jumonji coordinates cellular senescence including secretion of neural stem cell-attracting cytokines. Mol Cancer Res: MCR. 2015 doi: 10.1158/1541-7786.MCR-13-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce M, Scholer HR. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells (Dayton, Ohio) 2001;19:271–8. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–83. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadoss S, Chen X, Wang CY. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J Biol Chem. 2012;287:44508–17. doi: 10.1074/jbc.M112.424903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotili D, Tomassi S, Conte M, Benedetti R, Tortorici M, Ciossani G, et al. Pan-histone demethylase inhibitors simultaneously targeting Jumonji C and lysine-specific demethylases display high anticancer activities. J Med Chem. 2014;57:42–55. doi: 10.1021/jm4012802. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K, Hiltunen M, Kauppinen A. Histone demethylase Jumonji D3 (JMJD3/KDM6B) at the nexus of epigenetic regulation of inflammation and the aging process. J Mol Med (Berlin, Germany) 2014;92:1035–43. doi: 10.1007/s00109-014-1182-x. [DOI] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immun. 2010;11:936–44. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal morphogens. Cold Spring Harb Perspect Biol. 2009;1:a003459. doi: 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–6. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10:881–6. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Guo X, Wang Y, Qiu W, Chang Y, Zhang A, et al. Expression and significance of histone H3K27 demethylases in renal cell carcinoma. BMC Cancer. 2012;12:470. doi: 10.1186/1471-2407-12-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Svotelis A, Bianco S, Madore J, Huppe G, Nordell-Markovits A, Mes-Masson AM, et al. H3K27 demethylation by JMJD3 at a poised enhancer of anti-apoptotic gene BCL2 determines ERalpha ligand dependency. EMBO J. 2011;30:3947–61. doi: 10.1038/emboj.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- Tang Y, Li T, Li J, Yang J, Liu H, Zhang XJ, et al. Jmjd3 is essential for the epigenetic modulation of microglia phenotypes in the immune pathogenesis of Parkinson’s disease. Cell Death Differ. 2014;21:369–80. doi: 10.1038/cdd.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Deng H, Tang X, Hu H, Liu X, Luo F. Effect of Jumonji domain-containing protein-3 on the proliferation and migration of lung cancer cell line. Sheng wu yi xue gong cheng xue za zhi = J Biomed Eng = Shengwu yixue gongchengxue zazhi. 2012;29:514–8. [PubMed] [Google Scholar]
- Tokunaga R, Nakagawa S, Sakamoto Y, Karashima R, Ida S, Imamura Y, et al. Abstract 5150: JMJD3 suppresses progression of colorectal carcinoma by regulating cell cycle and anti-apoptosis. Cancer Res. 2014;74:5150. [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Van den Bossche J, Neele AE, Hoeksema MA, de Winther MP. Macrophage polarization: the epigenetic point of view. Curr Opin Lipidol. 2014;25:367–73. doi: 10.1097/MOL.0000000000000109. [DOI] [PubMed] [Google Scholar]
- Van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science (New York, NY) 2004;306:279–83. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- Wang Q, Xiao Y, Shi Y, Luo Y, Li Y, Zhao M, et al. Overexpression of JMJD3 may contribute to demethylation of H3K27me3 in CD4+ T cells from patients with systemic sclerosis. Clin Immun. 2015 doi: 10.1016/j.clim.2015.03.006. [DOI] [PubMed] [Google Scholar]
- Wei Y, Chen R, Dimicoli S, Bueso-Ramos C, Neuberg D, Pierce S, et al. Global H3K4me3 genome mapping reveals alterations of innate immunity signaling and overexpression of JMJD3 in human myelodysplastic syndrome CD34+ cells. Leukemia. 2013a;27:2177–86. doi: 10.1038/leu.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Dimicoli S, Bueso-Ramos C, Chen R, Yang H, Neuberg D, et al. Toll-like receptor alterations in myelodysplastic syndrome. Leukemia. 2013b;27:1832–40. doi: 10.1038/leu.2013.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Rappsilber J, Nielsen AL, Johansen JV, Helin K. The histone lysine demethylase JMJD3/KDM6B is recruited to p53 bound promoters and enhancer elements in a p53 dependent manner. PLOS ONE. 2014;9:e96545. doi: 10.1371/journal.pone.0096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46:444–50. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17:850–7. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- Xu J, Yu B, Hong C, Wang CY. KDM6B epigenetically regulates odontogenic differentiation of dental mesenchymal stem cells. Int J Oral Sci. 2013;5:200–5. doi: 10.1038/ijos.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Tateishi K, Kudo Y, Sato T, Yamamoto S, Miyabayashi K, et al. Loss of histone demethylase KDM6B enhances aggressiveness of pancreatic cancer through downregulation of C/EBPalpha. Carcinogenesis. 2014;35:2404–14. doi: 10.1093/carcin/bgu136. [DOI] [PubMed] [Google Scholar]
- Yan Q, Sun L, Zhu Z, Wang L, Li S, Ye RD. Jmjd3-mediated epigenetic regulation of inflammatory cytokine gene expression in serum amyloid A-stimulated macrophages. Cell Signal. 2014;26:1783–91. doi: 10.1016/j.cellsig.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Guo H, Zhang Y, Chen L, Ying D, Dong S. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS ONE. 2011;6:e21679. doi: 10.1371/journal.pone.0021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui T, Hirose J, Tsutsumi S, Nakamura K, Aburatani H, Tanaka S. Epigenetic regulation of osteoclast differentiation: possible involvement of Jmjd3 in the histone demethylation of Nfatc1. J Bone Miner Res: Off J Am Soc Bone Miner Res. 2011;26:2665–71. doi: 10.1002/jbmr.464. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Long H, Liao J, Zhao M, Liang G, Wu X, et al. Inhibited expression of hematopoietic progenitor kinase 1 associated with loss of jumonji domain containing 3 promoter binding contributes to autoimmunity in systemic lupus erythematosus. J Autoimmun. 2011;37:180–9. doi: 10.1016/j.jaut.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Zhang F, Xu L, Xu L, Xu Q, Li D, Yang Y, et al. JMJD3 promotes chondrocyte proliferation and hypertrophy during endochondral bone formation in mice. J Mol Cell Biol. 2015;7:23–34. doi: 10.1093/jmcb/mjv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Li Q, Ayers S, Gu Y, Shi Z, Zhu Q, et al. Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination. Cell. 2013;152:1037–50. doi: 10.1016/j.cell.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Zhang Y, Gao Y, Geng P, Lu Y, Liu X, et al. JMJD3 promotes SAHF formation in senescent WI38 cells by triggering an interplay between demethylation and phosphorylation of RB protein. Cell Death Differ. 2015 doi: 10.1038/cdd.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]


