Abstract
The development of disease-modifying pharmacologic therapy for osteoarthritis currently faces major obstacles largely because the pathogenetic mechanisms for development of osteoarthritis remain unclear. Previous studies suggest that the alterations in expression of catabolic and anabolic genes in articular chondrocytes may be involved in the pathogenesis of osteoarthritis. However, the regulatory mechanisms for gene expression in osteoarthritic chondrocytes are largely unknown.
The objective of this review is to highlight the recent studies on epigenetic regulation of gene expression in the development of osteoarthritis. The review will begin with current understanding of epigenetic mechanisms, especially the newly emerging areas including the regulatory role of non-coding RNAs in gene expression and crosstalk among the epigenetic mechanisms. The main content of this review focuses on the significance of epigenetic regulation of the expression of catabolic and anabolic genes in osteoarthritic chondrocytes, including the regulatory roles of various epigenetic mechanisms in the expression of genes for specific matrix-degrading proteinases, cytokines, and extracellular matrix proteins. Recent novel findings on the epigenetic regulation of specific transcription factor genes are particularly important for the understanding of osteoarthritis pathogenesis, as these transcription factors may act as upstream regulators of multiple catabolic and anabolic genes.
In conclusion, these recent advances in epigenetic studies have shed light on the importance of epigenetic regulation of gene expression in the development of osteoarthritis, leading to a better understanding of the epigenetic mechanisms underlying the pathogenesis of osteoarthritis. This may promote the development of new epigenetics-based strategies for the treatment of osteoarthritis.
Keywords: epigenetics, osteoarthritis, gene expression, proteinase, transcription factor, cytokine
1. Introduction
Osteoarthritis (OA) is the most common form of arthritis and the leading cause of chronic disability in middle-aged and older populations (Murphy and Helmick, 2012). As OA mainly occurs in load-bearing joints, such as the knee and hip, OA has long been thought of as a mechanical issue (Carter et al., 2004). However, there is a growing body of evidence supporting the notion that OA is a result of the interaction between mechanical and molecular events in the affected joint (Chen et al., 2013). There is no single specific cause that has been identified for OA to date. Some risk factors, including age, gender, obesity, joint injury, genetics, and mechanical abnormalities, have been shown to be associated with the development of OA (Blagojevic et al., 2010). However, how these risk factors trigger the onset of OA is not fully understood. While OA is a disease of the whole joint and may affect all of the joint tissues, articular cartilage degradation is a major hallmark of OA (Loeser et al., 2012). Aberrant gene expression in articular chondrocytes (ACs) of OA joints has been well documented in both animals and humans. Nevertheless, the underlying regulatory mechanism for the aberrant gene expression in OA cartilage remains to be elucidated.
Classically, “epigenetics” is referred to as changes in gene transcription caused by mechanisms other than changes in the underlying DNA sequences. Recently, non-coding RNAs (ncRNAs) which possess epigenetic-like properties in the regulation of gene expression have also been considered as one of the epigenetic mechanisms (Dawson and Kouzarides, 2012, Saetrom et al., 2007). In addition to the importance of epigenetics in normal development and tumorigenesis (Dawson and Kouzarides, 2012), recent studies on epigenetic changes in ACs have provided new insights into the pathogenesis of OA and potential therapeutic strategies for OA.
As the general concept of epigenetics has been previously reviewed elsewhere (Jenuwein and Allis, 2001, Jones, 2012, Mattick and Makunin, 2006), this review article will start with a brief discussion on the current understanding of epigenetic mechanisms, including DNA methylation, histone modification, and the emerging new areas: regulatory role of ncRNAs in gene expression and crosstalk among the epigenetic mechanisms. The main content of this review will focus on the epigenetic regulation of catabolic and anabolic genes and its significance in the pathogenesis of OA.
2. Current understanding of epigenetic mechanisms
2.1. DNA methylation
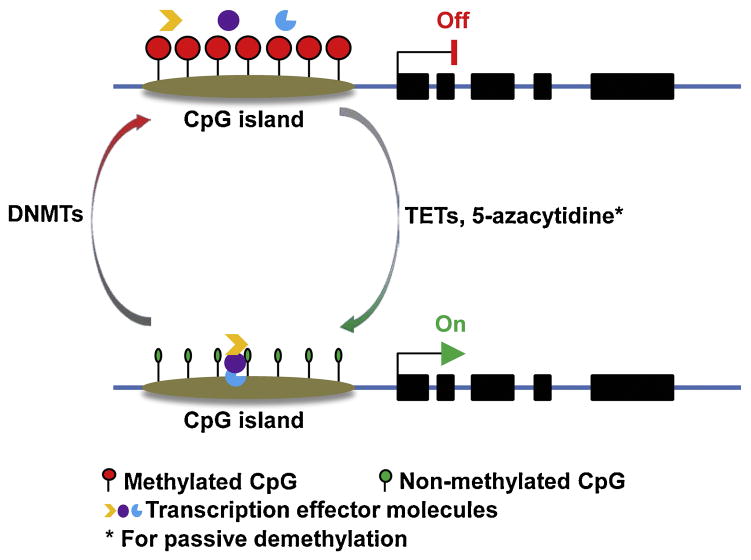
DNA methylation is a biochemical process whereby a methyl group is added to the cytosine or adenine, mainly at the C5 position of CpG dinucleotides, by DNA methyltransferases (DNMTs). At least four DNMTs (DNMT1, DNMT3A, DNMT3B and DNMT3L) have been identified for the de novo or maintenance of DNA methylation (Jin and Robertson, 2013). Methylated DNA can be passively or actively demethylated. 5-azacytidine is one of the chemical compounds for passive DNA demethylation, whereas three ten-eleven translocation proteins (TETs) and 5-methycytosine oxidases have been discovered to be responsible for active DNA demethylation (Wu and Zhang, 2014) (Figure 1). Theoretically, DNA methylation should be widespread in the genome; however, the first global methylomes analysis showed that CpG methylation follows a bimodal distribution, especially in the CpG islands (CGIs, which are averagely 1,000 base pairs long with increased GC density) in the promoter regions (Weber et al., 2007). To date, most of our knowledge of DNA methylation in the regulation of gene expression has come from the functional studies of DNA methylation in gene promoter region. DNA hypermethylation has been frequently described as a silent mark for gene transcription, while DNA hypomethylation is associated with active gene transcription. Although the detailed mechanisms by which DNA methylation suppresses gene expression are fairly well understood, two modes have been envisaged by Bird to explain the mechanism of DNA methylation-mediated transcriptional repression. The first mode involves direct interference of the methyl group in binding of a protein to its cognate DNA sequence; while the second mode involves proteins that are attracted to, rather than repelled by, methyl-CpG (Bird, 2002). Basically, DNA methylation alters protein-DNA interactions, especially blocking the binding of transcription effector molecules to promoter region (Figure 1).
Figure 1.
DNA methylation and gene expression. DNA methylation at a CpG island in the promoter region is mediated by DNA methyltransferases (DNMTs), which blocks the binding of transcription effector molecules to DNA and powers off gene transcription. Once methylated CpGs are reversibly demethylated by 5-methylcytosine oxidase ten-eleven translocation proteins (TETs) or 5-azacytidine, the promoter region becomes accessible for transcription effector molecules to bind and turn on gene transcription.
2.2. Histone modifications
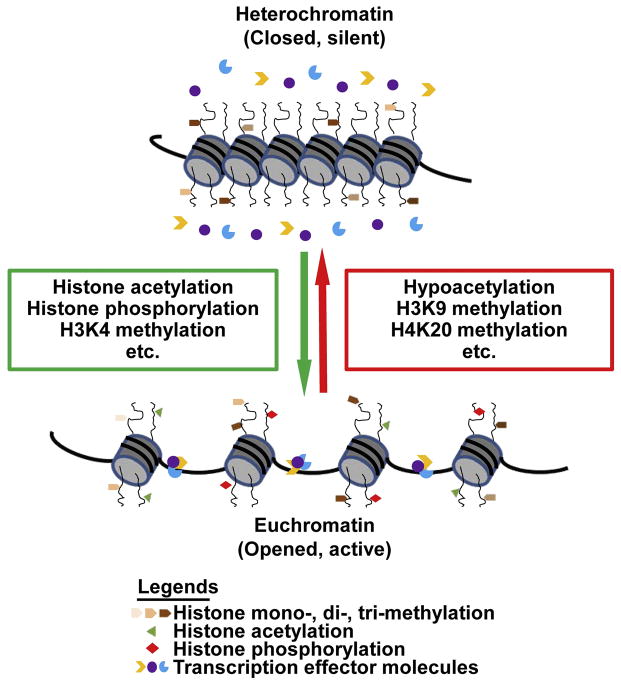
DNA is the ultimate template of heredity, which is in a tightly condensed form called chromatin. The role of distinct histone amino-terminal modifications in dictating dynamic transition between transcriptionally active or silent chromatin states has long been noticed. The notion of “histone code” has been proposed to explain the role of histone modifications in extending the information potential of the genetic code from DNA (Jenuwein and Allis, 2001). Histone modifications are enzymatic post-translational modifications which include methylation, acetylation, phosphorylation, sumoylation, ubiquitination, etc. (Bird, 2007, Vaquero et al., 2003). These modifications primarily occur within the amino-terminal tails of histone proteins which regulate gene expression by changing the chromatin structure (Cosgrove et al., 2004). Histone modifications are highly dynamic processes regulated by the opposing action of two families of enzymes, such as histone acetyltransferases (HATs) and histone deacetylases. Herein, we take well-studied histone methylation as an example. Histone methylation mainly occurs on lysine residues at positions 4, 9, 20, 27, 36 and 79 of histone 3. In contrast to other histone modification forms, lysine can be mono-, di- or tri-methylated. Histone lysine methyltransferases (HKMTs) and histone demethylase dynamically regulate the lysine methylation status. Lysine-specific demethylase 1 (LSD1) is the first identified lysine demethylase which has been found to target mono- and di-methylation of H3K4 (Shi et al., 2004), while lysine-specific demethylase 4A (KDM4A) demethylates tri-methylated lysines (Whetstine et al., 2006). The effects of histone modification on regulation of gene expression lie in two aspects: influencing the overall structure of chromatin and/or regulating the binding of transcription effector molecules (Bannister and Kouzarides, 2011). With the deposition of transcriptionally active histone codes such as H3K4 methylation, histone acetylation and phosphorylation, the structure of chromatin will be opened as euchromatin, which facilitates the binding of transcription effector molecules to promoter region and initiates gene expression. When the histone is modified by repressive codes, such as H3K9 and H4K20 methylation, chromatins will be condensed into heterochromatin, which closes the DNA binding sites for transcription effector molecules and turns off the gene expression (Figure 2).
Figure 2.
Histone modification and gene expression. While the histone is modified by repressive codes, such as H3K9 and H4K20 methylation, chromatins are condensed as heterochromatin, which close transcription effector molecules binding sites and turn off the gene transcription. Upon deposition of transcriptionally active histone codes, such as H3K4 methylation, histone acetylation and phosphorylation, the structure of chromatin will be changed into euchromatin, which opens the DNA binding sites for transcription effector molecules to initiate gene transcription.
2.3. Non-coding RNAs (ncRNAs)
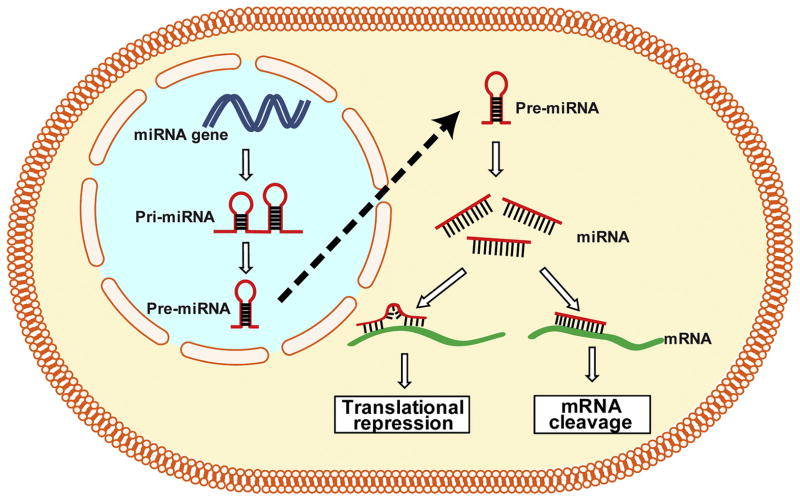
A gene is traditionally assumed to be transcribed into messager RNA (mRNA) and then translated into proteins; however, the finding of ncRNAs encoding genes extended the definition of a gene. The ncRNA genes produce transcripts functioning as structural, catalytic or regulatory RNAs rather than being translated into proteins. ncRNAs can be mainly divided into short ncRNAs (<30 nucleotides) and long ncRNAs (lncRNAs, >200 nucleotides). Short ncRNAs include microRNAs (miRNAs), short interfering RNAs (siRNAs) and piwi-interacting RNAs (piRNAs) (Mattick and Makunin, 2006). As for miRNAs, they are transcribed from miRNA genes as long primary transcripts (pri-miRNAs) characterized by a hairpin structure and are processed as pre-miRNAs (around 70-nucleotides long) in the nucleus. After being transported into the cytoplasm, pre-miRNAs are cleaved by Dicer and then matured into miRNA of 22-24 nucleotides. Generally, miRNAs modify protein expression mainly at the post-transcriptional level in cytoplasm by binding to a specific target mRNA with a complimentary sequence to induce cleavage, degradation or block translation (Bartel, 2004) (Figure 3). Recent progress in the study of ncRNAs has revealed the importance of ncRNAs in development and disease (Esteller, 2011, Stefani and Slack, 2008). The sequence-specific working manner of miRNAs makes miRNA an ideal target for the development of gene-specific therapeutic strategies for certain diseases, including OA (Shi et al., 2015).
Figure 3.
Biogenesis of miRNA and gene expression. miRNAs are transcribed from miRNA genes to long primary transcripts (pri-miRNAs) characterized by a hairpin structure, and are then processed to pre-miRNAs (around 70-nucleotide long) in the nucleus. After being transported into the cytoplasm, pre-miRNAs are cleaved by Dicer and finally matured into miRNA of 22-24 nucleotides. A partial binding of miRNAs to their complementary mRNAs leads to translational repression; a complete binding of miRNAs to their complementary mRNAs results in the cleavage of the targeted mRNAs.
2.4. Crosstalk among the epigenetic mechanisms
Although tremendous progress has been made to advance our understanding of epigenetic mechanisms in the past decade, the crosstalk among these epigenetic mechanisms in the regulation of gene expression adds another layer of complexity to epigenetics (Matzke and Mosher, 2014). For example, DNA methylation may dictate histone methylation (Okitsu and Hsieh, 2007) and DNA methylation of miRNA genes regulates miRNA expression (Vrba et al., 2010); miRNAs have also been shown to control the DNA methylation (Garzon et al., 2009). Moreover, miRNA-140 and miRNA-222 have been found to target histone deacetylases 4 in articular cartilage; miRNA-455 may target deacetylase Sirtuin 1, indicating miRNAs may control histone modifications (Songa et al., 2015, Swingler et al., 2015, Tuddenham et al., 2006). Furthermore, inactive unmethylated CpG island promoters have shown elevated levels of dimethylation of Lysine 4 of histone H3, suggesting that this chromatin mark may protect DNA from methylation (Weber, Hellmann, 2007). Histone deacetylase inhibitors, SAHA (vorinostat) and LBH589 (panobinostat), may elevate miRNA-146a expression and enhance negative regulation of interlukin 1-β (IL-1β) signaling in OA fibroblast-like synoviocytes (Wang et al., 2013).
3. Epigenetic mechanisms underlying the aberrant metabolic activities of OA chondrocytes
Adult articular cartilage is an avascular tissue in which chondrocytes are the unique cellular component for the maintenance of low-turnover of the extracellular matrix (ECM) via the delicate expression balance of catabolic and anabolic genes. Degradation of articular cartilage ECM is a major feature of OA. Increased expression of catabolic genes and decreased expression of anabolic genes are usually observed in OA chondrocytes, which disrupt the metabolic balance in articular cartilage. Epigenetic mechanisms may play an important role in the aberrant gene expression during the development of OA (Hollander et al., 1995, Huang and Wu, 2008). Early epigenetic studies using OA chondrocytes began with epigenetic regulation of catabolic genes, such as matrix-degrading proteinases. More recently, OA researchers have found that the expression of several transcription factor genes is also regulated by epigenetic mechanisms during the development of OA. This is a significant advance in epigenetic research because a transcription factor may regulate multiple target genes.
3.1. Epigenetic regulation of catabolic genes
3.1.1. Aggrecanases
ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs)-4 and -5 are two major aggrecanases which have been shown to play important role in development of OA (Glasson et al., 2005, Rogerson et al., 2008, Stanton et al., 2005, Tortorella et al., 2001). Increased ADAMTS-4 expression may be mediated by the loss of DNA methylation at specific CpG sites in the ADAMTS-4 promoter in OA chondrocytes (Cheung et al., 2009). In OA cartilage, specifically and highly expressed lncRNA-CIRs not only control the expression of collagen and aggrecan but also regulate the expression of matrix metalloproteinase 13 (MMP13) and ADAMTS-5 (Liu et al., 2014). Moreover, several miRNAs have also been found to regulate ADAMTS-5 expression in human OA cartilage (Ukai et al., 2012). In a study of cultured SW1353 chondrosarcoma cells and primary human chondrocytes, Young et al. demonstrated that histone deacetylase inhibitors decrease the level of collagenolytic enzymes in conditioned culture medium by down-regulating the expression of MMPs and ADAMTSs (Young et al., 2005).
3.1.2. Collagenases
MMP13, a major type II collagen (COL2A1)-degrading collagenase, not only contributes to the onset of OA, but also contributes to irreversible joint damage during the progression of OA (Little et al., 2009, Neuhold et al., 2001). In late-stage OA chondrocytes, loss of methylation at CpG sites in the promoter region has been found to be associated with increased expression of MMP-3, -9, -13, and ADAMTS-4 (Harper et al., 2015, Roach et al., 2005). The elevated histone deacetylase 7 expression in cartilage from OA patients was associated with up-regulated MMP13 gene expression (Higashiyama et al., 2010). Moreover, a recent study reported that miRAN-222 regulates MMP13 expression by targeting histone deacetylase 4 during the progression of OA (Songa, Jina, 2015). These studies suggest that all three epigenetic mechanisms (DNA methylation, histone modification, and ncRNA) play a role in regulation of expression of those matrix-degrading proteinases during the progression of OA.
3.1.3. Pro-inflammatory cytokines
It has been well-documented that cytokines play important roles in the development of rheumatoid arthritis (RA) which is a typical autoimmune disease in human joints (Feldmann et al., 1996). A new concept that OA is a joint disease of inflammation involving immune reaction has recently been proposed based on the findings of aberrant expression of pro-inflammatory cytokines in OA cartilage from humans and animals (Goldring and Otero, 2011). Although many cytokines have been implicated in OA, IL-1β, IL-6 and tumor necrosis factor-α (TNF-α) are the three main pro-inflammatory cytokines contributing to the degradation of articular cartilage (Goldring, 2000, Kapoor et al., 2011).
The epigenetic regulation of IL-1β expression in OA cartilage has been extensively studied. Specific CpG site at -299 bp of the IL-1β promoter has a significant impact on its promoter activity, and methylation of this site results in marked suppression of its transcriptional activity in human ACs (Hashimoto et al., 2013). Demethylation of this site increases the transcriptional response of IL-1β to other inflammatory cytokines in human ACs (Hashimoto et al., 2009). In addition, miRNA-149 is down-regulated in OA chondrocytes, and a functional study showed that this miRNA regulates the production of TNF-α, IL-1β and IL-6 (Santini et al., 2014). In addition to being regulated by epigenetic mechanisms, IL-1β also modulates epigenetic events in OA cartilage because stimulation of OA chondrocytes with IL-1β can affect miRNA production (Akhtar et al., 2010). Moreover, the overall methylation status of ACs in different histological zones of human cartilage was found to be different upon IL-1β stimulation (Akhtar and Haqqi, 2012).
The role of pro-inflammatory IL-6 in the development of OA has been extensively studied. IL-6 polymorphism has been found to be associated with the risk of hip OA (Pola et al., 2005). Moreover, the mRNA and protein levels of IL-6 were up-regulated in human and mouse OA cartilage (Ryu et al., 2011). Histone acetylation of the IL6 promoter induces an increase in IL-6 production in synovial fibroblasts of RA joints (Wada et al., 2014). Both DNA methylation and histone modification are involved in the control of TNF-α expression in cultured cell lines (Sullivan et al., 2007). However, the epigenetic status of IL-6 and TNF-α in osteoarthritic chondrocytes remains to be elucidated.
3.1.4. Runx2
Runt-related transcription factor 2 (Runx2), also known as core-binding factor subunit alpha-1 (Cbfa1), is a key transcription factor in osteogenesis (Kundu et al., 2002, Yoshida et al., 2002). Runx2 is required for chondrocyte maturation and osteoblast differentiation during skeletal development; global deletion of Runx2 results in a complete lack of bone formation in mice (Enomoto et al., 2000, Otto et al., 1997). In adult mice, Runx2 contributes to the pathogenesis of OA by promoting chondrocyte hypertrophy and matrix breakdown in articular cartilage. Runx2+/− mice exhibit decreased cartilage destruction and osteophyte formation, along with reduced type X collagen and MMP-13 expression, as compared with wild-type mice (Kamekura et al., 2006). Recently, differential methylation in RUNX2 has been identified in a genome-wide DNA methylation study in human OA cartilage (Jeffries et al., 2014). Decreased histone deacetylase 4 is associated with human cartilage degeneration by up-regulation of RUNX2 (Cao et al., 2014). Furthermore, two miRNAs, miRNA-602 and miRNA-608, have been shown to regulate sonic hedgehog (SHH) expression in human OA cartilage (Akhtar et al., 2015). Since the hedgehog signaling interacts with the Runx2 signaling pathway during the development of OA (Lin et al., 2009), these two miRNAs may play a significant role in the pathogenesis of OA..
3.2. Epigenetic regulation of anabolic genes
Collagen and proteoglycans are the major ECM protein components of articular cartilage. The maintenance of the normal amount and architecture of these components are required for articular cartilage to fulfill its mechanical properties (Kempson et al., 1973, Rizkalla et al., 1992). Therefore, aberrant expression of ECM genes (usually down-regulation) is one of the molecular characteristics of OA. Additionally, transcription factors possessing anabolic activities, such as SOX9, a member of the sex-determining region Y-type high mobility group box family of DNA binding proteins, play critical roles in the regulation of gene expression of ECM proteins in articular cartilage (Cucchiarini et al., 2007).
3.2.1. Aggrecan
Aggrecan is the major proteoglycan in articular cartilage, and loss of aggrecan is one of the characteristics of OA. In addition to the degradation by cartilage aggrecanases which play a critical role in the development of OA (Malfait et al., 2002, Song et al., 2007), decreased aggrecan expression is often evident in OA cartilage (Chambers et al., 2002, Eid et al., 2006). The expression of SIRT1, a member of HDACs, is decreased in human OA cartilage, and inhibition of SIRT1 significantly decreases the expression of aggrecan in both normal and OA human chondrocytes (Fujita et al., 2011). This finding indicates that histone acetylation may regulate the aggrecan expression in OA development. miRNA-146a has been found highly expressed in OA cartilage (Yamasaki et al., 2009). In a functional study, Li et al found that miRNA-146a functions in an anti-catabolic manner in articular cartilage by antagonizing the IL-1 induced expression of cartilage-degrading enzymes MMP13 and ADAMTS5, while simultaneously antagonizing IL-1 induced suppression of ECM proteins such as aggregan and collagen type II (Li et al., 2011). Nevertheless, a study on the correlation between gene methylation and expression of aggrecan (ACAN) in chondrocytes failed to find a significant correlation of ACAN mRNA expression levels and DNA methylation status in normal aged and osteoarthritic chondrocytes. This result suggests that DNA methylation does not play a central role in switching off ACAN promoter activity in human adult ACs (Poschl et al., 2005).
3.2.2. Collagens
Collagen type II is one of the major extracellular matrix components of articular cartilage. Transgenic mice bearing a small deletion mutation in type II collagen gene developed OA-like lesions (Saamanen et al., 2000). Histone acetyl-transferase CBP/P300 and the Class III NAD-dependent histone deacetylase Sirtuin 1 (SirT1) have been shown to co-regulate COL2A1 mRNA expression in cooperation with SOX9 (Dvir-Ginzberg et al., 2008, Tsuda et al., 2003). A recent study using human chondrocytes found that histone methyltransferase Set7/9 elevated trimethylated lysine 4 on histone 3 in the COL2A1 promoter, resulting in increased COL2A1 expression (Oppenheimer et al., 2014). Several miRNAs, such miRNA-146a, -140 and -675 have been found to regulate COL2A1 expression in articular chondrocytes (Dudek et al., 2010, Li, Gibson, 2011, Miyaki et al., 2010). Recently, Imagawa et al. found that there was no relationship between DNA methylation changes in the 309-bp COL2A1 enhancer and increased COL2A1 mRNA levels in OA chondrocytes which was a compensatory response to the absence of Type IX collagen (COL9A1) gene (Imagawa et al., 2014).
Although Type IX collagen is not as much as Type II collagen in articular cartilage, mice lacking Col9a1 develop normally but display OA-like cartilage degradation in the knee joints as they aged (Fassler et al., 1994). DNA methylation has also been found to control the decreased expression of COL9A1 mRNA in OA chondrocytes; CpG sites in the COL9A1 promoter were hypermethylated, and hypermethylated CpG sites attenuated SOX9 binding to the COL9A1 promoter, resulting in down-regulation of COL9A1 expression in OA cartilage (Imagawa, de Andres, 2014).
3.2.3. Sox9
SOX9 is a master transcription factor for chondrogenesis during the development of the skeletal system, in cooperation with SOX5 and SOX6 (Bi et al., 1999, Lefebvre et al., 1998). Although mice with conditional postnatal deletion of Sox9 in articular cartilage did not develop OA even by the age of 18 months (Henry et al., 2012), later OA usually is associated with decreased SOX9 expression (Lee and Im, 2011). Kim et al. recently reported that down-regulated SOX9 expression in advanced hip OA chondrocytes is mediated by DNA methylation and histone modification, including histone methylation and acetylation (Kim et al., 2013). In mouse ACs, we found Sox9 mRNA and protein were highly expressed in ACs during joint development but significantly decreased after 2 months of age. No histopathological features of osteoarthritis were observed in examined joints by 18 months. Epigenetic study revealed that the reduction of SOX9 expression in ACs of adult mice is primarily regulated by H3K4me2 (a histone modification for transcriptional activation) (Zhang et al., 2015a). miRNA-145 has been identified as an inhibitor of SOX9 expression in human cartilage and chondrosarcoma (Mak et al., 2015); increased miRNA-145 directly represses SOX9 expression, causing reduced expression of COL2A1 and aggrecan with an increased level of MMP13 (Martinez-Sanchez et al., 2012). In addition, miRAN-199a-3p and miRNA-193b have been found to down-regulate SOX9 expression (Ukai, Sato, 2012). While SOX9 is considered a typical anabolic factor in articular cartilage, the response of cultured chondrocytes to forced expression of SOX9 has been controversial. Kypriotou et al. found that overexpression of SOX9 itself was unable to restore the chondrocyte phenotype in dedifferentiated osteoarthritic chondrocytes (Kypriotou et al., 2003), whereas Cucchiarini et al. reported that r-AAV mediated SOX9 gene transfer up-regulated the expression levels of proteoglycans and type II collagen in normal and OA ACs (Cucchiarini, Thurn, 2007).
3.2.4. Nfat1
NFAT1 (Nfat1/NFATc2) is a member of the Nuclear Factor of Activated T-cells (NFAT) transcription factor family originally identified as a regulator of the expression of cytokine genes during the immune response (Hodge et al., 1996, Xanthoudakis et al., 1996). NFAT1 has recently been shown to play an important role in maintaining the permanent cartilage phenotype in adult mice. Nfat1 knockout (Nfat1−/−) mice exhibit normal skeletal development, but display over-expression of numerous matrix-degrading proteinases and pro-inflammatory cytokines and loss of collagen-2 and aggrecan during the initiation stage of OA. These initial changes are followed by articular chondrocyte clustering, formation of chondro-osteophytes, progressive articular surface destruction, formation of subchondral bone cysts, and exposure of thickened subchondral bone, all of which resemble human OA (Wang et al., 2009).
Our recent studies have demonstrated that NFAT1 regulates the chondrocyte function through its age-dependent expression in mouse articular cartilage. NFAT1 expression in wild-type articular chondrocytes was low in the embryonic, but high in the adult stage (2 to 6 months old). Our epigenetic studies (Rodova et al., 2011) revealed that an increase in NFAT1 expression in articular chondrocytes is associated with increased H3K4me2, while a decrease in NFAT1 expression in articular chondrocytes is correlated with increased H3K9me2 (a histone modification linked to transcriptional repression). Knockdown of Lsd1 in embryonic ACs up-regulates NFAT1 expression concomitant with increased H3K4me2 at the Nfat1 promoter. Knockdown of Jmjc-containing histone demethylase-2a (Jhdm2a) in 6-month ACs down-regulates NFAT1 expression concomitant with increased H3K9me2 at the Nfat1 promoter. These results suggest that the age-dependent NFAT1 expression in ACs is regulated by dynamic histone methylation (Rodova, Lu, 2011). Further study should be directed to investigate the expression of NFAT1 in aged articular cartilage and its underlying epigenetic mechanisms.
3.3. Dysregulation of gene expression in initiation and progression of OA
Even though the pathogenesis of OA is not fully understood, the development of OA can be generally divided into two stages: initiation and progression. Aberrant gene expression mediated by various epigenetic mechanisms has been found to be involved in the development of OA. Although it is not well defined in which stage a specific gene is involved, genetic and epigenetic studies suggest that OA could be initiated by overexpression of Mmp-13 (Neuhold, Killar, 2001), deletion of Col9a1 (Hu et al., 2006), or deletion of Nfat1 (Wang, Gardner, 2009); these three genes are known to be involved in OA initiation. Therefore, epigenetic mechanisms that regulate the expression of these genes in ACs may play an essential role in the initiation of OA. Mutation of COL2A1 has been linked to the development of human OA (Ala-Kokko et al., 1990, Knowlton et al., 1990, Ritvaniemi et al., 1995); however, the onset of OA seems secondary to the developmental defect and chondrodysplasia induced by COL2A1 mutation. Therefore, COL2A1 deficiency is not considered an initiator of OA. The progression of OA is usually a slow and irreversible process. A large number of OA-associated genes identified to date may be involved in the progression of OA, including some of the genes for the initiation of OA. The progression of OA from the early-stage to the end-stage OA involves multi-phase pathophysiological changes in multiple joint tissues. Previous studies have revealed that multiple epigenetic mechanisms are involved in the regulation of gene expression during the progression of OA. However, which epigenetic mechanism plays a key role in a phase-specific gene expression profile and in a specific joint tissue remains to be elucidated.
4. Future perspectives
The recent advances in epigenetic studies have greatly enhanced our understanding of the pathogenesis of multifactorial diseases, such as cancer and OA. With the use of new techniques, especially the application of new sequencing technology in the study of epigenetics (Schones and Zhao, 2008), the era of epigenetic study in the pathogenesis of OA is coming (Blanco and Rego-Perez, 2014). Genome wide analysis of epigenetic alterations in OA-related cells is being undertaken and more detailed epigenetic alterations in OA will be identified (Akhtar and Haqqi, 2012, Aref-Eshghi et al., 2015, den Hollander et al., 2015, Fu et al., 2015, Jeffries, Donica, 2014, Moazedi-Fuerst et al., 2014, Rushton et al., 2014, Shi, Wei, 2015, Zhang et al., 2015b). Clearly, further research is required to determine specific upstream regulators of methyltransferases and demethylases. It is also important to further elucidate the interactions among the different epigenetic mechanisms. The upcoming epigenetic findings may not only broaden our knowledge to appreciate the molecular mechanisms underlying the pathogenesis of OA (Kobayashi et al., 2015, Rushton et al., 2015), but also promote the development of new therapeutic strategies, such as epigenetic reprogramming of mesenchymal stem cells (Ezura et al., 2009) and miRNAs (Nakamachi et al., 2015), for the treatment of OA.
5. Conclusion
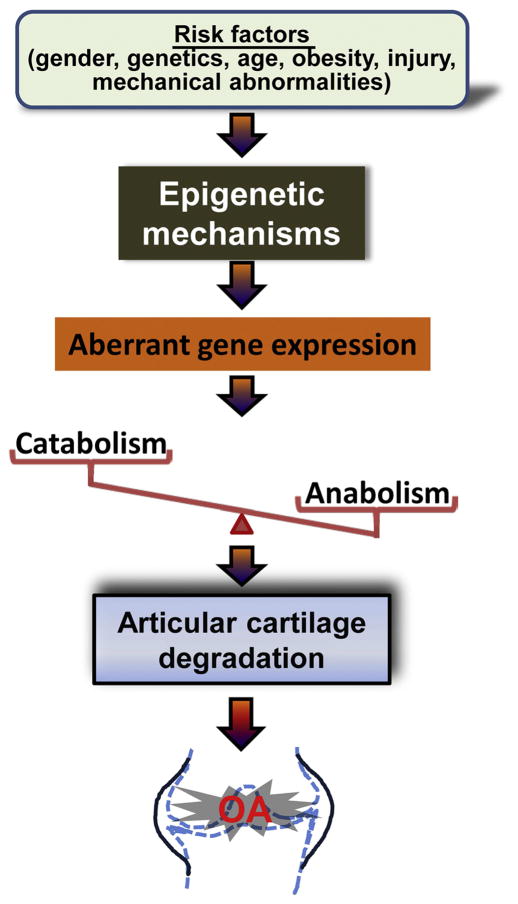
The recent advances in epigenetic studies have shed light on the importance of epigenetic regulation of gene expression in the pathogenesis of OA. Accumulative influence of risk factors which trigger epigenetic events such as DNA methylation, histone modifications and miRNAs in chondrocytes, may result in aberrant expression of specific genes for matrix-degrading proteinases, cytokines, transcription factors, collagens, and proteoglycans in articular cartilage. Abnormal expression of these genes may compromise the balance of catabolic and anabolic activity and disrupt cartilage homeostasis, leading to cartilage degradation, which is the key step of the development of OA (Figure 4). Further studies are required to determine specific risk factors triggering epigenetic events and interactions among the different epigenetic mechanisms during the initiation and progression of OA.
Figure 4.
Possible roles of epigenetic changes in the pathogenesis of OA. Under the accumulative effect of risk factors, chondrocytes undergo epigenetic events that result in aberrant expression of genes for specific transcription factors, cytokines, matrix-degrading proteinases, and ECM structural proteins in articular cartilage. Abnormal expression of these factors may disrupt the balance of catabolic and anabolic activities and compromise cartilage homeostasis, leading to articular cartilage degradation and the development of OA.
Acknowledgments
This work was supported by the U.S. National Institute of Health (NIH) Grant R01 AR059088 (to J. Wang), the U.S. Department of Defense Research Grant W81XWH-12-1-0304 (to J. Wang), and the Mary A. and Paul R. Harrington Distinguished Professorship Endowment (to J. Wang). The authors thank Mr. Theodore Budden, M.S. for his editorial assistance.
Abbreviations
- OA
osteoarthritis
- ACs
articular chondrocytes
- ncRNA
non-coding RNAs
- DNMTs
DNA methyltransferases
- TETs
ten-eleven translocation proteins
- CGIs
CpG islands
- HATs
histone acetyltransferases
- HKMTs
histone lysine methyltransferases
- LSD1
Lysine-specific demethylase 1
- KDM4A
while lysine-specific demethylase 4A
- mRNA
messager RNA
- miRNAs
microRNAs
- siRNAs
short interfering RNAs
- piRNAs
piwi-interacting RNAs
- IL-1β
interlukin 1-β
- ECM
extracellular matrix
- ADAMTS
a disintegrin and metalloproteinase with thrombospondin motifs
- MMP13
matrix metalloproteinase 13
- COL2A1
type II collagen
- COL9A1
type IX collagen
- RA
rheumatoid arthritis
- TNF-α
tumor necrosis factor-α
- Runx2
Runt-related transcription factor 2
- Cbfa1
core-binding factor subunit alpha-1
- SHH
sonic hedgehog
- ACAN
aggrecan
- NFAT
Nuclear Factor of Activated T-cells
- Jhdm2a
Jmjc-containing histone demethylase-2a
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- Akhtar N, Haqqi TM. Level of IL-1-Induced Epigenetic Modifications Differ in Chondrocytes From Different Histological Zones of Human Cartilage [abstract] Arthritis and rheumatism. 2012;64:29. [Google Scholar]
- Akhtar N, Makki MS, Haqqi TM. MicroRNA-602 and MicroRNA-608 Regulate Sonic Hedgehog Expression via Target Sites in the Coding Region in Human Chondrocytes. Arthritis Rheumatol. 2015;67:423–34. doi: 10.1002/art.38952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar N, Rasheed Z, Ramamurthy S, Anbazhagan AN, Voss FR, Haqqi TM. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis and rheumatism. 2010;62:1361–71. doi: 10.1002/art.27329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ala-Kokko L, Baldwin CT, Moskowitz RW, Prockop DJ. Single base mutation in the type II procollagen gene (COL2A1) as a cause of primary osteoarthritis associated with a mild chondrodysplasia. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:6565–8. doi: 10.1073/pnas.87.17.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aref-Eshghi E, Zhang Y, Liu M, Harper P, Martin G, Furey A, et al. Genome-wide DNA methylation study of hip and knee cartilage reveals embryonic organ and skeletal system morphogenesis as major pathways involved in osteoarthritis. Osteoarthritis and Cartilage. 2015;23 (Suppl):194. doi: 10.1186/s12891-015-0745-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–9. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–8. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Blagojevic M, Jinks C, Jeffery A, Jordan KP. Risk factors for onset of osteoarthritis of the knee in older adults: a systematic review and meta-analysis. Osteoarthritis and cartilage/OARS, Osteoarthritis Research Society. 2010;18:24–33. doi: 10.1016/j.joca.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Blanco FJ, Rego-Perez I. Editorial: Is it time for epigenetics in osteoarthritis? Arthritis Rheumatol. 2014;66:2324–7. doi: 10.1002/art.38710. [DOI] [PubMed] [Google Scholar]
- Cao K, Wei L, Zhang Z, Guo L, Zhang C, Li Y, et al. Decreased histone deacetylase 4 is associated with human osteoarthritis cartilage degeneration by releasing histone deacetylase 4 inhibition of runt-related transcription factor-2 and increasing osteoarthritis-related genes: a novel mechanism of human osteoarthritis cartilage degeneration. Arthritis Res Ther. 2014;16:491. doi: 10.1186/s13075-014-0491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter DR, Beaupre GS, Wong M, Smith RL, Andriacchi TP, Schurman DJ. The mechanobiology of articular cartilage development and degeneration. Clin Orthop Relat Res. 2004:S69–77. doi: 10.1097/01.blo.0000144970.05107.7e. [DOI] [PubMed] [Google Scholar]
- Chambers MG, Kuffner T, Cowan SK, Cheah KS, Mason RM. Expression of collagen and aggrecan genes in normal and osteoarthritic murine knee joints. Osteoarthritis and cartilage/OARS, Osteoarthritis Research Society. 2002;10:51–61. doi: 10.1053/joca.2001.0481. [DOI] [PubMed] [Google Scholar]
- Chen C, Tambe DT, Deng L, Yang L. Biomechanical properties and mechanobiology of the articular chondrocyte. American journal of physiology Cell physiology. 2013;305:C1202–8. doi: 10.1152/ajpcell.00242.2013. [DOI] [PubMed] [Google Scholar]
- Cheung KS, Hashimoto K, Yamada N, Roach HI. Expression of ADAMTS-4 by chondrocytes in the surface zone of human osteoarthritic cartilage is regulated by epigenetic DNA de-methylation. Rheumatol Int. 2009;29:525–34. doi: 10.1007/s00296-008-0744-z. [DOI] [PubMed] [Google Scholar]
- Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol. 2004;11:1037–43. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- Cucchiarini M, Thurn T, Weimer A, Kohn D, Terwilliger EF, Madry H. Restoration of the extracellular matrix in human osteoarthritic articular cartilage by overexpression of the transcription factor SOX9. Arthritis and rheumatism. 2007;56:158–67. doi: 10.1002/art.22299. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- den Hollander W, Ramos YF, Bomer N, Elzinga S, van der Breggen R, Lakenberg N, et al. Transcriptional associations of osteoarthritis mediated loss of epigenetic control in articular cartilage. Arthritis & Rheumatology. 2015 doi: 10.1002/art.39162. inpress. [DOI] [PubMed] [Google Scholar]
- Dudek KA, Lafont JE, Martinez-Sanchez A, Murphy CL. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. The Journal of biological chemistry. 2010;285:24381–7. doi: 10.1074/jbc.M110.111328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir-Ginzberg M, Gagarina V, Lee EJ, Hall DJ. Regulation of cartilage-specific gene expression in human chondrocytes by SirT1 and nicotinamide phosphoribosyltransferase. J Biol Chem. 2008;283:36300–10. doi: 10.1074/jbc.M803196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid K, Thornhill TS, Glowacki J. Chondrocyte gene expression in osteoarthritis: Correlation with disease severity. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2006;24:1062–8. doi: 10.1002/jor.20137. [DOI] [PubMed] [Google Scholar]
- Enomoto H, Enomoto-Iwamoto M, Iwamoto M, Nomura S, Himeno M, Kitamura Y, et al. Cbfa1 is a positive regulatory factor in chondrocyte maturation. J Biol Chem. 2000;275:8695–702. doi: 10.1074/jbc.275.12.8695. [DOI] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Ezura Y, Sekiya I, Koga H, Muneta T, Noda M. Methylation status of CpG islands in the promoter regions of signature genes during chondrogenesis of human synovium-derived mesenchymal stem cells. Arthritis and rheumatism. 2009;60:1416–26. doi: 10.1002/art.24472. [DOI] [PubMed] [Google Scholar]
- Fassler R, Schnegelsberg PN, Dausman J, Shinya T, Muragaki Y, McCarthy MT, et al. Mice lacking alpha 1 (IX) collagen develop noninflammatory degenerative joint disease. Proc Natl Acad Sci U S A. 1994;91:5070–4. doi: 10.1073/pnas.91.11.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann M, Brennan FM, Maini RN. Role of cytokines in rheumatoid arthritis. Annu Rev Immunol. 1996;14:397–440. doi: 10.1146/annurev.immunol.14.1.397. [DOI] [PubMed] [Google Scholar]
- Fu M, Huang G, Zhang Z, Liu J, Zhang Z, Huang Z, et al. Expression profile of long noncoding RNAs in cartilage from knee osteoarthritis patients. Osteoarthritis and cartilage/OARS, Osteoarthritis Research Society. 2015;23:423–32. doi: 10.1016/j.joca.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Fujita N, Matsushita T, Ishida K, Kubo S, Matsumoto T, Takayama K, et al. Potential involvement of SIRT1 in the pathogenesis of osteoarthritis through the modulation of chondrocyte gene expressions. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2011;29:511–5. doi: 10.1002/jor.21284. [DOI] [PubMed] [Google Scholar]
- Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–8. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–8. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- Goldring MB. Osteoarthritis and cartilage: the role of cytokines. Curr Rheumatol Rep. 2000;2:459–65. doi: 10.1007/s11926-000-0021-y. [DOI] [PubMed] [Google Scholar]
- Goldring MB, Otero M. Inflammation in osteoarthritis. Curr Opin Rheumatol. 2011;23:471–8. doi: 10.1097/BOR.0b013e328349c2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper P, Liu M, Aref-Eshghi E, Martin G, Furey A, Green R, et al. Gene expression analysis and DNA methylation in the promoter region of matrix metalloproteinase–13 (MMP-13) and osteoarthritis. Osteoarthritis and Cartilage. 2015;23 (Suppl):305. [Google Scholar]
- Hashimoto K, Oreffo RO, Gibson MB, Goldring MB, Roach HI. DNA demethylation at specific CpG sites in the IL1B promoter in response to inflammatory cytokines in human articular chondrocytes. Arthritis and rheumatism. 2009;60:3303–13. doi: 10.1002/art.24882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Otero M, Imagawa K, de Andres MC, Coico JM, Roach HI, et al. Regulated transcription of human matrix metalloproteinase 13 (MMP13) and interleukin-1beta (IL1B) genes in chondrocytes depends on methylation of specific proximal promoter CpG sites. J Biol Chem. 2013;288:10061–72. doi: 10.1074/jbc.M112.421156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry SP, Liang S, Akdemir KC, de Crombrugghe B. The postnatal role of Sox9 in cartilage. J Bone Miner Res. 2012;27:2511–25. doi: 10.1002/jbmr.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama R, Miyaki S, Yamashita S, Yoshitaka T, Lindman G, Ito Y, et al. Correlation between MMP-13 and HDAC7 expression in human knee osteoarthritis. Mod Rheumatol. 2010;20:11–7. doi: 10.1007/s10165-009-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge MR, Ranger AM, Charles de la Brousse F, Hoey T, Grusby MJ, Glimcher LH. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity. 1996;4:397–405. doi: 10.1016/s1074-7613(00)80253-8. [DOI] [PubMed] [Google Scholar]
- Hollander AP, Pidoux I, Reiner A, Rorabeck C, Bourne R, Poole AR. Damage to type II collagen in aging and osteoarthritis starts at the articular surface, originates around chondrocytes, and extends into the cartilage with progressive degeneration. J Clin Invest. 1995;96:2859–69. doi: 10.1172/JCI118357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Xu L, Cao L, Flahiff CM, Brussiau J, Ho K, et al. Pathogenesis of osteoarthritis-like changes in the joints of mice deficient in type IX collagen. Arthritis and rheumatism. 2006;54:2891–900. doi: 10.1002/art.22040. [DOI] [PubMed] [Google Scholar]
- Huang K, Wu LD. Aggrecanase and aggrecan degradation in osteoarthritis: a review. J Int Med Res. 2008;36:1149–60. doi: 10.1177/147323000803600601. [DOI] [PubMed] [Google Scholar]
- Imagawa K, de Andres MC, Hashimoto K, Itoi E, Otero M, Roach HI, et al. Association of Reduced Type IX Collagen Gene Expression in Human Osteoarthritic Chondrocytes With Epigenetic Silencing by DNA Hypermethylation. Arthritis Rheumatol. 2014;66:3040–51. doi: 10.1002/art.38774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries MA, Donica M, Baker LW, Stevenson ME, Annan AC, Humphrey MB, et al. Genome-wide DNA methylation study identifies significant epigenomic changes in osteoarthritic cartilage. Arthritis Rheumatol. 2014;66:2804–15. doi: 10.1002/art.38762. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jin B, Robertson KD. DNA methyltransferases, DNA damage repair, and cancer. Advances in experimental medicine and biology. 2013;754:3–29. doi: 10.1007/978-1-4419-9967-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kamekura S, Kawasaki Y, Hoshi K, Shimoaka T, Chikuda H, Maruyama Z, et al. Contribution of runt-related transcription factor 2 to the pathogenesis of osteoarthritis in mice after induction of knee joint instability. Arthritis and rheumatism. 2006;54:2462–70. doi: 10.1002/art.22041. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 2011;7:33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- Kempson GE, Muir H, Pollard C, Tuke M. The tensile properties of the cartilage of human femoral condyles related to the content of collagen and glycosaminoglycans. Biochim Biophys Acta. 1973;297:456–72. doi: 10.1016/0304-4165(73)90093-7. [DOI] [PubMed] [Google Scholar]
- Kim KI, Park YS, Im GI. Changes in the epigenetic status of the SOX-9 promoter in human osteoarthritic cartilage. J Bone Miner Res. 2013;28:1050–60. doi: 10.1002/jbmr.1843. [DOI] [PubMed] [Google Scholar]
- Knowlton RG, Katzenstein PL, Moskowitz RW, Weaver EJ, Malemud CJ, Pathria MN, et al. Genetic linkage of a polymorphism in the type II procollagen gene (COL2A1) to primary osteoarthritis associated with mild chondrodysplasia. The New England journal of medicine. 1990;322:526–30. doi: 10.1056/NEJM199002223220807. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Papaioannou G, Mirzamohammadi F, Kozhemyakina E, Zhang M, Blelloch R, et al. Early postnatal ablation of the microRNA-processing enzyme, Drosha, causes chondrocyte death and impairs the structural integrity of the articular cartilage. Osteoarthritis and Cartilage. 2015 doi: 10.1016/j.joca.2015.02.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Javed A, Jeon JP, Horner A, Shum L, Eckhaus M, et al. Cbfbeta interacts with Runx2 and has a critical role in bone development. Nat Genet. 2002;32:639–44. doi: 10.1038/ng1050. [DOI] [PubMed] [Google Scholar]
- Kypriotou M, Fossard-Demoor M, Chadjichristos C, Ghayor C, de Crombrugghe B, Pujol JP, et al. SOX9 exerts a bifunctional effect on type II collagen gene (COL2A1) expression in chondrocytes depending on the differentiation state. DNA Cell Biol. 2003;22:119–29. doi: 10.1089/104454903321515922. [DOI] [PubMed] [Google Scholar]
- Lee JS, Im GI. SOX trio decrease in the articular cartilage with the advancement of osteoarthritis. Connect Tissue Res. 2011;52:496–502. doi: 10.3109/03008207.2011.585409. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. Embo j. 1998;17:5718–33. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gibson G, Kim JS, Kroin J, Xu S, van Wijnen AJ, et al. MicroRNA-146a is linked to pain-related pathophysiology of osteoarthritis. Gene. 2011;480:34–41. doi: 10.1016/j.gene.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Seeto BL, Bartoszko JM, Khoury MA, Whetstone H, Ho L, et al. Modulating hedgehog signaling can attenuate the severity of osteoarthritis. Nat Med. 2009;15:1421–5. doi: 10.1038/nm.2055. [DOI] [PubMed] [Google Scholar]
- Little CB, Barai A, Burkhardt D, Smith SM, Fosang AJ, Werb Z, et al. Matrix metalloproteinase 13-deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis and rheumatism. 2009;60:3723–33. doi: 10.1002/art.25002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang X, Dai L, Hu X, Zhu J, Li L, et al. Long noncoding RNA related to cartilage injury promotes chondrocyte extracellular matrix degradation in osteoarthritis. Arthritis Rheumatol. 2014;66:969–78. doi: 10.1002/art.38309. [DOI] [PubMed] [Google Scholar]
- Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: a disease of the joint as an organ. Arthritis and rheumatism. 2012;64:1697–707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak IW, Singh S, Turcotte R, Ghert M. The epigenetic regulation of SOX9 by miR-145 in human chondrosarcoma. J Cell Biochem. 2015;116:37–44. doi: 10.1002/jcb.24940. [DOI] [PubMed] [Google Scholar]
- Malfait AM, Liu RQ, Ijiri K, Komiya S, Tortorella MD. Inhibition of ADAM-TS4 and ADAM-TS5 prevents aggrecan degradation in osteoarthritic cartilage. The Journal of biological chemistry. 2002;277:22201–8. doi: 10.1074/jbc.M200431200. [DOI] [PubMed] [Google Scholar]
- Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145) J Biol Chem. 2012;287:916–24. doi: 10.1074/jbc.M111.302430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes & development. 2010;24:1173–85. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazedi-Fuerst FC, Hofner M, Gruber G, Weinhaeusel A, Stradner MH, Angerer H, et al. Epigenetic differences in human cartilage between mild and severe OA. J Orthop Res. 2014;32:1636–45. doi: 10.1002/jor.22722. [DOI] [PubMed] [Google Scholar]
- Murphy L, Helmick CG. The impact of osteoarthritis in the United States: a population-health perspective. Am J Nurs. 2012;112:S13–9. doi: 10.1097/01.NAJ.0000412646.80054.21. [DOI] [PubMed] [Google Scholar]
- Nakamachi Y, Ohnuma K, Uto K, Noguchi Y, Saegusa J, Kawano S. MicroRNA-124 inhibits the progression of adjuvant-induced arthritis in rats. Ann Rheum Dis. 2015 doi: 10.1136/annrheumdis-2014-206417. in press. [DOI] [PubMed] [Google Scholar]
- Neuhold LA, Killar L, Zhao W, Sung ML, Warner L, Kulik J, et al. Postnatal expression in hyaline cartilage of constitutively active human collagenase-3 (MMP-13) induces osteoarthritis in mice. J Clin Invest. 2001;107:35–44. doi: 10.1172/JCI10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okitsu CY, Hsieh CL. DNA methylation dictates histone H3K4 methylation. Mol Cell Biol. 2007;27:2746–57. doi: 10.1128/MCB.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer H, Kumar A, Meir H, Schwartz I, Zini A, Haze A, et al. Set7/9 impacts COL2A1 expression through binding and repression of SirT1 histone deacetylation. J Bone Miner Res. 2014;29:348–60. doi: 10.1002/jbmr.2052. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Pola E, Papaleo P, Pola R, Gaetani E, Tamburelli FC, Aulisa L, et al. Interleukin-6 gene polymorphism and risk of osteoarthritis of the hip: a case-control study. Osteoarthritis and cartilage/OARS, Osteoarthritis Research Society. 2005;13:1025–8. doi: 10.1016/j.joca.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Poschl E, Fidler A, Schmidt B, Kallipolitou A, Schmid E, Aigner T. DNA methylation is not likely to be responsible for aggrecan down regulation in aged or osteoarthritic cartilage. Ann Rheum Dis. 2005;64:477–80. doi: 10.1136/ard.2004.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritvaniemi P, Korkko J, Bonaventure J, Vikkula M, Hyland J, Paassilta P, et al. Identification of COL2A1 gene mutations in patients with chondrodysplasias and familial osteoarthritis. Arthritis and rheumatism. 1995;38:999–1004. doi: 10.1002/art.1780380717. [DOI] [PubMed] [Google Scholar]
- Rizkalla G, Reiner A, Bogoch E, Poole AR. Studies of the articular cartilage proteoglycan aggrecan in health and osteoarthritis. Evidence for molecular heterogeneity and extensive molecular changes in disease. J Clin Invest. 1992;90:2268–77. doi: 10.1172/JCI116113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach HI, Yamada N, Cheung KS, Tilley S, Clarke NM, Oreffo RO, et al. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis and rheumatism. 2005;52:3110–24. doi: 10.1002/art.21300. [DOI] [PubMed] [Google Scholar]
- Rodova M, Lu Q, Li Y, Woodbury BG, Crist JD, Gardner BM, et al. Nfat1 regulates adult articular chondrocyte function through its age-dependent expression mediated by epigenetic histone methylation. J Bone Miner Res. 2011;26:1974–86. doi: 10.1002/jbmr.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogerson FM, Stanton H, East CJ, Golub SB, Tutolo L, Farmer PJ, et al. Evidence of a novel aggrecan-degrading activity in cartilage: Studies of mice deficient in both ADAMTS-4 and ADAMTS-5. Arthritis and rheumatism. 2008;58:1664–73. doi: 10.1002/art.23458. [DOI] [PubMed] [Google Scholar]
- Rushton M, Reynard L, Young D, Shepherd C, Darlay R, Cordell H, et al. Methylation of cartilage DNA is a mediator of genetic risk at several OA susceptibility loci. Osteoarthritis and Cartilage. 2015;23 (Suppl):71–2. [Google Scholar]
- Rushton MD, Reynard LN, Barter MJ, Refaie R, Rankin KS, Young DA, et al. Characterization of the cartilage DNA methylome in knee and hip osteoarthritis. Arthritis Rheumatol. 2014;66:2450–60. doi: 10.1002/art.38713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, Yang S, Shin Y, Rhee J, Chun CH, Chun JS. Interleukin-6 plays an essential role in hypoxia-inducible factor 2alpha-induced experimental osteoarthritic cartilage destruction in mice. Arthritis and rheumatism. 2011;63:2732–43. doi: 10.1002/art.30451. [DOI] [PubMed] [Google Scholar]
- Saamanen AK, Salminen HJ, Dean PB, De Crombrugghe B, Vuorio EI, Metsaranta MP. Osteoarthritis-like lesions in transgenic mice harboring a small deletion mutation in type II collagen gene. Osteoarthritis and cartilage/OARS, Osteoarthritis Research Society. 2000;8:248–57. doi: 10.1053/joca.2000.0298. [DOI] [PubMed] [Google Scholar]
- Saetrom P, Snove O, Jr, Rossi JJ. Epigenetics and microRNAs. Pediatr Res. 2007;61:17r–23r. doi: 10.1203/pdr.0b013e318045760e. [DOI] [PubMed] [Google Scholar]
- Santini P, Politi L, Vedova PD, Scandurra R, Scotto d’Abusco A. The inflammatory circuitry of miR-149 as a pathological mechanism in osteoarthritis. Rheumatol Int. 2014;34:711–6. doi: 10.1007/s00296-013-2754-8. [DOI] [PubMed] [Google Scholar]
- Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–91. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wei Y, Xia J, Wang S, Wu J, Chen F, et al. MicroRNAs are potential prognostic and therapeutic targets in diabetic osteoarthritis. J Bone Miner Metab. 2015;33:1–8. doi: 10.1007/s00774-014-0628-0. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Song RH, Tortorella MD, Malfait AM, Alston JT, Yang Z, Arner EC, et al. Aggrecan degradation in human articular cartilage explants is mediated by both ADAMTS-4 and ADAMTS-5. Arthritis and rheumatism. 2007;56:575–85. doi: 10.1002/art.22334. [DOI] [PubMed] [Google Scholar]
- Songa J, Jina EH, Kima D, Kimb KY, Chunc CH, Jin EJ. MicroRNA-222 regulates MMP-13 via targeting HDAC-4 during osteoarthritis pathogenesis. BBA clinical. 2015;3:79–89. doi: 10.1016/j.bbacli.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton H, Rogerson FM, East CJ, Golub SB, Lawlor KE, Meeker CT, et al. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434:648–52. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–30. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Sullivan KE, Reddy AB, Dietzmann K, Suriano AR, Kocieda VP, Stewart M, et al. Epigenetic regulation of tumor necrosis factor alpha. Mol Cell Biol. 2007;27:5147–60. doi: 10.1128/MCB.02429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingler T, Wormstone Y, Lott M, Barter M, Young D, Clark I. MicroRNA-455 targets Sirt1. Osteoarthritis and Cartilage. 2015;23 (Suppl):275–6. [Google Scholar]
- Tortorella MD, Malfait AM, Deccico C, Arner E. The role of ADAM-TS4 (aggrecanase-1) and ADAM-TS5 (aggrecanase-2) in a model of cartilage degradation. Osteoarthritis and cartilage/OARS, Osteoarthritis Research Society. 2001;9:539–52. doi: 10.1053/joca.2001.0427. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Takahashi S, Takahashi Y, Asahara H. Transcriptional co-activators CREB-binding protein and p300 regulate chondrocyte-specific gene expression via association with Sox9. J Biol Chem. 2003;278:27224–9. doi: 10.1074/jbc.M303471200. [DOI] [PubMed] [Google Scholar]
- Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–7. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- Ukai T, Sato M, Akutsu H, Umezawa A, Mochida J. MicroRNA-199a-3p, microRNA-193b, and microRNA-320c are correlated to aging and regulate human cartilage metabolism. J Orthop Res. 2012;30:1915–22. doi: 10.1002/jor.22157. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Loyola A, Reinberg D. The constantly changing face of chromatin. Sci Aging Knowledge Environ. 2003;2003:Re4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada TT, Araki Y, Sato K, Aizaki Y, Yokota K, Kim YT, et al. Aberrant histone acetylation contributes to elevated interleukin-6 production in rheumatoid arthritis synovial fibroblasts. Biochem Biophys Res Commun. 2014;444:682–6. doi: 10.1016/j.bbrc.2014.01.195. [DOI] [PubMed] [Google Scholar]
- Wang J, Gardner BM, Lu Q, Rodova M, Woodbury BG, Yost JG, et al. Transcription factor Nfat1 deficiency causes osteoarthritis through dysfunction of adult articular chondrocytes. J Pathol. 2009;219:163–72. doi: 10.1002/path.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Shih KS, Wu YW, Wang AW, Yang CR. Histone deacetylase inhibitors increase microRNA-146a expression and enhance negative regulation of interleukin-1beta signaling in osteoarthritis fibroblast-like synoviocytes. Osteoarthritis and cartilage/OARS, Osteoarthritis Research Society. 2013;21:1987–96. doi: 10.1016/j.joca.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–81. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Viola JP, Shaw KT, Luo C, Wallace JD, Bozza PT, et al. An enhanced immune response in mice lacking the transcription factor NFAT1. Science. 1996;272:892–5. doi: 10.1126/science.272.5263.892. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Nakasa T, Miyaki S, Ishikawa M, Deie M, Adachi N, et al. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis and rheumatism. 2009;60:1035–41. doi: 10.1002/art.24404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, et al. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat Genet. 2002;32:633–8. doi: 10.1038/ng1015. [DOI] [PubMed] [Google Scholar]
- Young DA, Lakey RL, Pennington CJ, Jones D, Kevorkian L, Edwards DR, et al. Histone deacetylase inhibitors modulate metalloproteinase gene expression in chondrocytes and block cartilage resorption. Arthritis Res Ther. 2005;7:R503–12. doi: 10.1186/ar1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Lu Q, Miller A, Barnthouse N, Wang J. Developmental switch in SOX9 expression in articular cartilage is regulated by epigenetic histone methylation. Osteoarthritis and Cartilage. 2015a;23 (Suppl):195–6. [Google Scholar]
- Zhang Y, Fukui N, Yahata M, Lee M. Genome-wide DNA methylation profiling of osteoarthritic articular cartilage. Osteoarthritis and Cartilage. 2015b;23 (Suppl):194–5. doi: 10.1016/j.joca.2015.12.013. [DOI] [PubMed] [Google Scholar]