Abstract
The cyclic AMP response element binding protein (CREB) is a signal-dependent transcription factor that exerts its positive effects on gene transcription of a broad range of genes by recruiting coactivators including CREB-binding protein (CBP), its paralog p300, and the family of CRTC (CREB-regulated transcriptional coactivators) proteins. Whereas recruitment of CBP/p300 is dependent on CREB phosphorylation at Ser133, recruitment of CRTCs is not. Here we describe how both mechanisms could concurrently drive transcription of CREB targets in a subset of head and neck cancers featuring chromosomal translocations that fuse portions of CRTC1/3 genes with that of the Mastermind like transcriptional coactivator MAML2. We show that a peptide derived from the transactivation domain 1 (TAD1) of MAML2 binds to the CBP KIX domain with micromolar affinity. A ~20-residue segment within this peptide, conserved in MAML2 orthologs and paralogs, binds directly to a KIX surface previously shown to bind to MLL1. The 20-residue MAML2 segment shares sequence similarity with MLL1, especially at those positions in direct contact with KIX, and like MLL1, the segment is characterized by the presence of a ~10-residue helix. Since CRTC1/3-MAML2 fusion proteins are constitutively nuclear, like CREB, our results suggest constitutive recruitment of CBP/p300 to CREB targets that could be further enhanced by signals that cause CREB Ser133 phosphorylation.
Introduction
CREB is the prototypical signal-dependent, sequence-specific DNA-binding transcriptional activator that regulates expression of a broad range of genes in response to increases in intracellular cAMP and Ca2+ levels caused by extracellular signals such as hormones and nutrients.(1, 2) CREB exerts its positive effects on gene transcription by recruiting two families of transcriptional coactivators with intrinsic or associated lysine acetyltransferase (KAT) activities via distinct molecular mechanisms. Recruitment of the CBP/p300 family of coactivators relies on cAMP-activated, protein kinase A-mediated phosphorylation of a specific serine residue (Ser133) located in the kinase inducible transactivation domain (KID) of CREB.(3) Ser133 phosphorylation potentiates CBP/p300 recruitment via direct interactions of the KID with the CBP/p300 KIX domain.(4, 5)
Recruitment of the CRTC family and their associated KAT, PCAF/KAT2B, occurs through a mechanism independent of CREB phosphorylation.(6–8) Members of the CRTC family comprising CRTC1, CRTC2, and CRTC3 are retained in an inactive, hyper-phosphorylated form in complex with 14-3-3 proteins in the cytoplasm under basal conditions.(9) Elevations in the intracellular levels of cAMP and Ca2+ trigger the de-phosphorylation and release of the CRTC proteins from 14-3-3 complexes. Following nuclear entry, the CRTC proteins associate via a conserved N-terminal helical segment with the DNA-bound basic leucine zipper (bZip) domain of CREB.(10)
Besides their well-established roles in promoting long-term memory and glucose homeostasis,(11–13) CRTC1 and CRTC3 have been implicated in a subset of mucoepidermoid carcinomas (head and neck cancers).(14, 15) These cancers are characterized by recurrent chromosomal translocations that fuse the segment encoding the N-terminal CREB-binding domain (CBD) of CRTC1/3 with substantial portions of the coding regions of another coactivator called MAML2.
MAML2 is homologous to the Mastermind protein in Drosophila and along with MAML1 and MAML3 comprise the Mastermind-like family of transcriptional coactivators that play an important role in Notch signaling, regulating multiple developmental pathways.(16) Upon Notch activation, which results in the cleavage of the Notch receptor followed by translocation of the intracellular domain to the nucleus, these coactivators bind via a conserved N-terminal basic domain to Notch as well as to the CSL family of DNA-binding factors, forming a ternary complex.(17) The MAML proteins harbor two acidic transactivation domains C-terminal to the basic domain, both of which are essential for the activation of Notch-regulated genes. Although the molecular target of the C-terminal TAD (TAD2) is presently unknown, TAD1 harbors a binding site for CBP/p300.
The CRTC1-MAML2 fusion lacks the N-terminal 171 residues including the basic domain of MAML2 important for binding to Notch and CSL and all but the N-terminal 42 residues of CRTC1 corresponding to the CBD.(14) The protein thus lacks the nuclear export signals and regulatory domains of CRTC1 that are required for CRTC sequestration in the cytoplasm. Like the MAML proteins, the resulting fusion protein is constitutively nuclear.(18) Previous studies have shown that CRTC1-MAML2 can function as a potent coactivator of CREB and that the transforming activity of the oncoprotein is due in large part to aberrant activation of CREB targets.(6, 18) Additionally, one of the studies mapped a 175-residue segment within MAML2 TAD1 for efficient interactions with CBP/p300.(18) Deletion of this segment corresponding to residues 44–222 of CRTC1-MAML2 resulted in significantly reduced induction of CREB target genes and transforming ability compared to the full-length protein, implying a central role for CBP/p300 in both processes.
Experimental Procedures
Production of CBP KIX and CRTC1-MAML2 TAD polypeptides
Mouse CBP KIX (residues 586–683) and CRTC1-MAML2 TAD1 (spanning residues 150–190 and 172–207; residue numbering follows the fusion protein and not native MAML2) constructs were produced by sub-cloning the respective segments in pMCSG7 and pMCSG23 vectors as His6-tagged or His6-MBP-tagged constructs. 15N/13C-labeled and unlabeled proteins were expressed in E. coli and purified using Ni2+-affinity chromatography followed by removal of the tag by TEV protease and reversed-phase HPLC as described previously.(19) The identity and purity of as well as the extent of isotope incorporation in the preparations were confirmed by mass spectrometry and SDS-PAGE.
NMR sample preparation and spectroscopy
NMR samples in the 0.7–0.75 mM range were prepared in 50 mM sodium phosphate buffer (pH 6.0) containing 50 mM NaCl, 0.2% NaN3 and 10% D2O. Spectra were recorded at 35 °C on an Agilent DirectDrive 600 MHz spectrometer equipped with a pulsed-field gradient triple-resonance cold probe. NMR data processing and analysis were performed using Felix 98.0 (Felix NMR) and Sparky,(20) respectively. Backbone 1H, 15N, and 13C resonance assignments for KIX and TAD150–190 were made by analyzing 2D 1H-15N HSQC, 3D CBCA(CO)NH, HNCACAB, and HNCO spectra; assignments for apo KIX were from ref. (21). To gain insights into the architecture of the complex, 3D C(CO)NH-TOCSY, 3D 15N-edited NOESY, and 3D 13C-filtered, 13C-edited NOESY were analyzed. Chemical shift perturbations (CSPs) induced by various ligands were calculated using the formula: Δδav = √(0.5*((δHN,bound – δ HN,free)2 + 0.04*(δN,bound – δN,free)2)). NMR titrations for measuring dissociation constants were performed using a 0.15 mM CBP KIX sample at 25 °C. The CSPs induced as a function of added TAD150–190 peptide for each peak were fitted independently using non-linear regression assuming a single binding site using Grace software employing the fitting function y = A1*(((x + 1 + A0) - sqrt ((x + 1 + A0)^2 – 4*x))/2), where y is the observed CSP at molar ratio x and A1 and A0 are the fitted parameters related to the dissociation constant and the CSP at x=∞, respectively. Naphthol AS-E phosphate was purchased from Sequoia Research Products and used in NMR titrations without further purification. Two equivalents of the compound from a 5 mM stock prepared in D2O were added to the NMR samples in the concentration range of 0.7–0.75 mM and CSPs were computed as described above.
Isothermal titration calorimetry
ITC experiments were carried out on a MicroCal iTC200 calorimeter. Titrations were performed in triplicate at 20 °C in 20 mM sodium phosphate buffer (pH 7.2) and 150 mM NaCl. Proteins were dialyzed overnight against the buffer used for the titrations. CBP KIX was in the cell while the CRTC1-MAML2 TAD150–190 was in the syringe at initial concentrations of 0.1 mM and 2.3 mM, respectively. The titration curves were fitted using a sequential two-site binding model in the Origin 7.0 software.
Co-Immunoprecipitation assays
The experiments were conducted exactly as previously described following overexpression of full-length HA-tagged CBP with full-length Flag-tagged CRTC1-MAML2 or the CRTC1-MAML2 L180P mutant in HEK293T cells (22). The plasmids encoding the wild-type proteins were generous gifts of Drs. Marc Montminy and Michael Conkright, respectively. The L180P mutation was introduced using the QuikChange II XL kit (Agilent) and the presence of the mutation was confirmed via DNA sequencing.
Results and Discussion
CRTC1-MAML2 TAD1 targets the KIX domain of CBP/p300
Although previous studies identified a segment within CRTC1-MAML2 TAD1 in CBP/p300 recruitment,(18) the region of CBP/p300 involved in TAD1-binding was not known. Sequence analysis of CRTC1-MAML2 TAD1 suggested some similarity between the segment spanning residues 150–185 to the two helical regions that comprise the minimal KIX-binding domain of CREB pKID (Supplementary Figure S1A). Two CRTC1-MAML2 TAD constructs, with one construct spanning residues 150–190 and the other spanning residues 172–207 were generated and tested for binding to CBP KIX. Both peptides produced similar changes in the NMR spectrum of KIX (Supplementary Figure S1B), implying that each peptide harbored the essential determinants for binding to KIX. Since the magnitude of the perturbations was slightly greater for TAD150–190 than for TAD172–207 and since the former peptide could be readily produced on a large scale, all subsequent analyses were conducted with TAD150–190 (unless explicitly indicated otherwise, we shall refer to TAD150–190 below as simply TAD).
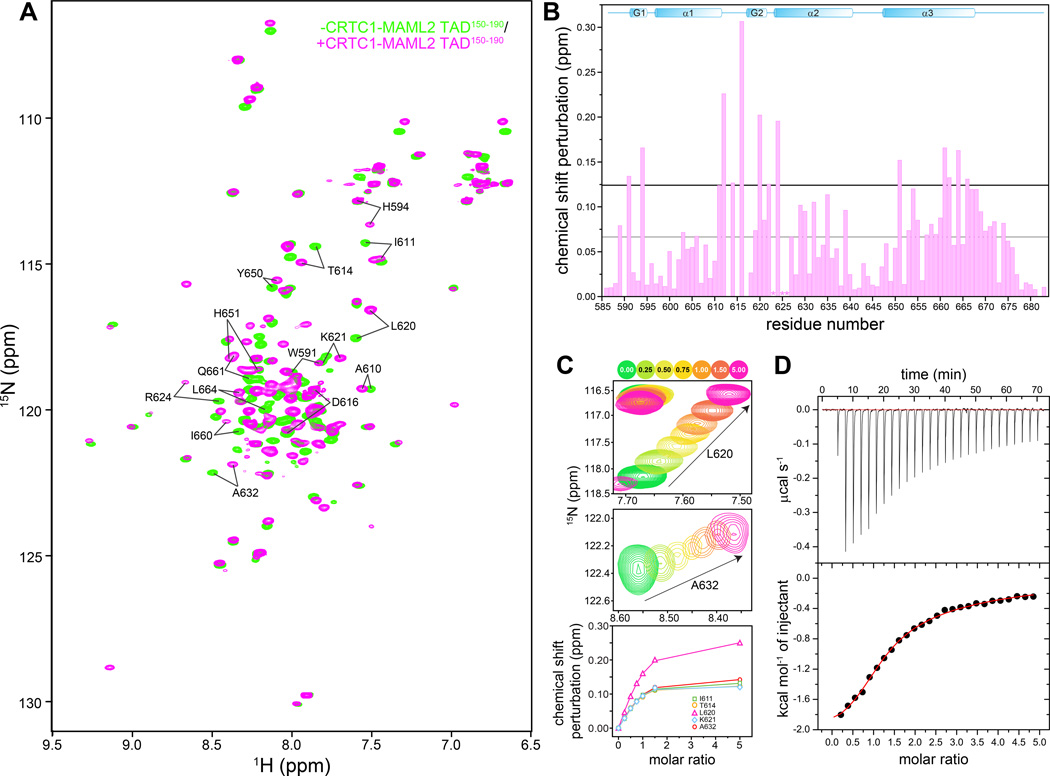
To gain further insights into the interaction, backbone resonances for the TAD-loaded form of KIX were assigned, which allowed for the quantification of backbone chemical shift perturbations (CSPs) and revealed the identities of the associated resonances (Figure 1A & 1B). These perturbations were non-uniform and significant (>>0.025 ppm), indicative of a specific association. Titrations of KIX with TAD conducted over a wide range of molar ratios caused ‘shifting’ of resonances to new positions as a function of the amount of added ligand, characteristic of a protein-peptide complex with fast dissociation kinetics (Figure 1C). Non-linear least-squares fitting of the extent of CSP observed as a function of added ligand for five well-resolved, significantly perturbed resonances including I611, T614, L620, K621, and A632 yielded an average equilibrium dissociation constant (Kd) for the interaction of 21 ± 13 µM (Figure 1C & Supplementary Figure S3). The KIX-binding affinity for the TAD peptide is comparable to those measured for c-Myb (15 µM),(23) although they are weaker than phosphorylated KID (pKID; 0.7 µM) of CREB and MLL1 (2–3 µM).(24, 25)
Figure 1. A MAML2 transactivation domain interacts with the KIX domain of CBP.
(A) 1H-15N correlated spectra of mouse CBP KIX recorded in the absence (green) and presence (orange) of CRTC1-MAML2 TAD150–190. (B) Backbone amide chemical shift perturbations (CSP) induced upon addition of one equivalent of CRTC1-MAML2 TAD150–190 polypeptide to a 0.75 mM NMR sample of KIX. The grey and black horizontal lines denote the average CSP and the average+1 standard deviation CSP induced in KIX, respectively. The asterisks denote residues with severely broadened resonances in the presence of the TAD. (C) Expanded plots of 1H-15N correlated spectra depicting changes to the positions of the L620 and A632 backbone resonances (top and middle panels) as a function of added TAD. Titrations were performed with 0.15 mM CBP KIX at 25 °C. Peaks are colored according to the number of added equivalents of TAD150–190 (see key on top). The raw data (symbols; bottom panel) following quantification of the chemical shift changes are shown along with the fitted data (solid lines). (D) A representative binding isotherm from an ITC experiment showing association between the KIX and TAD polypeptides. KIX was in the cell while the TAD150–190 was in the syringe.
CRTC1-MAML2 TAD1 binds KIX via a helical motif within a conserved region
To gain complementary insights into KIX binding by CRTC1-MAML2 TAD, we recorded NMR spectra of the TAD in the absence and presence of KIX. In the absence of KIX, the amide proton resonances of the TAD are narrow, poorly dispersed, and of uniform intensity, characteristic of a natively unfolded peptide (Figure 2A). The addition of one equivalent of KIX leads to significant perturbations in the position of a subset of resonances accompanied by a broadening of resonance linewidths, characteristic of a specific association with a much larger molecular entity (Figure 2A and Supplementary Figure S2). The increase in amide proton chemical shift dispersion implies folding of the peptide upon binding to KIX.
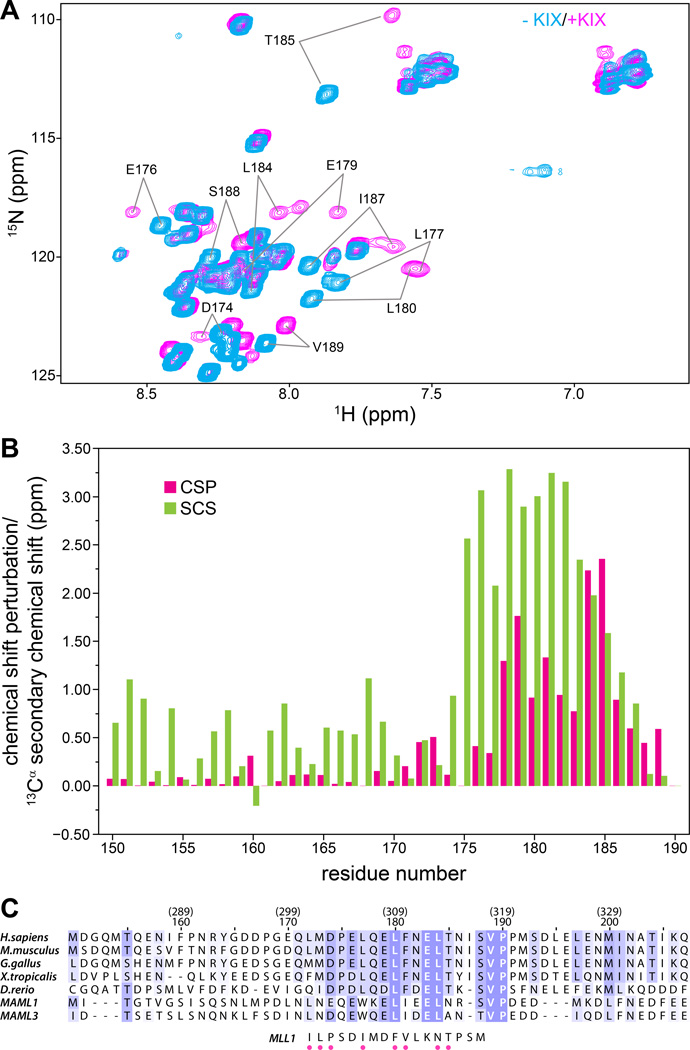
Figure 2. The CRTC1-MAML2 TAD1 binds KIX via a short conserved, helical segment.
(A) 1H-15N correlated spectra of 15N,13C-labeled CRCT1-MAML2 TAD150–190 recorded in the absence (cyan) and presence (magenta) of one equivalent of CBP KIX at 25 °C. (B) Backbone amide CSPs (green) for CRTC1-MAML2 TAD150–190 induced by the addition of one equivalent of KIX plotted as a function of residue number; the TAD concentration was 0.7 mM. CSPs were calculated as described in Figure 1B. Also graphed are 13Cα secondary chemical shifts (SCS; red) for TAD150–190 when bound to KIX as a function of residue number. (C) CLUSTAL Ω-guided multiple sequence alignment of MAML2 homologues from various species and comparison with the sequence of the MLL1 segment that engages KIX. The residue numbering in the context of human MAML2 is given in parenthesis on top of the numbering in the context of the CRTC1-MAML2 fusion. Sequences were shaded based on substitution scores from the BLOSUM62 scoring matrix in JalView.(31) MLL1 residues that make direct contacts with KIX in the NMR structure(25) are identified by dots below the sequence.
To quantify the perturbations and to map the KIX-binding interface, the backbone resonances of the TAD were assigned. Large-scale CSPs in the range of 0.5 ppm or well above this level were observed for several residues with the most significant perturbations mapping to the segment spanning residues 172–189 at the C-terminus of the peptide (Figure 2B), implicating this region in direct interactions with KIX. These results are consistent with our findings that the TAD172–207 construct could induce similar perturbations in KIX (Supplementary Figure S1B). To gain insight into the backbone conformation of the segment in direct contact with KIX, the 13Cα secondary chemical shifts were quantified (Figure 2B). Many (but not all) residues at the KIX interface exhibited secondary chemical shifts (SCSs) in the range of 2 ppm or well above this level, suggesting the presence of a helical segment extending from P175 to T185.
To test whether the KIX-binding segment identified from our analyses was conserved in MAML2 homologues, a multiple sequence alignment of MAML2 orthologs from a variety of species and the paralogous MAML1 and MAML3 proteins was constructed. This revealed a conserved region spanning residues 172–190 in CRTC1-MAML2 (Figure 2C). Residues at five positions within this 19-residue segment were invariant while five other positions were characterized by conservative substitutions. This implies that this motif is vital for the function of MAML proteins given that its conservation extends to over ~450 million years of evolution.
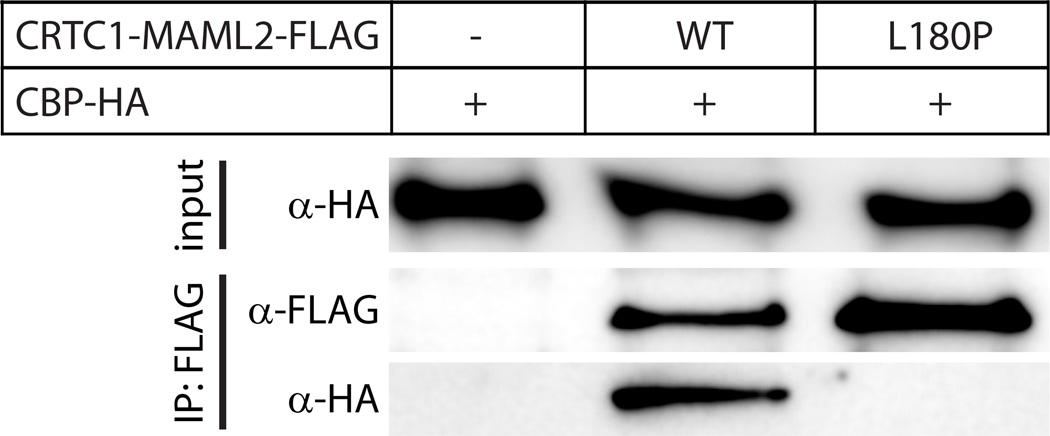
To evaluate the role of the conserved segment and, in particular, the involvement of a helix in promoting the interaction, we conducted immunoprecipitation experiments with full-length, Flag-tagged CRTC1-MAML2 and HA-tagged CBP following their expression in HEK293T cells. As expected, CBP failed to be immunoprecipitated when expressed alone but was efficiently immunoprecipitated when co-expressed with wild-type CRTC1-MAML2 (Figure 3). Since L180 in CRTC1-MAML2 is the centrally located residue within the helical segment that interacts with CBP besides being invariant in MAML2 orthologs (Figure 2C), we mutated this residue to proline with the goal of disrupting the helical structure while concomitantly introducing a non-conservative substitution. Unlike the wild-type protein, the L180P mutant failed to immunoprecipitate CBP (Figure 3), implying that L180 and/or the helical segment played a crucial role(s) in promoting efficient interactions with CBP.
Figure 3.
Analysis of the interaction between CRTC1-MAML2 and CBP in cells. Co-immunoprecipitation analyses of HA-tagged CBP protein by Flag-tagged, wild-type or mutant CRTC1-MAML2. Cell lysates (input) and the immunoprecipitated proteins (IP) were resolved by SDS-PAGE and visualized by Western blot using anti-HA or anti-Flag antibodies.
CRTC1-MAML2 TAD1 binds primarily to the MLL1-binding surface of KIX
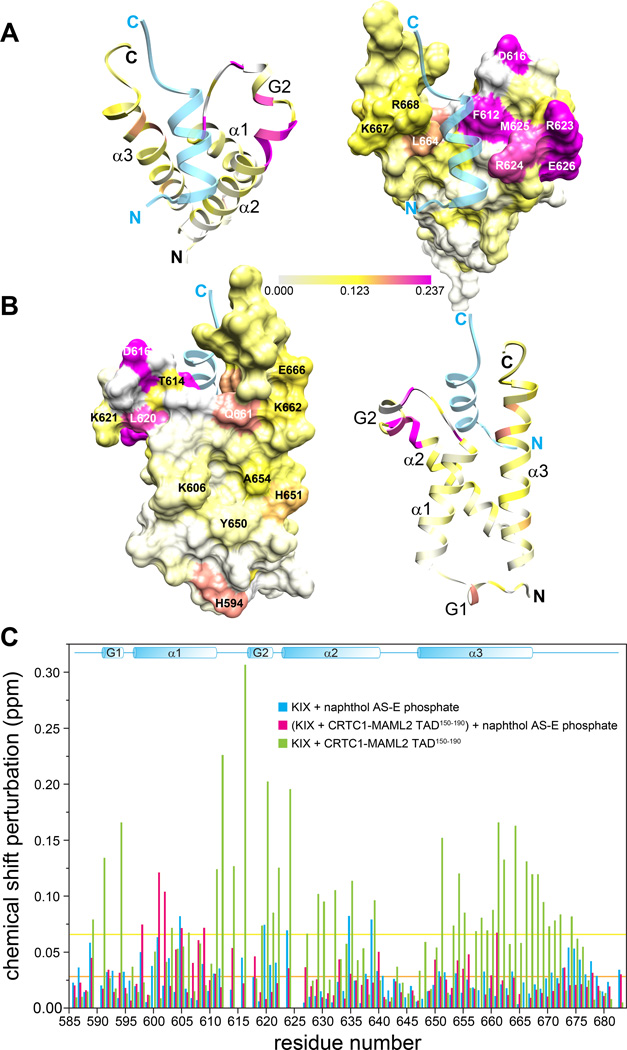
To identify the surface targeted by the CRTC1-MAML2 TAD, we mapped the CSPs on to the molecular surface of KIX. Some of the most strongly perturbed residues located in the loop connecting helices α1 and α2, the N-terminus of α2 and the C-terminus of α3 formed a surface that was previously shown to be targeted by MLL1 and FOXO3a (Figure 4A).(25, 26) This is confirmed by the observation of intermolecular NOEs between residues in this region of KIX and TAD. Interestingly, the segment of TAD that binds KIX shares some similarity to the MLL1 sequence, at least at those positions in the latter protein known to engage KIX (Figure 2C).(25) Our efforts to determine a high-resolution three-dimensional structure of the CRTC1-MAML2 TAD:CBP KIX complex were thwarted by severe resonance line broadening for several key side chains at the protein-protein interface and the pattern of intermolecular NOEs that suggested more than one binding site for the TAD.
Figure 4. The CRTC1-MAML2 TAD1 binds to the MLL1-binding surface of KIX.
(A) Backbone amide CSPs induced by CRTC1-MAML2 TAD150–190 presented in Figure 1B mapped on to the backbone and molecular surface of KIX (PDB ID: 2AGH).(25) The coloring scheme follows the legend shown at the bottom of the panel. Note that KIX residues that show extreme line broadening are rendered with the same color as the maximally perturbed residue. The segment of MLL1 corresponding to residues 844–860 that shows similarity to CRTC1-MAML2 TAD150–190 (Figure 2C) is rendered as a semi-transparent ribbon. (B) Views of the principal CREB pKID-binding surface of KIX showing the same data as in panel A. (C) CSPs of KIX induced by naphthol AS-E phosphate in the absence (blue) and presence (red) of one equivalent of CRTC1-MAML2 TAD150–190; CSPs induced by CRTC1-MAML2 TAD150–190 are included for comparison (green). Protein concentrations as well as solution and experimental conditions were the same as those described in Figure 1A. The orange and yellow horizontal lines denote the average perturbations induced in KIX by naphthol AS-E phosphate (0.027 ppm for both KIX samples) and by CRTC1-MAML2 TAD150–190 (0.066 ppm).
Closer analysis of the CSP maps revealed a secondary binding surface that is distinct from the MLL1 site, comprising residues in the middle and N-terminal portions of the α3 helix and near the N-terminus of the protein (Figure 4B). The latter surface is targeted by CREB pKID and c-Myb and the pattern of CSPs is reminiscent of that observed for CREB pKID, although the degree of perturbation is much lower in the case of CRTC1-MAML2 TAD.(21) Interestingly, MLL1 and FOXO3a have also been found to bind to this secondary site but with much lower affinity.(26, 27) That this represents a lower affinity site for CRTC1-MAML2 TAD is suggested by isothermal titration calorimetric analysis (ITC) conducted with KIX and TAD (Figure 1D). The titration data could be readily fitted assuming a 2-site binding model with values of Kd1 = 20 ± 5 µM and Kd2 = 370 ± 60 µM. Since the affinity for Site 1 is very similar to the one measured by NMR (note that all five residues used for NMR measurements are located at or in the vicinity of the MLL1-binding site), we assign the higher affinity site to the MLL1-binding surface and the lower affinity site to the pKID-binding surface. This is further supported by the curvilinear trajectories in NMR titrations traced by resonances belonging to residues that straddle the two interfaces including A610 and Q661 (at both the main chain and side chain levels; Supplementary Figure S3). These types of curvilinear trajectories are observed when there are multiple binding sites in the vicinity with widely different binding affinities.(27) Finally, the TAD and pKID share striking similarity at the sequence level, especially in the segment corresponding to the αB helix of pKID (Figure 1A) that engages KIX in this region. We note that the phosphorylated form of a peptide spanning only the αB helix binds KIX with a Kd of 80 µM (>100-fold lower affinity than a phosphopeptide spanning both helices).(23)
To further confirm whether the pKID-binding surface of KIX represents a weaker binding site for the TAD, titrations of naphthol AS-E phosphate, a known allosteric inhibitor of CREB pKID and c-Myb, which binds to an overlapping surface in KIX, were conducted in the absence and presence of the TAD.(28, 29) As expected, the compound induced CSPs when titrated with KIX with trends that were similar to those described previously, indicative of a direct association (Figure 4C). The pattern of CSPs suggested that it bound to the surface defined by the α1 and α2 helices distinct from both CREB- and MLL1-binding sites, as described previously.(28) However, in the presence of CRTC1-MAML2 TAD, the CSPs induced by the compound were strikingly different with significantly diminished perturbations for residues at or in the vicinity of the MLL1-binding site (residues 620–624) although perturbations for residues in helix α1 comprising its primary binding site were noticeably enhanced (Figure 4C). Importantly, no large-scale CSPs were noted for KIX residues affected by CRTC1-MAML2 TAD binding, especially at or in the vicinity of the MLL1-binding site, implying that the TAD stayed bound to KIX even in the presence of naphthol AS-E phosphate. This was also verified directly by recording spectra for CRTC1-MAML2 TAD in the absence and presence of naphthol AS-E phosphate that resulted in little or no changes in the NMR spectrum (Supplementary Figure S4).
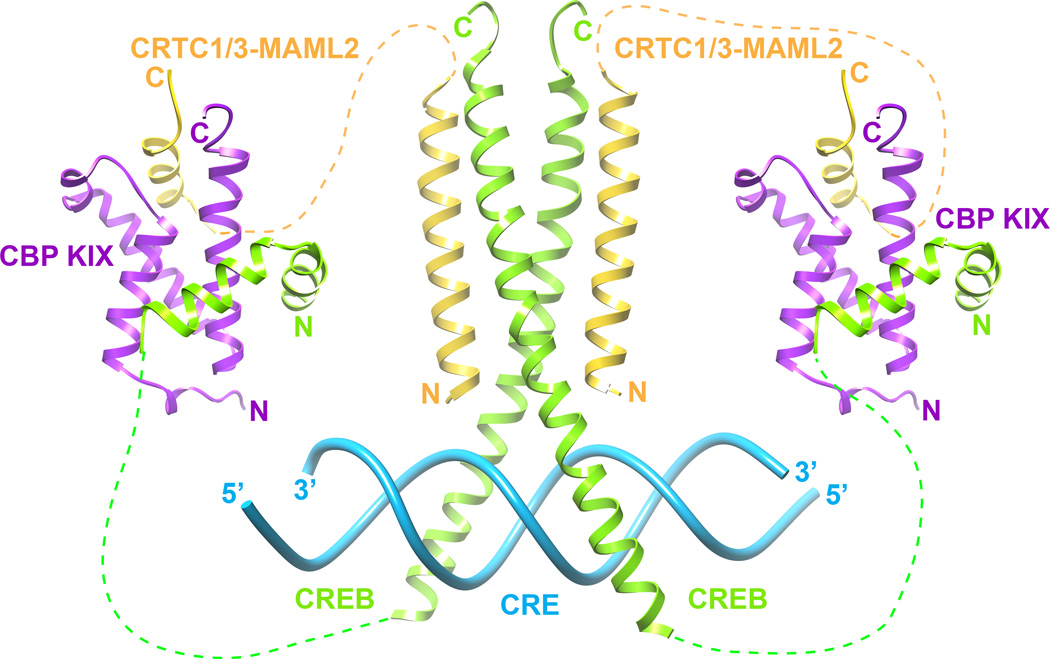
Molecular model for collaborative recruitment of CBP/p300 by CREB and CRTC1/3-MAML2
The results of our studies lead us to propose a molecular model for CBP/p300 recruitment by CRTC1/3-MAML2 and CREB (Figure 5). In this model, the N-terminal CBD of CRTC1/3-MAML2 associates with the CRE-bound bZip domain of CREB.(10, 30) The CRTC1/3-MAML2 TAD1 recruits CBP/p300 KIX by engaging a surface that is distinct from the CREB pKID binding surface. We note that CREB KID can associate with KIX even when unphosphorylated but with significantly diminished affinity (~110 µM, which is over 100-fold weaker affinity relative to pKID).(23) In the absence of Ser133 phosphorylation, CREB KID could thus collaborate and potentially synergize with CRTC1/3-MAML2 TAD1 to associate with KIX. Indeed, certain KIX-interactors including c-Myb and MLL1 have previously been shown to interact with higher affinity with KIX in this manner.(24) CREB Ser133 phosphorylation by various stimuli including increases in intracellular cAMP levels could further enhance the recruitment of CBP/p300 KIX to the promoters of CREB targets. Further studies are needed to explore these possibilities.
Figure 5.
A molecular model for collaborative recruitment of CBP/p300 KIX (purple) at CREs (blue) by CREB (green) and CRTC1-MAML2 (gold). The model is based on the structures of CREB bound to CRE (PDB: 1DH3),(30) the CREB binding domain of CRTC2 (PDB: 4HTM),(10) the MLL1-CBP KIX (PDB: 2AGH)(25) and CREB-CBP KIX (PDB: 1KDX)(5) complexes.
Supplementary Material
Acknowledgements
We are grateful to Drs. Marc Montminy and Michael Conkright for generously sharing CBP and CRTC1-MAML2 mammalian expression constructs. We are grateful to the Robert H. Lurie Comprehensive Cancer Center at Northwestern for supporting structural biology research.
Funding Source. This work was supported by grants from the American Diabetes Association (#1-12-BS-168) and the NIH (1S10 OD012016) (I.R.). M.D.C. was supported by a Northwestern University Weinberg College of Arts and Sciences Summer Research Grant and R.M. was supported by the Molecular Biophysics Training Grant (T32 GM008382).
Abbreviations
- bZip
basic leucine zipper
- CREB
cyclic AMP response element binding protein
- CBD
CREB-binding domain
- CBP
CREB binding protein
- CRTC
CREB regulated transcriptional coactivators
- CSP
chemical shift perturbation
- KID
kinase inducible transactivation domain
- pKID
phosphorylated KID
- MAML2
Mastermind like coactivator 2
- MBP
maltose binding protein
- MECT
mucoepidermoid carcinoma translocation protein
- MLL
mixed lineage leukemia
- PCAF
p300/CBP-associated factor
- SCS
secondary chemical shift
- TAD
transactivation domain
- TEV
Tobacco Etch Virus
Footnotes
Supporting Information. Four additional figures are available as supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montminy M. Transcriptional regulation by cyclic AMP. Annu. Rev. Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 3.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 4.Parker D, Jhala US, Radhakrishnan I, Yaffe MB, Reyes C, Shulman AI, Cantley LC, Wright PE, Montminy M. Analysis of an activator:coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol. Cell. 1998;2:353–359. doi: 10.1016/s1097-2765(00)80279-8. [DOI] [PubMed] [Google Scholar]
- 5.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 6.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol. Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Iourgenko V, Zhang W, Mickanin C, Daly I, Jiang C, Hexham JM, Orth AP, Miraglia L, Meltzer J, Garza D, Chirn GW, McWhinnie E, Cohen D, Skelton J, Terry R, Yu Y, Bodian D, Buxton FP, Zhu J, Song C, Labow MA. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravnskjaer K, Hogan MF, Lackey D, Tora L, Dent SY, Olefsky J, Montminy M. Glucagon regulates gluconeogenesis through KAT2B- and WDR5-mediated epigenetic effects. J. Clin. Invest. 2013;123:4318–4328. doi: 10.1172/JCI69035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, 3rd, Takemori H, Okamoto M, Montminy M. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 10.Luo Q, Viste K, Urday-Zaa JC, Senthil Kumar G, Tsai WW, Talai A, Mayo KE, Montminy M, Radhakrishnan I. Mechanism of CREB recognition and coactivation by the CREB-regulated transcriptional coactivator CRTC2. Proc. Natl. Acad. Sci. U. S. A. 2012;109:20865–20870. doi: 10.1073/pnas.1219028109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, Takemori H, Montminy M. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 12.Kovacs KA, Steullet P, Steinmann M, Do KQ, Magistretti PJ, Halfon O, Cardinaux JR. TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4700–4705. doi: 10.1073/pnas.0607524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y, Wu H, Li S, Chen Q, Cheng XW, Zheng J, Takemori H, Xiong ZQ. Requirement of TORC1 for late-phase long-term potentiation in the hippocampus. PLoS ONE [Electronic Resource] 2006;1:e16. doi: 10.1371/journal.pone.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tonon G, Modi S, Wu L, Kubo A, Coxon AB, Komiya T, O'Neil K, Stover K, El-Naggar A, Griffin JD, Kirsch IR, Kaye FJ. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat. Genet. 2003;33:208–213. doi: 10.1038/ng1083. [DOI] [PubMed] [Google Scholar]
- 15.Fehr A, Roser K, Heidorn K, Hallas C, Loning T, Bullerdiek J. A new type of MAML2 fusion in mucoepidermoid carcinoma. Genes Chromosomes Cancer. 2008;47:203–206. doi: 10.1002/gcc.20522. [DOI] [PubMed] [Google Scholar]
- 16.McElhinny AS, Li JL, Wu L. Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene. 2008;27:5138–5147. doi: 10.1038/onc.2008.228. [DOI] [PubMed] [Google Scholar]
- 17.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 18.Wu L, Liu J, Gao P, Nakamura M, Cao Y, Shen H, Griffin JD. Transforming activity of MECT1-MAML2 fusion oncoprotein is mediated by constitutive CREB activation. EMBO J. 2005;24:2391–2402. doi: 10.1038/sj.emboj.7600719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar GS, Xie T, Zhang Y, Radhakrishnan I. Solution structure of the mSin3A PAH2-Pf1 SID1 complex: a Mad1/Mxd1-like interaction disrupted by MRG15 in the Rpd3S/Sin3S complex. J. Mol. Biol. 2011;408:987–1000. doi: 10.1016/j.jmb.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goddard TD, Kneller DG. Sparky 3. 2004 http://www.cgl.ucsf.edu/home/sparky/. [Google Scholar]
- 21.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Structural analyses of CREB-CBP transcriptional activator-coactivator complexes by NMR spectroscopy: implications for mapping the boundaries of structural domains. J. Mol. Biol. 1999;287:859–865. doi: 10.1006/jmbi.1999.2658. [DOI] [PubMed] [Google Scholar]
- 22.Clark MD, Marcum R, Graveline R, Chan CW, Xie T, Chen Z, Ding Y, Zhang Y, Mondragon A, David G, Radhakrishnan I. Structural insights into the assembly of the histone deacetylase-associated Sin3L/Rpd3L corepressor complex. Proc. Natl. Acad. Sci. U. S. A. 2015 doi: 10.1073/pnas.1504021112. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zor T, Mayr BM, Dyson HJ, Montminy MR, Wright PE. Roles of phosphorylation and helix propensity in the binding of the KIX domain of CREB-binding protein by constitutive (c-Myb) and inducible (CREB) activators. J. Biol. Chem. 2002;277:42241–42248. doi: 10.1074/jbc.M207361200. [DOI] [PubMed] [Google Scholar]
- 24.Goto NK, Zor T, Martinez-Yamout M, Dyson HJ, Wright PE. Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP). The mixed lineage leukemia protein (MLL) activation domain binds to an allosteric site on the KIX domain. J. Biol. Chem. 2002;277:43168–43174. doi: 10.1074/jbc.M207660200. [DOI] [PubMed] [Google Scholar]
- 25.De Guzman RN, Goto NK, Dyson HJ, Wright PE. Structural basis for cooperative transcription factor binding to the CBP coactivator. J. Mol. Biol. 2006;355:1005–1013. doi: 10.1016/j.jmb.2005.09.059. [DOI] [PubMed] [Google Scholar]
- 26.Wang F, Marshall CB, Yamamoto K, Li GY, Gasmi-Seabrook GM, Okada H, Mak TW, Ikura M. Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment. Proc. Natl. Acad. Sci. U. S. A. 2012;109:6078–6083. doi: 10.1073/pnas.1119073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arai M, Dyson HJ, Wright PE. Leu628 of the KIX domain of CBP is a key residue for the interaction with the MLL transactivation domain. FEBS Lett. 2010;584:4500–4504. doi: 10.1016/j.febslet.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Best JL, Amezcua CA, Mayr B, Flechner L, Murawsky CM, Emerson B, Zor T, Gardner KH, Montminy M. Identification of small-molecule antagonists that inhibit an activator: coactivator interaction. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17622–17627. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uttarkar S, Dukare S, Bopp B, Goblirsch M, Jose J, Klempnauer KH. Naphthol AS-E phosphate inhibits the activity of the transcription factor Myb by blocking the interaction with the KIX domain of the coactivator p300. Mol. Cancer Ther. 2015 doi: 10.1158/1535-7163.MCT-14-0662. [DOI] [PubMed] [Google Scholar]
- 30.Schumacher MA, Goodman RH, Brennan RG. The structure of a CREB bZIP.somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding. J. Biol. Chem. 2000;275:35242–35247. doi: 10.1074/jbc.M007293200. [DOI] [PubMed] [Google Scholar]
- 31.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.