Abstract
A number of different epidemiological studies have measured the association between the risk of different cancers and polymorphism at promoter region of 5′ untranslated region (5′-UTR) of the Ataxia-telangiectasia mutated (ATM) gene. However the results were contentious rather than conclusive. The current study was aimed at evaluating the association between the SNP (rs189037 G>A) and the risk of head and neck cancer and lung cancer by conducting a meta-analysis. A total of 9 case–control studies were considered for this quantitative analysis. Stats Direct Statistical software (version 2.7.2) was used to evaluate the crude odds ratio (OR) with their 95% confidence interval (CI). The dominant model (GG vs. GA + AA) showed no heterogeneity and the fixed effects pooled OR was found to be significant (OR = 1.14, 95% CI = 1.05–1.25) at p = 0.003. The pooled OR for fixed effects of heterozygote and homozygote mutant allele (GA vs. AA) model was significant (OR = 1.17, 95% CI = 1.04–1.30, p = 0.006) and no heterogeneity was observed for this model. The current meta-analysis manifested that ATM rs189037 G>A genetic polymorphism may contribute increased risk of head and neck and lung cancer. Moreover, the AA mutant allele was found to be related significantly with the prognosis of lung cancer and head and neck cancer.
Keywords: ATM (rs189037 G>A), Head and neck cancer, Lung cancer, Meta-analysis
1. Introduction
Cancer is an aberrant, uncontrolled growth of cells caused by myriad damage or mutations in the genetic material of the cells due to hereditary or environmental factors, which become immune to many signals controlling cellular growth and death often having the potentiality of invading or spreading to varied body parts (Perez-Herrero & Fernandez-Medarde, 2015). DNA double strand breaks (DSBs) are the most injurious among different types of DNA lesions caused by exogenous toxins such as environmental mutagens and chemical carcinogens in conjunction with endogenous sources resulting from reactive oxygen species (ROS) assembly during cellular respiration etc. (Di Domenico et al., 2014; Khalil et al., 2012). It has been proposed that a single unrepaired DSB might be enough to evoke cell death (Bennett et al., 1993) whereas misrepaired DSBs can result in loss of genetic information, harmful mutation and chromosomal rearrangements which can lead to cancer development (Weber & Ryan, 2015).
One of the frequently mutated genes in cancer that functions upstream of TP53 in the DNA damage response (DDR) pathway and conceivably linked to the escape from apoptosis/senescence hallmark is the ataxia-telangiectasia mutated (ATM) located on the long arm of chromosome 11 (11q22–23) and covers around 160 Kb of genomic DNA (Khalil et al., 2012; Macheret & Halazonetis, 2015). The ATM protein is a large 370 kDa serine/threonine kinase that activates over a hundred proteins involved in DDR and other cellular responses like DSB repair, cell cycle regulation, chromatin remodeling, apoptosis etc. and its loss of function contributes to the major features of rare neurodegenerative autosomal recessive disorder ataxia-telangiectasia (AT) including increased cancer risk (Di Domenico et al., 2014; Weber & Ryan, 2015; Chaudhary & Al-Baradie, 2014).
The ATM gene has 66 exons and the first four exons lie in the 5′-UTR which undergo extensive alternative splicing thereby giving rise to different mRNA transcripts with varying sequences and lengths having different regulatory roles via the formation of different secondary structures and varying number of start codons (Khalil et al., 2012; Rotman & Shiloh, 1998). Of the majority of the ATM single nucleotide polymorphisms (SNPs), the rs189037 (G > A), located at the 5'UTR of the promoter region of ATM gene is one of the vital SNPs associated with the risk of several cancers.
Epidemiological studies around the globe confirmed the association between various ATM polymorphisms in different cancer risks (Cancer Genome Atlas Network, 2012; Cancer Genome Atlas Research Network, 2012a; Cancer Genome Atlas Research Network, 2012b; Cancer Genome Atlas Research Network, 2014; Bea et al., 2013; Landau & Wu, 2013).Various articles related to rs189037 SNP in promoter region of the ATM gene and the risk of different cancers have also been published (Liu et al., 2014; Gu et al., 2014a; Shen et al., 2014; Bau et al., 2010; Damiola et al., 2014; Song et al., 2015; Lo et al., 2010; Kim et al., 2006; Hsia et al., 2013). However, the results were not conclusive. In order to evaluate the association between rs 189037 SNP and cancer risk, we have performed a meta-analysis from relevant scientific literatures.
2. Materials and methods
2.1. Identification and eligibility of relevant literatures
A range of electronic databases (PubMed, Google Scholar, SNPedia, OMIM, Cochrane library) were searched (last search update was 2015 March) using the search terms ‘ATM’, ‘polymorphism’, ‘cancer’, and ‘rs189037 in ATM’. Manual search of references identified in relevant publication was also performed to find other relevant literatures.
Studies included in our meta-analysis followed the inclusion criteria mentioned below: (1) All were case–control studies related to rs189037 (G > A) polymorphism, (2) Control group were in accordance with Hardy–Weinberg equilibrium, (3) Details of genotype frequencies of ATM rs189037G > A genetic polymorphisms, and (4) Only studies in English literature were considered for this meta-analysis. The major exclusion criteria: (1) No control group, and (2) No details of genotype frequencies of desired polymorphism.
2.2. Data extraction & quality assessment
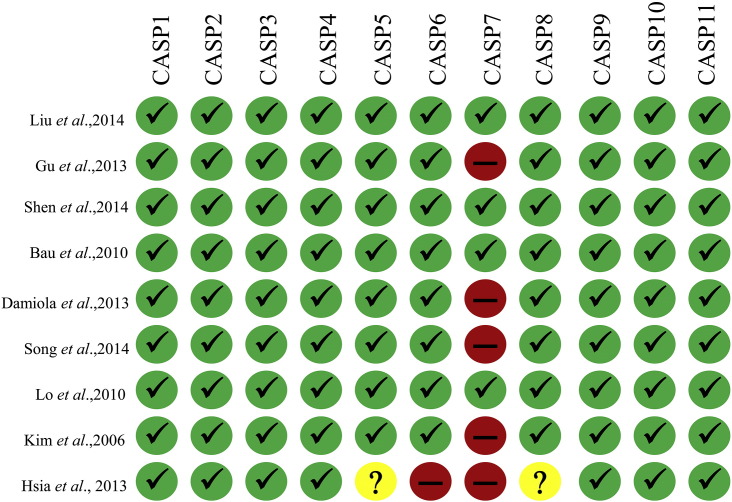
Data were carefully extracted from all eligible publications independently by two of the authors, according to the inclusion criteria mentioned above. The data were collected from each relevant study according to the following details: the first author's full name, publication year, ethnicity of studied population, design of study, genotyping methods, genotype frequencies etc. The critical appraisal skills program (CASP) for case–control studies (http://www.casp-uk.net/) was performed for quality assurance of the selected eligible studies considered for the quantitative analysis. The scoring pattern for the CASP criteria is as follows: the study addresses a clearly focused issue (CASP1); an appropriate research design answers the research problem (CASP2); the selected cases were inscribed acceptably (CASP3); the controls were selected acceptably (CASP4); the measurement for exposure factors is precise to minimize classification bias (CASP5); the potential confounding factors are considered in the study design/analysis (CASP6); the research results are accomplished (CASP7); the research results are accurate (CASP8); the research results are authentic (CASP9); the research results are applicable to the local population (CASP10); and the research results fit with other available evidence like systematic reviews, case–control studies, cohort studies etc. (CASP11).
2.3. Statistical analysis
Meta-analysis was performed using the StatsDirect Statistical software (version 2.7.2). The intensity of the association between SNP and cancers was calculated from odds ratios (ORs) with 95% confidence intervals (CIs). The Z test was performed to estimate the statistical significance of pooled OR. Genotype frequencies of control group were tested for HWE using χ2 test. Cochran's Q statistic and I2 tests were used to test the heterogeneity between studies (Zintzaras & Ioannidis, 2005). If Q test shows a p < 0.05 or I2 demonstrates > 50% which indicates significant heterogeneity, the random effect model was chosen otherwise the fixed effects model was used. Sensitivity analysis was performed to check the potential outliers each time omitting a single study. Funnel plots and Egger's linear regression test were conducted to investigate the significant publication bias (Peters et al., 2006). In funnel plot standard error of log (OR) of each study was plotted against its log (OR) and an asymmetric plot suggested possible publication bias. On other hand the degree of asymmetry was tested using Egger's test where p < 0.05 indicates significant publication bias. The pooled OR was considered to be significant at p < 0.05.
3. Results
3.1. Characteristics of studies
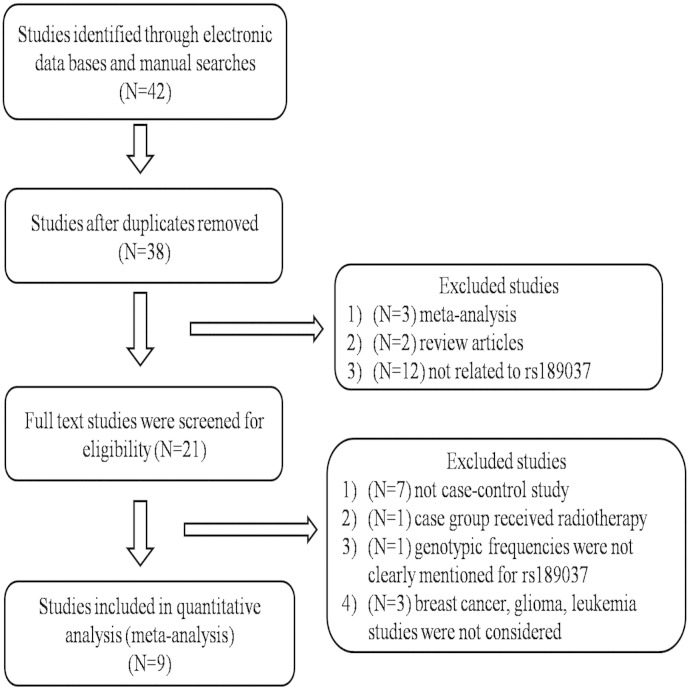
Initially we identified total 42 studies from different database and manual searches. After reviewing the titles and abstracts, we excluded 4 studies. Out of 38 remaining studies, 29 were excluded further. Finally, 9 case–control studies containing of 4507 cancer patients and 4778 control subjects, met our inclusion criteria for quantitative data analysis (Fig. 1). Overall, the studies were carried out among 8 Asians and 1 Caucasian population according to the inclusion criteria. CASP scores and the characteristics of all the studies are summarized in Fig. 2 and Table 1, respectively (Liu et al., 2014; Gu et al., 2014a; Shen et al., 2014; Bau et al., 2010; Damiola et al., 2014; Song et al., 2015; Lo et al., 2010; Kim et al., 2006; Hsia et al., 2013).
Fig. 1.
Flow chart of literature search and relevant study selection. Total 9 case–control studies were included in this meta-analysis.
Fig. 2.
CASP scores for 9 eligible studies for the relationship between ATM rs189037 polymorphism and susceptibility to lung, head and neck cancer.
Table 1.
Characteristics of relevant studies included in meta-analysis and genotypic frequencies of rs189037 G > A.
| Study | Ethnicity | Type of cancer | Source of control | Genotyping method | Rs189037 G > A (case/control) | pa value | Case |
Control |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | GG | GA | AA | |||||||
| Liu et al. (2014) | Asians | Lung | HB | TaqMan real-time PCR | 852/852 | 0.29 | 217 | 435 | 200 | 264 | 434 | 154 |
| Gu et al. (2013) | Asians | Papillary thyroid carcinoma | HB | MALDI-TOF-MS | 355/360 | 0.26 | 90 | 196 | 69 | 102 | 189 | 69 |
| Shen et al. (2014) | Asians | Lung | HB | TaqMan real-time PCR | 487/516 | 0.12 | 148 | 240 | 99 | 152 | 272 | 92 |
| Bau et al. (2010) | Asians | Oral | HB | PCR-RFLP | 620/620 | 0.47 | 181 | 277 | 162 | 239 | 285 | 96 |
| Damiola et al. (2013) | Caucasians | Papillary thyroid carcinoma | PB | Illumina GoldenGate Genotyping Assay | 68/201 | 0.31 | 13 | 32 | 23 | 35 | 106 | 60 |
| Song et al. (2014) | Asians | Papillary thyroid carcinoma | HB | TaqMan assay | 428/182 | 0.34 | 134 | 211 | 83 | 56 | 84 | 42 |
| Lo et al. (2010) | Asians | Lung | HB | MassARRAY assay | 728/717 | 0.72 | 238 | 345 | 145 | 239 | 354 | 124 |
| Kim et al. (2006) | Asians | Lung | HB | SNP-IT assays | 611/614 | 0.71 | 190 | 316 | 105 | 195 | 306 | 113 |
| Hsia et al. (2013) | Asians | Lung | HB | PCR-RFLP | 358/716 | 0.61 | 118 | 176 | 64 | 255 | 339 | 122 |
pa value > 0.05 indicates control group is in Hardy–Weinberg equilibrium equation.
3.2. Meta-analysis result
The pooled ORs for ATM rs189037G > A and cancer risk is listed in Table 2. The homozygote model (GG vs. AA) showed significant heterogeneity (p = 0.01, I2 = 63.6%) and the random effects pooled OR was 1.23 (95% CI = 1.00–1.51) at p = 0.053. The dominant model (GG vs. GA + AA) showed no heterogeneity and the fixed effects pooled OR was found to be significant (OR = 1.14, 95% CI = 1.05–1.25) at p = 0.003. On other hand recessive model (GG + GA vs. AA) showed significant heterogeneity (p = 0.01, I2 = 60.7%) and the random effects pooled OR did not show any significant result (OR = 1.17, 95% CI = 1.00–1.40) (p = 0.068). The association between A allele and the risk of developing cancer relative to the allele G revealed significant heterogeneity (p = 0.01, I2 = 63.7%) but the random effects pooled OR did not show significant result (OR = 1.11, 95% CI = 1.00–1.23, p = 0.046). Homozygote wild and heterozygote alleles (GG vs. GA) showed no heterogeneity and the pooled OR for fixed effects was not significant (OR = 1.09, 95% CI = 1.00–1.20, p = 0.066). However, the pooled OR for fixed effects of heterozygote and homozygote mutant allele (GA vs. AA) model was significant (OR = 1.17, 95% CI = 1.04–1.30, p = 0.006) and no heterogeneity was observed for this model (Fig. 3).
Table 2.
Main result of pooled ORs of ATM rs189037 G > A polymorphism.
| Genotype rs189037 G > A | OR | 95% CI | pb value | p value for Cochran Q test | I2 | Combination method |
|---|---|---|---|---|---|---|
| GG vs. AA | 1.23 | 1.00–1.51 | 0.053 | 0.01 | 63.6% | Random effect |
| GG vs. (GA + AA) | 1.14 | 1.05–1.25 | 0.003 | 0.17 | 31.6% | Fixed effect |
| (GG + GA) vs. AA | 1.17 | 1.00–1.40 | 0.068 | 0.01 | 60.7% | Random effect |
| G vs. A | 1.11 | 1.00–1.23 | 0.046 | 0.01 | 63.7% | Random effect |
| GG vs. GA | 1.09 | 1.00–1.20 | 0.066 | 0.66 | 0% | Fixed effect |
| GA vs. AA | 1.17 | 1.04–1.30 | 0.006 | 0.63 | 45.8% | Fixed effect |
b — p heterogeneity.
Fig. 3.
Figure showing forest plots of ATM rs189037 (G > A) polymorphism in association with head and neck and lung cancer: a) GG vs. AA b) GG vs. GA + AA, c) GG + GA vs. AA d) G vs. A e) GG vs. GA f) GA vs. AA.
Sensitivity test was assigned to check the influence of each studies on overall pooled OR. Begg–Mazumdar's funnel plot (Fig. 4) did not show any publication asymmetry which was confirmed by Egger's test. There was no publication bias for ATM rs189037G > A (Homozygote model: tau = − 0.11, p = 0.24; Dominant model: tau = − 0.11, p = 0.33; Recessive model: tau = − 0.39, p = 0.26; Allele model: tau = − 0.06, p = 0.34; Homozygote wild type vs. Heterozygote model: tau = − 0.22, p = 0.39; Heterozygote vs. Homozygote mutant model: tau = − 0.39, p = 0.33).
Fig. 4.
Funnel plot results of different models used in meta-analysis: a) GG vs. AA b) GG vs. GA + AA, c) GG + GA vs. AA d) G vs. A e) GG vs. GA f) GA vs. AA.
4. Discussion
ATM is one of the central kinases in cellular responses to DNA DSBs. ATM is recruited by the MRN complex (MRE11-RAD50-NBS1) to the site of DNA DSBs. The C-terminus of NBS1 acts as the binding site for ATM, thereby enhancing its kinase activity. Immediately after recruitment, ATM phosphorylates the histone variant H2AX on serine 139, which further recruits additional proteins in DNA damage response pathway and helps ATM to amplify its signal and form discrete ATM foci at DNA DSBs (Khalil et al., 2012). In response to DNA damage, ATM also interacts with different downstream effectors and induces different signaling functions like cell cycle check point arrest with p53, Mdm2 and Chk2 at G1, damage induced S-phase arrest with NBS1, BRCA1, FancD2 and SMC1, G2/M arrest with BRCA1 and hRad17 and apoptosis with E2F1, Chk2, P53, P73 and Bax (Khalil et al., 2012).
The polymorphism rs189037 is located at the 5′UTR of the promoter region of ATM gene (Gu et al., 2014b). It was evident from different studies that polymorphisms in the promoter region of a gene may change the binding sites of transcription factors, which affects gene expression. The heterozygote of rs189037 has been observed to be significantly related to longevity through an effect on ATM transcript like AP-2α, a transcription factor that participates in many important life processes (Rotman & Shiloh, 1998; Chen et al., 2010; Piaceri et al., 2013). There are other possibilities of one or more transcription factors which are regulated by rs189037, and thus different genotypes may up-regulate or down-regulate the expression of ATM gene, which may lead to cancer formation.
Different case–control studies have reported the association of rs189037 polymorphism and risk of different cancers (Liu et al., 2014; Gu et al., 2014a; Shen et al., 2014; Bau et al., 2010; Damiola et al., 2014; Song et al., 2015; Lo et al., 2010; Kim et al., 2006). (Liu et al., 2014), (Shen et al., 2014) and (Bau et al., 2010) reported significantly increased risk between the association of lung cancer and oral cancer with the A allele of ATM (Liu et al., 2014; Shen et al., 2014; Bau et al., 2010) but the rest of the studies (Gu et al., 2014a; Damiola et al., 2014; Song et al., 2015; Lo et al., 2010; Kim et al., 2006; Hsia et al., 2013) could not find any association between the A allele and cancer risk. In our meta-analysis, it was clear that the variant A allele resulted in cancer risk compared to the G allele. Homozygote, recessive and allele models could not show significant cancer risk with the A allele compared to the G allele. Homozygote dominant vs. Heterozygote (GG vs. GA) model also did not show any risk of cancer having A allele. However, Dominant model (GG vs. GA + AA), heterozygote vs. homozygote mutant (GA vs. AA) model showed significant cancer risk associated with A allele in comparison to the G allele.
It would be interesting to determine the effect of interaction between the ATM rs189037 (G > A) polymorphism and various etiological habits such as smoking, alcohol consumption, betel-quid use etc. on lung cancer and head and neck cancer risk. However, the unavailability of required data in all the studies included in this meat-analysis restricted the evaluation.
5. Conclusion
This study provides for the first time conclusive evidence that ATM rs189037 (G > A) polymorphism is associated with risk of lung cancer and head and neck cancer, which may encourage further large scale population/hospital based case–control/cohort studies.
Conflict of interest
None declared.
Acknowledgments
This work was supported by the Department of Biotechnology, Ministry of Science and Technology, Government of India with sanction no. BT/CP/09/NE/TBP/2010 dated 20th October, 2011. The funders had no role in study design, data collection or analysis, decision to publish and preparation of the manuscript. The authors would like to appreciate the reviewers for their assistive comments on this meta-analysis.
References
- Bau D.T., Chang C.H., Tsai M.H., Chiu C.F., Tsou Y.A., Wang R.F., Tsai C.W., Tsai R.Y. Association between DNA repair gene ATM polymorphisms and oral cancer susceptibility. Laryngoscope. 2010;120:2417–2422. doi: 10.1002/lary.21009. [DOI] [PubMed] [Google Scholar]
- Bea S., Valdes-Mas R., Navarro A., Salaverria I., Martin-Garcia D., Jares P., Gine E., Pinyol M., Royo C., Nadeu F., Conde L., Juan M., Clot G., Vizan P., Di Croce L., Puente D.A., Lopez-Guerra M., Moros A., Roue G., Aymerich M., Villamor N., Colomo L., Martinez A., Valera A., Martin-Subero J.I., Amador V., Hernandez L., Rozman M., Enjuanes A., Forcada P., Muntanola A., Hartmann E.M., Calasanz M.J., Rosenwald A., Ott G., Hernandez-Rivas J.M., Klapper W., Siebert R., Wiestner A., Wilson W.H., Colomer D., Lopez-Guillermo A., Lopez-Otin C., Puente X.S., Campo E. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 2013;110:18250–18255. doi: 10.1073/pnas.1314608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.B., Lewis A.L., Baldwin K.K., Resnick M.A. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl. Acad. Sci. U. S. A. 1993;90:5613–5617. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary M.W., Al-Baradie R.S. Ataxia-telangiectasia: future prospects. Appl. Clin. Genet. 2014;7:159. doi: 10.2147/TACG.S35759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Dong B., Lu Z., Tian B., Zhang J., Zhou J., Wu H., Zhang Y., Wu J., Lin P., Xu H., Mo X. A functional single nucleotide polymorphism in promoter of ATM is associated with longevity. Mech. Ageing Dev. 2010;131:636–640. doi: 10.1016/j.mad.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Damiola F., Byrnes G., Moissonnier M., Pertesi M., Deltour I., Fillon A., Le Calvez-Kelm F., Tenet V., McKay-Chopin S., McKay J.D., Malakhova I., Masyakin V., Cardis E., Lesueur F., Kesminiene A. Contribution of ATM and FOXE1 (TTF2) to risk of papillary thyroid carcinoma in Belarusian children exposed to radiation. Int. J. Cancer. 2013 doi: 10.1002/ijc.28483. [DOI] [PubMed] [Google Scholar]
- Damiola F., Byrnes G., Moissonnier M., Pertesi M., Deltour I., Fillon A., Le Calvez-Kelm F., Tenet V., McKay-Chopin S., McKay J.D., Malakhova I., Masyakin V., Cardis E., Lesueur F., Kesminiene A. Contribution of ATM and FOXE1 (TTF2) to risk of papillary thyroid carcinoma in Belarusian children exposed to radiation. Int. J. Cancer. 2014;134:1659–1668. doi: 10.1002/ijc.28483. [DOI] [PubMed] [Google Scholar]
- Di Domenico E.G., Romano E., Del Porto P., Ascenzioni F. Multifunctional role of ATM/Tel1 kinase in genome stability: from the DNA damage response to telomere maintenance. Biomed Res Int. 2014;2014:787404. doi: 10.1155/2014/787404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Yu Y., Ai L., Shi J., Liu X., Sun H., Liu Y. Association of the ATM gene polymorphisms with papillary thyroid cancer. Endocrine. 2013 doi: 10.1007/s12020-013-0020-1. [DOI] [PubMed] [Google Scholar]
- Gu Y., Yu Y., Ai L., Shi J., Liu X., Sun H., Liu Y. Association of the ATM gene polymorphisms with papillary thyroid cancer. Endocrine. 2014;45:454–461. doi: 10.1007/s12020-013-0020-1. [DOI] [PubMed] [Google Scholar]
- Gu Y., Liu X., Yu Y., Shi J., Ai L., Sun H., Kanu J.S., Wang C., Liu Y. Association of ATM gene polymorphism with PTC metastasis in female patients. Int. J. Endocrinol. 2014;2014:370825. doi: 10.1155/2014/370825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia T.C., Tsai C.W., Liang S.J., Chang W.S., Lin L.Y., Chen W.C., Tu C.Y., Tsai C.H., Bau D.T. Effects of ataxia telangiectasia mutated (ATM) genotypes and smoking habits on lung cancer risk in Taiwan. Anticancer Res. 2013;33:4067–4071. [PubMed] [Google Scholar]
- Khalil H., Tummala H., Zhelev N. ATM in focus: A damage sensor and cancer target. Biodiscovery. 2012;5 [Google Scholar]
- Kim J.H., Kim H., Lee K.Y., Choe K.H., Ryu J.S., Yoon H.I., Sung S.W., Yoo K.Y., Hong Y.C. Genetic polymorphisms of ataxia telangiectasia mutated affect lung cancer risk. Hum. Mol. Genet. 2006;15:1181–1186. doi: 10.1093/hmg/ddl033. [DOI] [PubMed] [Google Scholar]
- Landau D.A., Wu C.J. Chronic lymphocytic leukemia: molecular heterogeneity revealed by high-throughput genomics. Genome Med. 2013;5:47. doi: 10.1186/gm451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wang X., Ren Y., Li X., Zhang X., Zhou B. Effect of single nucleotide polymorphism Rs189037 in ATM gene on risk of lung cancer in Chinese: a case–control study. PLoS One. 2014;9:e115845. doi: 10.1371/journal.pone.0115845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y.L., Hsiao C.F., Jou Y.S., Chang G.C., Tsai Y.H., Su W.C., Chen Y.M., Huang M.S., Chen H.L., Yang P.C., Chen C.J., Hsiung C.A. ATM polymorphisms and risk of lung cancer among never smokers. Lung Cancer. 2010;69:148–154. doi: 10.1016/j.lungcan.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Macheret M., Halazonetis T.D. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015;10:425–448. doi: 10.1146/annurev-pathol-012414-040424. [DOI] [PubMed] [Google Scholar]
- Perez-Herrero E., Fernandez-Medarde A. Advanced targeted therapies in cancer: drug nanocarriers, the future of chemotherapy. Eur. J. Pharm. Biopharm. 2015 doi: 10.1016/j.ejpb.2015.03.018. DOI S0939-6411(15)00151-4, [pii] 10.1016/j.ejpb.2015.03.018. [DOI] [PubMed] [Google Scholar]
- Peters J.L., Sutton A.J., Jones D.R., Abrams K.R., Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- Piaceri I., Bagnoli S., Tedde A., Sorbi S., Nacmias B. Ataxia-telangiectasia mutated (ATM) genetic variant in Italian centenarians. Neurol. Sci. 2013;34:573–575. doi: 10.1007/s10072-012-1188-5. [DOI] [PubMed] [Google Scholar]
- Rotman G., Shiloh Y. ATM: from gene to function. Hum. Mol. Genet. 1998;7:1555–1563. doi: 10.1093/hmg/7.10.1555. [DOI] [PubMed] [Google Scholar]
- Shen L., Yin Z., Wu W., Ren Y., Li X., Zhou B. Single nucleotide polymorphism in ATM gene, cooking oil fumes and lung adenocarcinoma susceptibility in Chinese female non-smokers: a case–control study. PLoS One. 2014;9:e96911. doi: 10.1371/journal.pone.0096911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.M., Kwon T.K., Park B.L., Ji Y.B., Tae K. Single Nucleotide Polymorphisms of Ataxia TelangiectasiaMutated and the Risk of Papillary Thyroid Carcinoma. Environ. Mol. Mutagenesis. 2014 doi: 10.1002/em.21898. [DOI] [PubMed] [Google Scholar]
- Song C.M., Kwon T.K., Park B.L., Ji Y.B., Tae K. Single nucleotide polymorphisms of ataxia telangiectasia mutated and the risk of papillary thyroid carcinoma. Environ. Mol. Mutagen. 2015;56:70–76. doi: 10.1002/em.21898. [DOI] [PubMed] [Google Scholar]
- Weber A.M., Ryan A.J. ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 2015;149:124–138. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Zintzaras E., Ioannidis J.P. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics. 2005;21:3672–3673. doi: 10.1093/bioinformatics/bti536. [DOI] [PubMed] [Google Scholar]