Abstract
Ebola (EBOV) is an enveloped, negative-sense RNA virus belonging to the family Filoviridae that causes hemorrhagic fever syndromes with high-mortality rates. To date, there are no licensed vaccines or therapeutics to control EBOV infection and prevent transmission. Consequently, the need to better understand the mechanisms that regulate virus transmission is critical to developing countermeasures. The EBOV VP40 matrix protein plays a central role in late stages of virion assembly and egress, and independent expression of VP40 leads to the production of virus-like particles (VLPs) by a mechanism that accurately mimics budding of live virus. VP40 late (L) budding domains mediate efficient virus-cell separation by recruiting host ESCRT and ESCRT-associated proteins to complete the membrane fission process. L-domains consist of core consensus amino acid motifs including PPxY, P(T/S)AP, and YPx(n)L/I, and EBOV VP40 contains overlapping PPxY and PTAP motifs whose interactions with Nedd4 and Tsg101, respectively, have been characterized extensively. Here, we present data demonstrating for the first time that EBOV VP40 possesses a third L-domain YPx(n)L/I consensus motif that interacts with the ESCRT-III protein Alix. We show that the YPx(n)L/I motif mapping to amino acids 18–26 of EBOV VP40 interacts with the Alix Bro1-V fragment, and that siRNA knockdown of endogenous Alix expression inhibits EBOV VP40 VLP egress. Furthermore, overexpression of Alix Bro1-V rescues VLP production of the budding deficient EBOV VP40 double PTAP/PPEY L-domain deletion mutant to wild-type levels. Together, these findings demonstrate that EBOV VP40 recruits host Alix via a YPx(n)L/I motif that can function as an alternative L-domain to promote virus egress.
Keywords: Alix, Ebola, VLPs, budding, virus-host interaction, L-domain, ESCRT, filovirus
Ebola virus (EBOV) and Marburg virus (MARV) are enveloped, negative-strand RNA viruses and members of the Filoviridae. These filoviruses cause severe hemorrhagic syndromes with high-mortality rates in humans and nonhuman primates [1, 2] and are categorized by guidelines of the Centers for Disease Control and Prevention and the National Institutes of Health (NIH) as BSL-4 agents. The current lack of approved vaccines or antiviral therapeutics for these pathogens is problematic because outbreaks and new strains continue to emerge, as exemplified by the ongoing Ebola (Zaire) outbreak in West Africa. Thus, there remains a vital need for the development of effective and safe therapeutics and vaccines against these emerging, high-priority pathogens.
It is well established that the filovirus VP40 matrix protein plays a key role in driving virion assembly and budding. Indeed, independent expression of EBOV VP40 (eVP40) or MARV VP40 (mVP40) leads to the production and budding of virus-like particles (VLPs) by a mechanism that accurately mimics that of live virus [3–7]. Using this VLP system, we and others have established that efficient filovirus budding is dependent on the subversion of specific host proteins associated with the cellular ESCRT machinery (eg, Tsg101 and Nedd4) by viral Late (L) domains within eVP40 as well as mVP40 and mNP proteins [3, 4, 6–21]. Viral L-domains consist of conserved consensus amino acid motifs such as PPxY, P(T/S)AP, YPx(n)L/I, or FPIV (x = any amino acid; for review, see [6, 8, 22, 23]) within matrix proteins of many RNA viruses. This conservation of structure suggests that L domains are generally required for efficient virus-cell separation [22, 23]. The 3 best characterized L-domain/host interactions that contribute to budding of many RNA viruses include the: (i) PTAP-Tsg101 interaction, (ii) PPxY-Nedd4 interaction, and (iii) YPx(n)L/I-Alix interaction [22, 23].
Alix is an ESCRT associated protein that facilitates virus egress by bridging ESCRT-I and ESCRT-III components via interactions with viral YPx(n)L/I L-domain motifs [24–42]. Alix consists of 3 major domains named Bro1 (aa 1–366), V (aa 367–701), and PRD (proline-rich domain, aa 702–868) and each mediates interactions with a multitude of cellular and viral proteins (for review, see [43]). A number of interactions mediated by these domains of Alix are important for retrovirus budding. For example, the Alix V domain binds to the viral YPx(n)L/I L-domain motif within human immunodeficiency virus type 1 (HIV-1) and EIAV Gag, the Bro1 domain interacts with the ESCRT-III protein CHMP4 and membrane lipid LBPA to promote membrane bending, and the PRD domain interacts with the ESCRT-I protein Tsg101 and calcium binding protein ALG-2 [43].
To date, there have been no reports suggesting a role for Alix in filovirus budding; however, a role for Alix in EBOV budding could account for the fact that an eVP40 mutant lacking both PTAP and PPEY L-domains (eVP40-ΔPT/PY) can still bud as a VLP, albeit at levels lower than those of VP40 wild type (WT). Similarly, recombinant EBOV containing mutations in both PTAP and PPEY L-domains is viable in cell culture, although budding of this double mutant is also reduced compared to WT EBOV [11]. These results indicate that additional host proteins/pathways can play a complementary, overlapping, or redundant role in EBOV budding. Interestingly, we have identified a YPx(n)L/I-type motif between amino acids 18–26 of eVP40, which is similar to an HIV-1 Gag motif that mediates interactions with Alix and can rescue budding of HIV-1 Gag PTAP deletion mutants [24, 26, 35]. In addition to EBOV VP40, the YPx(n)L/I-type motif is also present in SUDV and BDBV VP40 proteins but not in those of RESTV and TAFV.
Here we demonstrate for the first time that endogenous Alix, through its interactions with the eVP40 YPx(n)L/I-type motif, contributes to efficient egress of eVP40 VLPs. We also demonstrate that the Alix Bro1-V region physically interacts with eVP40-ΔPT/PY and that Bro1-V rescues deficient budding of eVP40-ΔPT/PY. Finally, we demonstrate that the Bro1-V/eVP40-ΔPT/PY interaction is dependent on the YPx(n)L/I-type motif spanning N-terminal amino acids 18–26 of eVP40. These findings have important implications for our complete understanding of the complex virus-host interactions required for efficient egress and spread of EBOV.
MATERIALS AND METHODS
Cell Lines, Plasmids, and Reagents
HEK293T and COS-1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), penicillin (100 U/mL)/streptomycin (100 µg/mL) at 37°C in a humidified 5% CO2 incubator. The pCAGGS based plasmids expressing eVP40-WT and eVP40-ΔPT/PY have been described elsewhere [5, 10]. The pCAGGS-based plasmid expressing eVP40-ΔPT/PY-A2 was generated by mutating 18YP19 amino acids in the eVP40-ΔPT/PY background to AA using standard polymerase chain reaction (PCR) and cloning techniques. pHM6-based plasmids expressing Alix-WT, Bro1, Bro1-V, and V-PRD were kindly provided by F. Bouamr (NIH/NIAID) and have been described previously [26].
VLP Budding Assay
Filovirus VLP budding assays using HEK293T cells have been described elsewhere [5, 10, 44–46]. Proteins in VLPs and cell lysates were detected by SDS-PAGE and Western blot using polyclonal rabbit anti-VP40 antibody for eVP40-WT, eVP40-ΔPT/PY, and eVP40-ΔPT/PY-A2; mouse anti-HA antibody for HA-tagged Alix-WT, Bro1, Bro1-V, V-PRD proteins; and anti-actin antibody (Sigma).
siRNA Transfection
Human Alix-specific or random siRNAs were purchased from Dharmacon. HEK293 T cells in collagen-coated 6-well plates were transfected twice with either random or Alix siRNAs at a final concentration of 200 nM using Lipofectamine (Invitrogen) at 2-day intervals. The final transfection included both siRNAs and 0.5 µg of eVP40 expression plasmid. VLPs and cell lysates were harvested at 24 hours following the last transfection as described above. eVP40 protein levels in VLPs and eVP40, and Alix levels in cell extracts, were analyzed by Western blotting and quantified using NIH Image-J software.
IP/Western Analysis
Cos-1 cells were transfected with the indicated plasmids using Lipofectamine reagent (Invitrogen) and the protocol of the supplier. Cells were harvested and lysed in nondenaturing buffer (20 mM Tris-HCl [pH = 8.0], 137 mM NaCl, 1.0% Nonidet P-40 [NP-40], 2.0 mM EDTA, and 10% glycerol) at 24 hours post-transfection. Cell lysates were clarified for 10 minutes and then incubated with anti-eVP40 or normal rabbit immunoglobuling G (IgG; Cell Signaling) overnight at 4°C. rProtein A/G agarose beads (Invitrogen) were added to the samples, which were incubated with agitation for 2 hours at 4°C. The beads were washed 5× in nondenaturing lysis buffer, suspended in loading buffer with boiling, and then fractionated by SDS-PAGE. HA-tagged Alix-WT or Bro1-V were detected in samples by Western blot using anti-HA antiserum.
RESULTS
Alix Regulates eVP40 VLP Budding
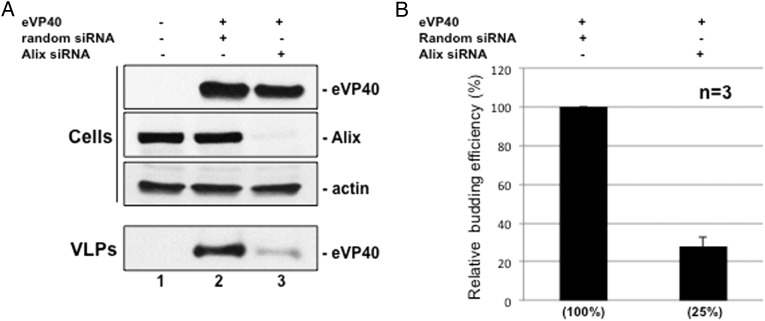
We evaluated the role of endogenous Alix in eVP40 VLP egress by suppressing its expression in HEK293T cells. Alix suppression (>90%, Figure 1A, lane 3) did not affect cellular eVP40 protein levels; however, eVP40 VLP budding was reduced in Alix-suppressed cells by approximately 4-fold (Figure 1A and 1B) relative to random siRNA controls (Figure 1A and 1B), indicating a role for Alix in efficient eVP40 VLP egress from HEK293T cells.
Figure 1.
siRNA knockdown of Alix inhibits egress of eVP40 VLPs. A, HEK293T cells were mock-transfected or transfected with eVP40 plus random or Alix-specific siRNAs. eVP40, endogenous Alix, and actin were detected in cell extracts, and eVP40 was detected in VLPs by Western blot. B, Quantification of the relative budding efficiency of eVP40 VLPs from cells receiving random siRNAs (set at 100%) or Alix-specific siRNAs. Each bar represents the average ±SEM of 3 independent experiments. Abbreviations: SEM, standard error of the mean; VLP, virus-like particle.
Alix Rescues eVP40 VLP Budding
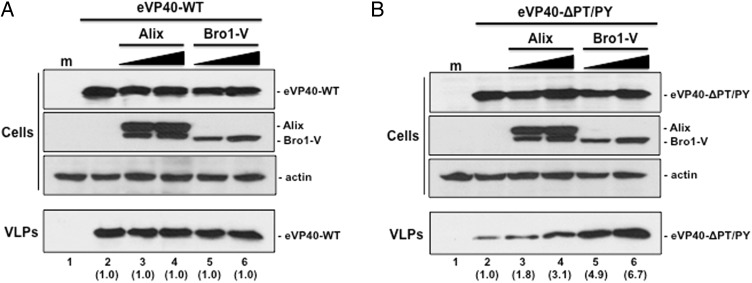
The Bro1, V, and PRD domains of Alix play critical roles in retrovirus-host interactions required to drive efficient budding [25, 27, 32, 33, 39, 41, 47, 48]. Indeed, overexpression of Alix rescues budding of PTAP L-domain mutants of HIV-1 Gag, which cannot interact with host Tsg101 [26, 35]. We first sought to determine whether overexpression of full-length WT or the Bro1-V fragment would affect budding of either eVP40-WT or eVP40-ΔPT/PY, a budding defective PTAP/PPEY double L-domain deletion mutant of eVP40 [9, 10]. Briefly, eVP40-WT budding from HEK293T cells (Figure 2A, lane 2) was approximately 10-fold more efficient than that of eVP40-ΔPT/PY (Figure 2B, lane 2). Expression of increasing amounts of full length or Bro1-V fragment of Alix in HEK293T had no effect on eVP40-WT VLP egress (Figure 2A); however, coexpression of WT Alix or Bro1-V with eVP40-ΔPT/PY enhanced release of eVP40-ΔPT/PY VLPs (Figure 2B). Indeed, expression of WT Alix enhanced eVP40-ΔPT/PY VLP budding by approximately 2–3 fold (Figure 2B, compare lanes 2–4) although unexpectedly, Bro1-V expression enhanced budding of eVP40-ΔPT/PY VLPs by approximately 4–7 fold compared to controls (Figure 2B, compare lanes 2, 5, and 6).
Figure 2.
Bro1-V fragment of Alix rescues budding of eVP40-ΔPT/PY L-domain mutant. A, HEK293T cells were mock-transfected (lane 1) or transfected with eVP40-WT alone (lane 2), or with increasing amounts of either Alix-WT (lanes 3 and 4) or Bro1-V (lanes 5 and 6). eVP40-WT, Alix-WT, Bro1-V, and actin were detected in cell extracts by Western blot, and eVP40-WT was detected in VLPs by Western blot. Numbers in parentheses represent fold change in levels of VLPs relative to control (lane 2). B, HEK293T cells were mock-transfected (lane 1) or transfected with eVP40-ΔPT/PY alone (lane 2), or with increasing amounts of either Alix-WT (lanes 3 and 4) or Bro1-V (lanes 5 and 6). eVP40-ΔPT/PY, Alix-WT, Bro1-V, and actin were detected in cell extracts by Western blot, and eVP40-ΔPT/PY was detected in VLPs by Western blot. Numbers in parentheses represent fold change in levels of VLPs relative to control (lane 2). Abbreviations: VLP, virus-like particle; WT, wild type.
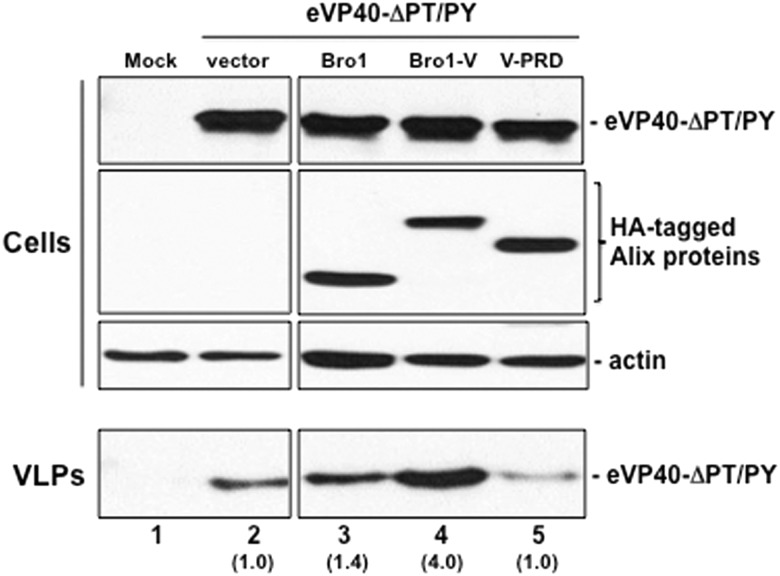
We used an identical approach to determine whether other Alix domains (Bro1 and V-PRD) could enhance budding of eVP40-ΔPT/PY VLP egress (Figure 3). Once again, we found that expression of Bro1-V enhanced eVP40-ΔPT/PY VLP release by approximately 4-fold (Figure 3, compare lanes 2 and 4); however, expression of the Alix Bro1 or V-PRD domains had little to no effect on eVP40-ΔPT/PY VLP egress (Figure 3, compare lanes 2, 3, and 5). Together, these data demonstrate that the Bro1-V fragment rescues budding of the double L-domain deletion mutant of eVP40 most efficiently and suggest that Alix functions as an auxiliary target for eVP40 engagement of the ESCRT pathway in the absence of functional VP40 PTAP and PPEY L-domains.
Figure 3.
Bro1-V region of Alix rescues budding of eVP40-ΔPT/PY L-domain mutant most efficiently. HEK293T cells were mock-transfected (lane 1) or transfected with eVP40-ΔPT/PY plus empty vector (lane 2), Bro1 domain (lane 3), Bro1-V domain (lane 4), or V-PRD domain (lane 5). eVP40-ΔPT/PY, Bro1, Bro1-V, V-PRD, and actin were detected in cell extracts by Western blot, and eVP40-ΔPT/PY was detected in VLPs by Western blot. Numbers in parentheses represent fold change in levels of VLPs relative to control (lane 2). Abbreviations: VLP, virus-like particle; WT, wild type.
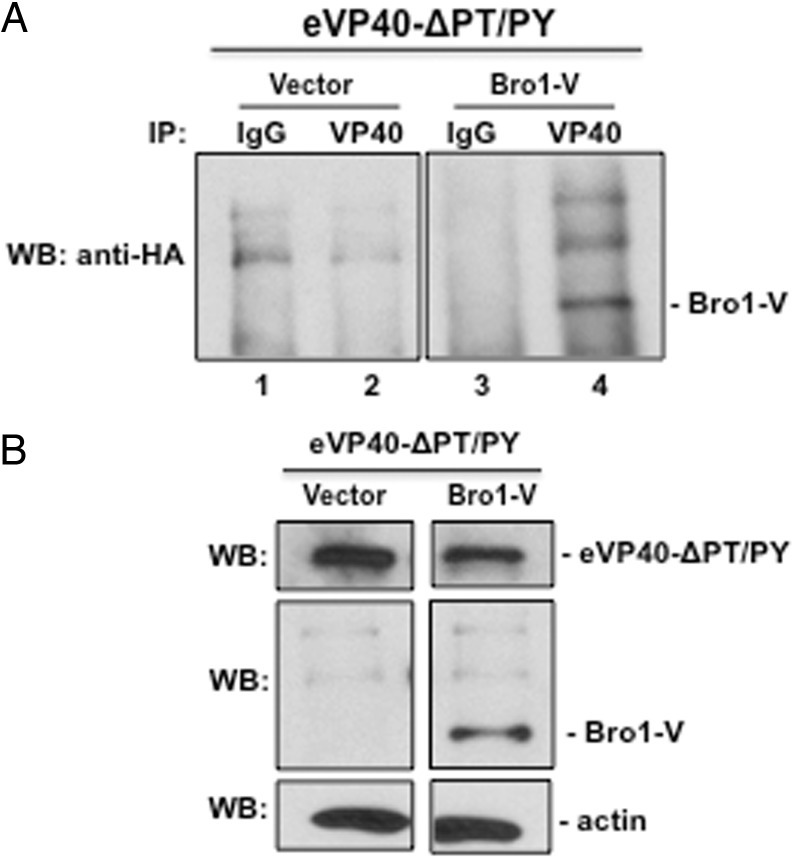
eVP40-ΔPT/PY Interacts With Alix Bro1-V
Because the Alix Bro1-V fragment rescues eVP40-ΔPT/PY VLP budding most efficiently and colocalized with eVP40-ΔPT/PY in confocal microscope images (data not shown), we asked whether the mechanism of rescue involved a direct interaction between these two proteins. To test this, eVP40-ΔPT/PY was expressed in COS-1 cells with vector alone or Bro1-V, and after 24 hours we quantified Bro1-V levels in eVP40-specific immune-precipitates (Figure 4A). Importantly, Alix Bro1-V was detected in eVP40-ΔPT/PY precipitates (lane 4), but was not detected in precipitates produced with control antiserum (Figure 4A, lane 3) or in precipitates from cells that expressed control vector (Figure 4A, lanes 1 and 2). We also demonstrate that expression levels of eVP40-ΔPT/PY, Bro1-V, and actin in appropriate samples are equivalent (Figure 4B). These results demonstrate that Bro1-V interacts with eVP40-ΔPT/PY, and it should be noted that Bro1-V was detected in eVP40-ΔPT/PY VLPs (Han and Harty, data not shown).
Figure 4.
eVP40-ΔPT/PY interacts with Bro1-V fragment of Alix. A, Total protein from cells transfected with eVP40-ΔPT/PY plus vector alone (lanes 1 and 2) or Bro1-V (lanes 3 and 4) was first immunoprecipitated with either rabbit pre-immune (IgG; lanes 1 and 3) serum, or anti-eVP40 (lanes 2 and 4) antiserum as indicated, and anti-HA antiserum was used to detect HA-tagged Bro1-V in the precipitated samples by Western analysis. B, Controls for expression of eVP40-ΔPT/PY, Bro1-V, and actin, are shown. Abbreviation: VLP, virus-like particle.
An eVP40 YPx(n)L/I Type L-domain Motif Controls Bro1-V Mediated Rescue of eVP40-ΔPT/PY VLPs
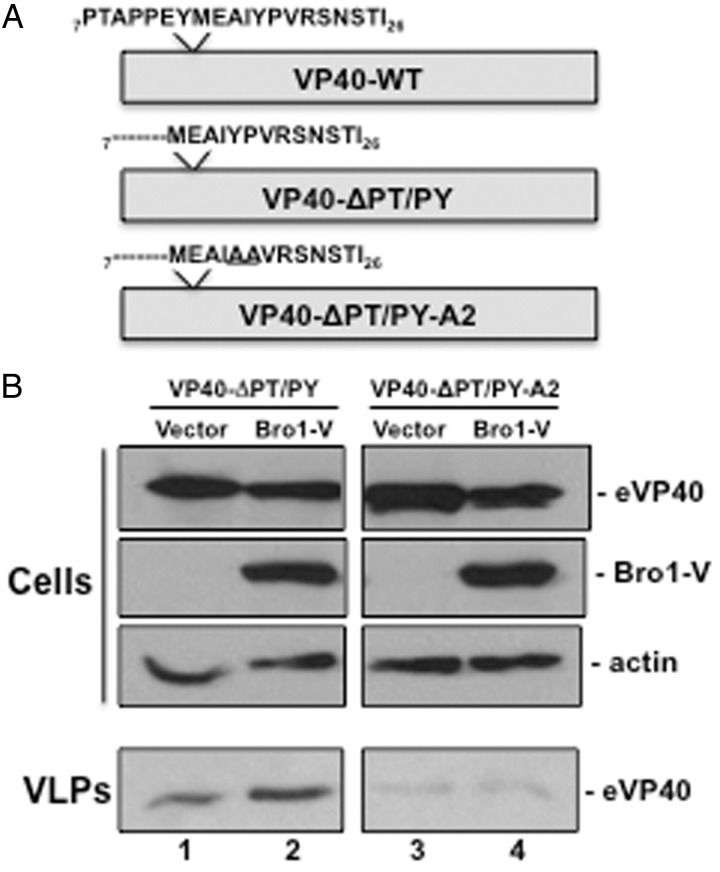
Interactions between Alix and other viral matrix proteins such as HIV-1 Gag are mediated by a viral YPx(n)L/I type L-domain motif. While not previously described, we have identified a similar motif near the eVP40 N-terminus (amino acids 18–26) (Figure 5A). We asked whether this putative L domain is required for the interaction we observed between Bro1-V and eVP40-ΔPT/PY and also the rescue of eVP40-ΔPT/PY VLP budding. To test this, we first mutated the YPx(6)I sequence to AAx(6)I in the eVP40-ΔPT/PY background to generate eVP40-ΔPT/PY-A2 (Figure 5A). Either eVP40-ΔPT/PY or eVP40-ΔPT/PY-A2 were expressed in HEK293T cells with either empty vector or Bro1-V (Figure 5B). Once again, we observed an enhancement in eVP40-ΔPT/PY VLP budding in the presence of Bro1-V vs vector control (Figure 5B, VLPs, lanes 1 and 2). In contrast, we did not observe any enhancement in budding of eVP40-ΔPT/PY-A2 VLPs in the presence of Bro1-V compared to vector control (Figure 5B, VLPs, lanes 3 and 4). It should also be noted that basal levels of budding of eVP40-ΔPT/PY-A2 VLPs in the presence of vector alone (Figure 5B, VLPs, lane 3) were lower than those of eVP40-ΔPT/PY VLPs (Figure 5B, VLPs, lane 1), further suggesting that the YPx(6)I may function as an alternative eVP40 L-domain motif. We also demonstrate that expression levels of eVP40-ΔPT/PY, eVP40-ΔPT/PY-A2, Bro1-V, and actin are equivalent in appropriate samples (Figure 5B).
Figure 5.
A cryptic YPx(n)L/I motif in eVP40-ΔPT/PY is required for VLP rescue by Bro1-V. A, Schematic diagram highlighting the amino acid sequence (aa 7–26) present at the N-termini of eVP40-WT, eVP40-ΔPT/PY, and eVP40-ΔPT/PY-A2. In addition to the deletion of L-domain amino acids 7-PTAPPEY-13, the eVP40-ΔPT/PY-A2 protein contains a YP>AA mutation at amino acids 18–19. B, HEK293 T cells were transfected with eVP40-ΔPT/PY or eVP40-ΔPT/PY-A2 plus empty vector or Bro1-V as indicated. eVP40-ΔPT/PY, eVP40-ΔPT/PY-A2, Bro1-V, and cellular actin were detected in cell extracts by Western blot, whereas eVP40-ΔPT/PY (lanes 1 and 2) and eVP40-ΔPT/PY-A2 (lanes 3 and 4) were detected in VLPs by Western blot. Abbreviations: VLP, virus-like particle; WT, wild type.
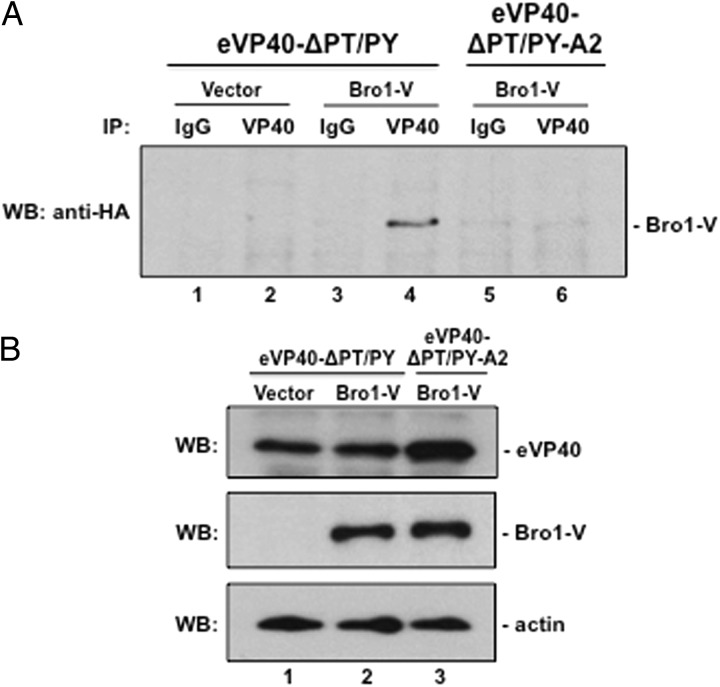
If, as our data suggest, the YPx(6)L/I motif in eVP40 represents a third viable L-domain motif, then we would predict that eVP40-ΔPT/PY, but not eVP40-ΔPT/PY-A2, should interact with Alix Bro1-V. To test this, eVP40-ΔPT/PY or eVP40-ΔPT/PY-A2 were expressed in COS-1 cells together with Bro1-V (or vector control) for 24 hours (Figure 6A). Although each of these proteins exhibited comparable levels of expression (Figure 6B), Bro1-V was detected in eVP40-ΔPT/PY but not eVP40-ΔPT/PY-A2 immune-precipitates (Figure 6A, compare lanes 4 and 6), Moreover, Bro1-V was not detected in vector (lane 2) or serum control immune-precipitates (lanes 1, 3, and 5). Together, these data suggest that the YPx(6)L/I motif mediates the interaction between VP40 and Bro1-V and establishes the YPx(6)L/I motif of eVP40 as a third functional L-domain motif capable of binding to the Bro1-V fragment of Alix and contributing to the egress of EBOV VLPs.
Figure 6.
eVP40-ΔPT/PY-A2 does not interact with Bro1-V. A, Total protein from cells transfected with eVP40-ΔPT/PY plus vector alone (lanes 1 and 2) or Bro1-V (lanes 3 and 4), and from cells transfected with eVP40-ΔPT/PY-A2 plus Bro1-V (lanes 5 and 6) were first immunoprecipitated with either rabbit pre-immune (IgG; lanes 1, 3, and 5) serum, or anti-eVP40 (lanes 2, 4, and 6) antiserum as indicated, and anti-HA antiserum was used to detect HA-tagged Bro1-V in the precipitated samples by Western analysis. B, Controls for expression of eVP40-ΔPT/PY (lanes 1 and 2), eVP40-ΔPT/PY-A2 (lane 3), Bro1-V, and actin are shown. Abbreviation: VLP, virus-like particle.
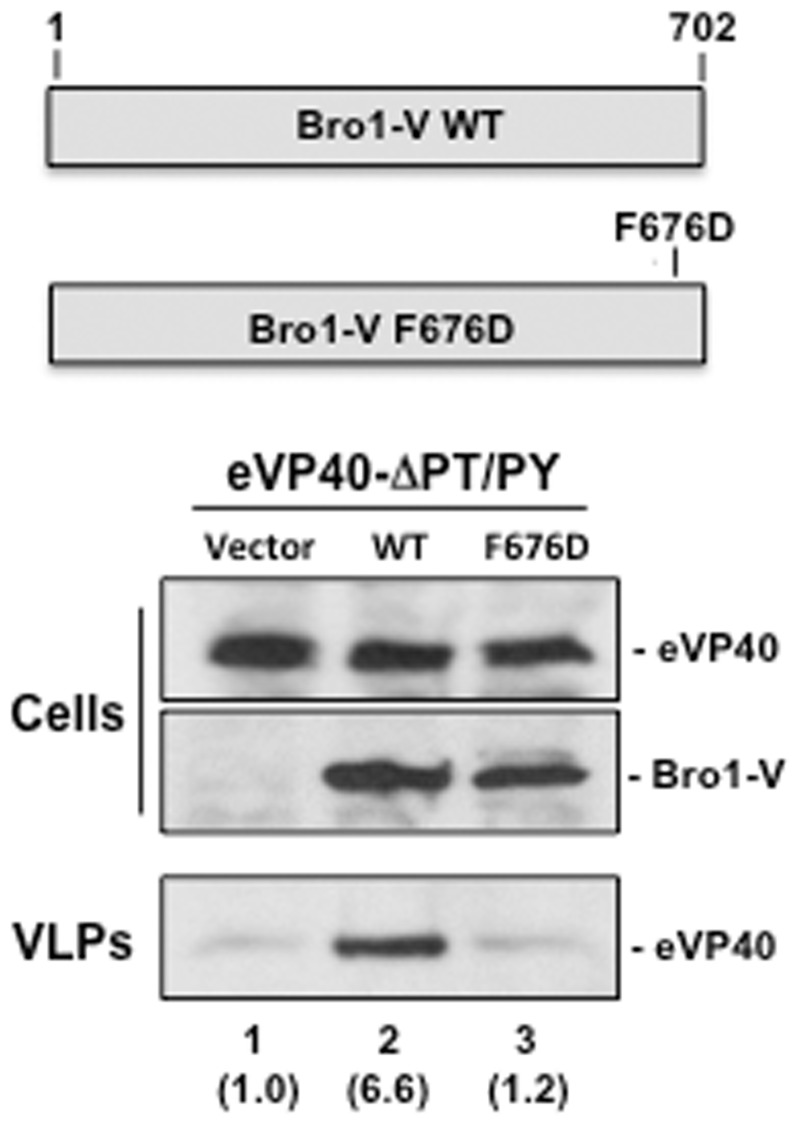
Alix F676 is Required for Rescue of eVP40-ΔPT/PY VLP Budding
Residue F676 within the Bro1-V region of Alix plays a critical role in the interaction between Alix and YPX(n)L/I L-domain motifs of EIAV p9 and HIV-1 p6 Gag proteins [24]. Indeed, a F676D mutation in Alix reduced its ability to bind to the HIV-1 p6 Gag L-domain and prevented Alix mediated rescue of budding of an HIV-1 Gag PTAP deletion mutant [24, 31, 33, 49]. Thus, we sought to determine whether the F676D mutation likewise prevented rescue of eVP40-ΔPT/PY VLP budding. Briefly, eVP40-ΔPT/PY was expressed in HEK293T cells with WT Bro1-V or F676D Bro1-V and VLPs in supernatants were quantified 24 hours post-transfection (Figure 7). Cells contained equivalent amounts of eVP40-ΔPT/PY and Bro1-V, and eVP40-ΔPT/PY VLP budding was rescued (>6-fold increase compared to control) by WT (Figure 7, VLPs, lane 2) but not F676D Bro1-V (Figure 7, VLPs, lane 3). These data suggest that the mechanism of Bro1-V rescue involves Bro1-V F676 recognition of and binding to the YPx(6)I motif of eVP40-ΔPT/PY.
Figure 7.
Bro1-V F676D fails to rescue budding of eVP40-ΔPT/PY VLPs. Schematic diagram of Bro1-V WT and Bro1-V F676D point mutant are shown. HEK293T cells were transfected with eVP40-ΔPT/PY plus empty vector (lane 1), Bro1-V WT (lane 2), or Bro1-V F676D mutant (lane 3). eVP40-ΔPT/PY, Bro1-V WT, and Bro1-V F676D were detected in cell extracts by Western blot, and eVP40-ΔPT/PY was detected in VLPs by Western blot. Numbers in parentheses represent fold change in levels of VLPs relative to control (lane 1). Abbreviations: VLP, virus-like particle; WT, wild type.
DISCUSSION
Host ESCRT protein Alix has a well-documented role in budding of retroviruses such as HIV-1 and EIAV [25, 33, 34, 38, 41, 43, 50], and Alix has also been implicated in egress of other RNA viruses [47, 51–54]. In this report, we present evidence for the first time showing that Alix has a role in budding of EBOV particles and have identified the structural domain of Alix that mediates its interaction with eVP40 required for budding. The V domain of Alix is critical for interactions with the YPx(n)L/I L-domain of HIV-1 and EIAV Gag [24, 25, 31, 33, 55], and overexpression of Alix rescues budding of PTAP and/or PPxY L-domain deletion mutants of HIV-1 and EIAV Gag to WT levels [26, 28, 39, 41, 56]. For HIV-1, the PTAP-Tsg101 interaction is thought to be the predominant relevant interaction for recruiting ESCRT components, while the retroviral YPx(n)L/I dependent interaction with Alix is thought to function as an alternate mechanism by which the virus can recruit ESCRT components to facilitate budding [38, 41, 42, 57].
Our results establish that expression of full-length Alix supports efficient egress of eVP40 VLPs, and specifically that the Bro1-V fragment of Alix is sufficient to rescue budding of a double PTAP/PPEY mutant of eVP40 to levels approaching those of eVP40-WT. Importantly, the Bro1-V rescue of eVP40-ΔPT/PY VLP budding involves a direct protein-protein interaction between the Alix V domain and the YPx(6)I motif at the N-terminus of eVP40. Moreover, a single F676D mutation within the Bro1-V domain, which is a residue required for its interactions with the HIV-1 p6 Gag YPx(n)L/I L-domain, abolished the ability of Bro1-V to rescue budding of eVP40-ΔPT/PY VLPs, as did a mutation that changed YPx(6)I to AAx(6)I within eVP40-ΔPT/PY.
Although expression of WT Alix enhances eVP40-ΔPT/PY VLP budding by approximately 2–3 fold, the Bro1-V fragment enhances eVP40-ΔPT/PY VLP budding by up to approximately 7-fold. This enhanced rescue by the shorter Bro1-V fragment may reflect a structural difference between the proteins. Indeed, intramolecular interactions are believed to lock Alix in a folded conformation [25, 58, 59], which is maintained by interactions between the PRD region and Bro1-V. Thus, in the absence of the PRD region, the Bro1-V fragment may adopt a more open conformation that facilitates its interaction with the YPx(6)I motif of eVP40-ΔPT/PY to promote more efficient budding. Additional studies will be required to fully elucidate the mechanisms that regulate Alix activation and facilitate its interaction with eVP40 to promote budding.
An important and intriguing unanswered question is why many enveloped RNA viruses like EBOV have redundant L-domain motifs. One possibility is that the requirement for each viral L-domain reflects cell specific differences in the expression levels of their interacting host protein(s). The EBOV VP40 PTAP/PPEY L-domains and their recruitment of host Tsg101 and Nedd4 are thought to be the primary driving force for EBOV budding, whereas the recruitment of Alix via the eVP40 YPx(6)I motif likely serves as a secondary or alternate mechanism for EBOV egress. It should be noted that the YPx(n)L/I-type motif is not conserved in RESTV and TAFV VP40 proteins, and perhaps this lack of conservation among the VP40 proteins reflects its role as an alternative L-domain motif.
In summary, we have identified and characterized a previously unrecognized third L-domain motif, YPx(6)I at aa 18–26 in EBOV VP40, which allows EBOV VP40 to interact with the host ESCRT protein Alix. The eVP40-Alix interaction can rescue a block in VLP formation that results from mutations in previously recognized eVP40 L-domain motifs and expands our current understanding of the role of ESCRT proteins in eVP40-mediated VLP formation. This new domain we have identified that mediates VP40-Alix interactions may represent a novel therapeutic target for control of EBOV infection. While these studies provide important new insight into the mechanisms of EBOV egress and host interactions, and while Alix may not be the primary ESCRT protein involved in budding, a better understanding of the interaction between Alix and EBOV VP40 may nonetheless pave the way for development of small molecule inhibitors of this virus-host interaction, that can be combined with antiviral therapeutics we have identified recently that target the PTAP-Tsg101 and PPxY-Nedd4 interactions [45, 46]. Indeed, drugs that target this YPx(6)I motif, if combined with PTAP and PPxY L-domain inhibitors, may prove to be more effective and less toxic than single L-domain inhibitors. The importance of identifying new targets and treatment modalities for this increasingly global public health challenge is highlighted by the extent of the current outbreak in West Africa and potential future outbreaks.
Notes
Acknowledgments. The authors wish to thank F. Bouamr for generously providing Alix plasmids/reagents. The authors also thank D. Argento for assistance in preparation of the article and figures, and members of the Harty and Freedman labs for fruitful discussion and suggestions.
Financial support. This work was supported in part by National Institutes of Health/National Institute of Allergy and Infectious Diseases grants AI102104 and AI103785 to R. N. H.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Feldmann H, Klenk HD. Filoviruses. In: Baron S, ed. Medical Microbiology. 4th ed Galveston, TX: 1996. [PubMed] [Google Scholar]
- 2.Feldmann H, Slenczka W, Klenk HD. Emerging and reemerging of filoviruses. Arch Virol Suppl 1996; 11:77–100. [DOI] [PubMed] [Google Scholar]
- 3.Hartlieb B, Weissenhorn W. Filovirus assembly and budding. Virology 2006; 344:64–70. [DOI] [PubMed] [Google Scholar]
- 4.Jasenosky LD, Kawaoka Y. Filovirus budding. Virus Res 2004; 106:181–8. [DOI] [PubMed] [Google Scholar]
- 5.Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci U S A 2000; 97:13871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Harty RN. Viral and host proteins that modulate filovirus budding. Future Virol 2010; 5:481–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson RF, Bell P, Harty RN. Effect of Ebola virus proteins GP, NP and VP35 on VP40 VLP morphology. Virol J 2006; 3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harty RN. No exit: targeting the budding process to inhibit filovirus replication. Antiviral Res 2009; 81:189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irie T, Licata JM, Harty RN. Functional characterization of Ebola virus L-domains using VSV recombinants. Virology 2005; 336:291–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Licata JM, Simpson-Holley M, Wright NT, Han Z, Paragas J, Harty RN. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J Virol 2003; 77:1812–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neumann G, Ebihara H, Takada A, et al. Ebola virus VP40 late domains are not essential for viral replication in cell culture. J Virol 2005; 79:10300–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noda T, Sagara H, Suzuki E, Takada A, Kida H, Kawaoka Y. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J Virol 2002; 76:4855–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urata S, Noda T, Kawaoka Y, Morikawa S, Yokosawa H, Yasuda J. Interaction of Tsg101 with Marburg virus VP40 depends on the PPPY motif, but not the PT/SAP motif as in the case of Ebola virus, and Tsg101 plays a critical role in the budding of Marburg virus-like particles induced by VP40, NP, and GP. J Virol 2007; 81:4895–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Timmins J, Schoehn G, Ricard-Blum S, et al. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J Mol Biol 2003; 326:493–502. [DOI] [PubMed] [Google Scholar]
- 15.Timmins J, Scianimanico S, Schoehn G, Weissenhorn W. Vesicular release of ebola virus matrix protein VP40. Virology 2001; 283:1–6. [DOI] [PubMed] [Google Scholar]
- 16.Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology 2006; 344:55–63. [DOI] [PubMed] [Google Scholar]
- 17.Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med 2001; 7:1313–9. [DOI] [PubMed] [Google Scholar]
- 18.Dolnik O, Kolesnikova L, Stevermann L, Becker S. Tsg101 is recruited by a late domain of the nucleocapsid protein to support budding of Marburg virus-like particles. J Virol 2010; 84:7847–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolesnikova L, Bohil AB, Cheney RE, Becker S. Budding of Marburgvirus is associated with filopodia. Cell Microbiol 2007; 9:939–51. [DOI] [PubMed] [Google Scholar]
- 20.Kolesnikova L, Strecker T, Morita E, et al. Vacuolar protein sorting pathway contributes to the release of Marburg virus. J Virol 2009; 83:2327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urata S, Yasuda J. Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola virus VP40. J Gen Virol 2010; 91:228–34. [DOI] [PubMed] [Google Scholar]
- 22.Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 2008; 372:221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossman JS, Lamb RA. Viral membrane scission. Annu Rev Cell Dev Biol 2013; 29:551–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat Struct Mol Biol 2008; 15:43–9. [DOI] [PubMed] [Google Scholar]
- 25.Zhai Q, Landesman MB, Chung HY, et al. Activation of the retroviral budding factor ALIX. J Virol 2011; 85:9222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dussupt V, Javid MP, Abou-Jaoude G, et al. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog 2009; 5:e1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sette P, Dussupt V, Bouamr F. Identification of the HIV-1 NC binding interface in Alix Bro1 reveals a role for RNA. J Virol 2012; 86:11608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sette P, Jadwin JA, Dussupt V, Bello NF, Bouamr F. The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4–1 to facilitate HIV-1 release through the LYPXnL L domain motif. J Virol 2010; 84:8181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sette P, Mu R, Dussupt V, et al. The Phe105 loop of Alix Bro1 domain plays a key role in HIV-1 release. Structure 2011; 19:1485–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 2003; 114:689–99. [DOI] [PubMed] [Google Scholar]
- 31.Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 2007; 128:841–52. [DOI] [PubMed] [Google Scholar]
- 32.Fujii K, Hurley JH, Freed EO. Beyond Tsg101: the role of Alix in ‘ESCRTing’ HIV-1. Nat Rev Microbiol 2007; 5:912–6. [DOI] [PubMed] [Google Scholar]
- 33.Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol 2007; 14:194–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin-Serrano J, Marsh M. ALIX catches HIV. Cell Host Microbe 2007; 1:5–7. [DOI] [PubMed] [Google Scholar]
- 35.Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol 2007; 81:6614–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lazert C, Chazal N, Briant L, Gerlier D, Cortay JC. Refined study of the interaction between HIV-1 p6 late domain and ALIX. Retrovirology 2008; 5:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou X, Pan S, Sun L, Corvera J, Lin SH, Kuang J. The HIV-1 p6/EIAV p9 docking site in Alix is autoinhibited as revealed by a conformation-sensitive anti-Alix monoclonal antibody. Biochem J 2008; 414:215–20. [DOI] [PubMed] [Google Scholar]
- 38.Fujii K, Munshi UM, Ablan SD, et al. Functional role of Alix in HIV-1 replication. Virology 2009; 391:284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Usami Y, Popov S, Popova E, Inoue M, Weissenhorn W, Göttlinger HG. The ESCRT pathway and HIV-1 budding. Biochem Soc Trans 2009; 37:181–4. [DOI] [PubMed] [Google Scholar]
- 40.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol 2011; 13:394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandrin V, Sundquist WI. ESCRT requirements for EIAV budding. Retrovirology 2013; 10:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma A, Bruns K, Roder R, et al. Solution structure of the equine infectious anemia virus p9 protein: a rationalization of its different ALIX binding requirements compared to the analogous HIV-p6 protein. BMC Struct Biol 2009; 9:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bissig C, Gruenberg J. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol 2014; 24:19–25. [DOI] [PubMed] [Google Scholar]
- 44.Lu J, Qu Y, Liu Y, et al. Host IQGAP1 and Ebola virus VP40 interactions facilitate virus-like particle egress. J Virol 2013; 87:7777–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han Z, Lu J, Liu Y, et al. Small-Molecule Probes Targeting the Viral PPxY-Host Nedd4 Interface Block Egress of a Broad Range of RNA Viruses. J Virol 2014; 88:7294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu J, Han Z, Liu Y, et al. A host-oriented inhibitor of Junin Argentine hemorrhagic fever virus egress. J Virol 2014; 88:4736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boonyaratanakornkit J, Schomacker H, Collins P, Schmidt A. Alix serves as an adaptor that allows human parainfluenza virus type 1 to interact with the host cell ESCRT system. PloS One 2013; 8:e59462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Usami Y, Popov S, Weiss ER, Vriesema-Magnuson C, Calistri A, Gottlinger HG. Regulation of CHMP4/ESCRT-III Function in Human Immunodeficiency Virus Type 1 Budding by CC2D1A. J Virol 2012; 86:3746–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fisher RD, Wang B, Alam SL, et al. Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem 2003; 278:28976–84. [DOI] [PubMed] [Google Scholar]
- 50.Gottlinger HG. How HIV-1 hijacks ALIX. Nat Struct Mol Biol 2007; 14:254–6. [DOI] [PubMed] [Google Scholar]
- 51.Patch JR, Han Z, McCarthy SE, et al. The YPLGVG sequence of the Nipah virus matrix protein is required for budding. Virol J 2008; 5:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dilley KA, Gregory D, Johnson MC, Vogt VM. An LYPSL late domain in the gag protein contributes to the efficient release and replication of Rous sarcoma virus. J Virol 2010; 84:6276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irie T, Inoue M, Sakaguchi T. Significance of the YLDL motif in the M protein and Alix/AIP1 for Sendai virus budding in the context of virus infection. Virology 2010; 405:334–41. [DOI] [PubMed] [Google Scholar]
- 54.Irie T, Shimazu Y, Yoshida T, Sakaguchi T. The YLDL sequence within Sendai virus M protein is critical for budding of virus-like particles and interacts with Alix/AIP1 independently of C protein. J Virol 2007; 81:2263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. Identification and structural characterization of the ALIX-binding late domains of simian immunodeficiency virus SIVmac239 and SIVagmTan-1. J Virol 2011; 85:632–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bello NF, Dussupt V, Sette P, et al. Budding of retroviruses utilizing divergent L domains requires nucleocapsid. J Virol 2012; 86:4182–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weissenhorn W, Gottlinger H. Essential ingredients for HIV-1 budding. Cell Host Microbe 2011; 9:172–4. [DOI] [PubMed] [Google Scholar]
- 58.Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. Structure of the Bro1 domain protein BROX and functional analyses of the ALIX Bro1 domain in HIV-1 budding. PloS One 2011; 6:e27466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou X, Si J, Corvera J, Gallick GE, Kuang J. Decoding the intrinsic mechanism that prohibits ALIX interaction with ESCRT and viral proteins. Biochem J 2010; 432:525–34. [DOI] [PMC free article] [PubMed] [Google Scholar]