Abstract
Marburg virus (MARV) and the ebolaviruses belong to the family Filoviridae (the members of which are filoviruses) that cause severe hemorrhagic fever. Infection requires fusion of the host and viral membranes, a process that occurs in the host cell endosomal compartment and is facilitated by the envelope glycoprotein fusion subunit, GP2. The N-terminal fusion loop (FL) of GP2 is a hydrophobic disulfide-bonded loop that is postulated to insert and disrupt the host endosomal membrane during fusion. Here, we describe the first structural and functional studies of a protein corresponding to the MARV GP2 FL. We found that this protein undergoes a pH-dependent conformational change, as monitored by circular dichroism and nuclear magnetic resonance. Furthermore, we report that, under low pH conditions, the MARV GP2 FL can induce content leakage from liposomes. The general aspects of this pH-dependent structure and lipid-perturbing behavior are consistent with previous reports on Ebola virus GP2 FL. However, nuclear magnetic resonance studies in lipid bicelles and mutational analysis indicate differences in structure exist between MARV and Ebola virus GP2 FL. These results provide new insight into the mechanism of MARV GP2–mediated cell entry.
Keywords: Marburg virus, viral membrane fusion, viral entry, envelope glycoprotein
The fusion of virus and host lipid membrane is a critical requirement for viral infection [1–3]. This process is likely favorable but is dependent on viral fusion proteins to decrease kinetic barriers associated with initial lipid mixing events. Envelope glycoproteins from different viruses accomplish the membrane fusion reaction by use of various mechanisms, but all share a common intermediate in which the glycoprotein fusion subunit spans the host and viral membranes (the so-called prehairpin or extended intermediate). In the structurally defined class I viruses (ie, those that contain α-helical content), it is believed that a triggering mechanism causes the prefusion trimeric envelope spike to undergo a conformational change in which a hydrophobic region at the N-terminus of the fusion subunit (fusion peptide, or fusion loop [FL]) inserts into the host cell membrane. This process leads to the extended intermediate conformation that is anchored into the host cell membrane by the N-terminal fusion peptide/loop and into the viral membrane by the C-terminal transmembrane domain. Collapse of the extended intermediate is believed to bring the 2 membranes into proximity; folding of the ectodomain into a highly thermostable 6-helix bundle provides the driving force for overcoming barriers associated with membrane fusion.
The family Filoviridae (the members of which are filoviruses) includes 5 species of ebolaviruses and Marburg virus (MARV); these pathogens cause a severe and rapidly progressing hemorrhagic fever [4]. Filoviruses are class I viruses, and the glycoprotein consists of 2 subunits, the surface subunit (GP1) and the transmembrane/fusion subunit (GP2) [5–9]; the domain organization of MARV GP1 and GP2 are specified in Figure 1A. The prefusion assembly consists of 3 copies each of GP1 and GP2, with the 2 subunits disulfide bonded to one another. In the current model for filovirus membrane fusion, the viral particle is first taken up into endosomal compartments, where host endoproteolytic processing removes major segments of GP1 [7, 10, 11]. This proteolysis step uncovers a receptor binding site that engages a critical host factor, Niemann-Pick type C1 [10–12]. Next, an unknown triggering mechanism causes insertion of the GP2 FL into the host cell membrane, leading to the GP2 extended intermediate. The GP2 ectodomains then collapse into the 6-helix bundle, which has been visualized for proteins from the Zaire ebolavirus species (EBOV) and MARV by X-ray crystallography [13–15], leading to initial lipid mixing events. Subsequent undefined events result in formation of a full fusion pore through which the viral contents are delivered to the host cytoplasm. Antibodies, peptides, and small molecules that disrupt any step of this process have antiviral activity in vitro and, in the case of antibodies, can provide postexposure protection of animals from viral challenge [16–20].
Figure 1.
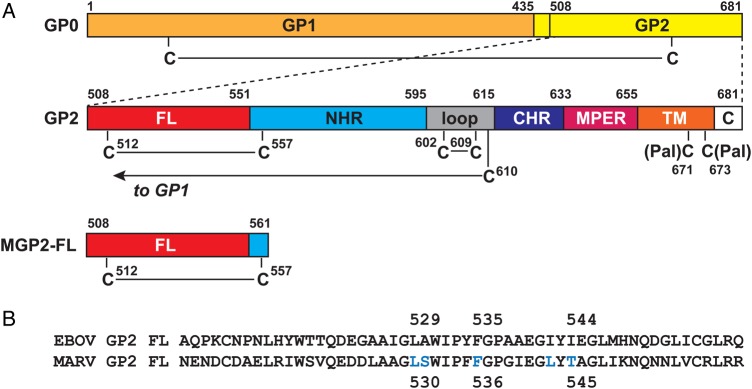
Schematic of the Marburg virus (MARV) GP2 fusion loop (FL). A, The MARV glycoprotein is produced as a single precursor (GP0) that is cleaved by furin during maturation into the 2 subunits GP1 and GP2. The primary sequence of GP2 contains the FL, N-heptad repeat region (NHR), loop, C-heptad repeat region (CHR), membrane proximal external region (MPER), transmembrane domain (TM; which includes 2 palmitoylated cysteines, as indicated), and C-terminal tail (C). The GP1 subunit is disulfide bonded to the GP2 subunit via an intramolecular disulfide bond from C610. The Marburg virus GP2 fusion loop (MGP2-FL) protein studied here consists of residues 508–561, with the native internal disulfide bond between C512 and C557. B, Amino acid sequence alignment between the EBOV GP2 FL and the MARV GP2 FL (note that residue numbering differs by 1 position). The positions of residues that form the core of the EBOV GP2 FL hydrophobic fist [9] are indicated. The aligning positions in MARV glycoprotein FL, as well as 2 others (all indicated in blue), were subjected to alanine scanning mutagenesis. Abbreviation: EBOV, Ebola virus.
From this fusion mechanism, it seems that the likely role of the GP2 FL is to anchor into the host cell membrane during formation of the extended intermediate conformation. However, the isolated EBOV GP2 FL exhibits lipid mixing activity that is dependent on a low pH–induced conformational change [9, 21]. These results suggest that the FL also plays a direct role in facilitating membrane fusion, perhaps by inducing general membrane instability at late-stage points in the host endosomal pathway. For EBOV GP2 FL, it has been demonstrated that this pH-dependent lipid perturbing activity is dependent on formation of a hydrophobic fist conformation that is stabilized by a core consisting of L529, F535, and I544. Mutation of these residues impaired lipid-perturbing activities of purified EBOV GP2 FL protein and entry by GP2-containing virus-like particles [9].
Here we describe studies on the FL region of MARV GP2, which has not been previously characterized in detail. Although we find that general aspects of the pH-dependent structure and membrane-disrupting activities are similar to those of the EBOV GP2 FL, nuclear magnetic resonance (NMR) and mutational analysis suggest that the structure and requirements for this activity differ. These results confirm a likely direct role for the FL region in filovirus membrane fusion and provide novel insight into MARV GP2 function.
MATERIALS AND METHODS
Protein Expression and Purification
A synthetic DNA fragment encoding the MARV GP2 FL followed by a C-terminal TEV cleavage site and polyhistidine (His) tag was obtained from a commercial supplier (Genewiz, South Plainfield, New Jersey). The gene was cloned into a pET-22b(+) vector (Novagen, Madison, Wisconsin), using NdeI and XhoI restriction sites, so that 2 His tags were encoded at the C-terminus of the coding sequence (the corresponding protein construct is referred to as “MGP2-FL”). The correct sequence of the constructed plasmid was confirmed by DNA sequencing (Genewiz). The FL protein was expressed in Escherichia coli strain BL21(DE3). The cells were induced with 1 mM isopropyl-β-d-thiogalactopyranoside at an A600 of 1.0 and harvested by centrifugation after shaking for 14–18 hours at 22°C. The cell pellet was lysed with Bugbuster (Merck Millipore, Billerica, Massachusetts) supplemented with deoxyribonuclease I (Invitrogen, Waltham, Massachusetts) in phosphate-buffered saline (PBS; pH 7.0), and the insoluble fraction, which contained MGP2-FL in inclusion bodies, was washed twice with PBS buffer. The pellet was then solubilized in 8 M urea in PBS. MGP2-FL was purified over a Ni–nitrilotriacetic acid agarose (Ni-NTA; Qiagen, Valencia, California) column, and refolded on-column by exchanging with n-dodecylphosphocholine (DPC; Avanti Polar Lipids, Alabaster, Alabama). Final purification was performed by gel filtration chromatography on a PD-10 column (GE Healthcare, Little Chalfont, United Kingdom) equilibrated with DPC to remove imidazole. The purified FL was then reconstituted into isotropic bicelles (q = 0.33) of 1,2-dimyristoyl-sn-glycero-3-phosphocholine/1,2-dihexanoyl-sn-glycero-3-phosphocholine (DMPC/DHPC; Avanti Polar Lipids) with a PD-10 column (GE Healthcare). To purify the protein in the absence of detergent, the FL was eluted from the Ni-NTA column with 8 M urea in PBS containing 250 mM imidazole and then refolded by stepwise dialysis, first into 10 mM sodium acetate (pH 4) with 2 mM tris(2-carboxyethyl)phosphine for 6 hours, followed by 10 mM sodium acetate (pH 4) for14 hours.
Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis method as directed by the manufacturer (Agilent Technologies, Santa Clara, California). The sequence of each mutant plasmid was confirmed by DNA sequencing. The expression and purification of each mutant followed the protocol described above for the wild-type FL.
Circular Dichroism (CD)
CD spectra of FL constructs were recorded at room temperature on a Jasco J-815 spectrometer with a 1-mm path-length cuvette. Protein concentrations for CD were 40 µM, as determined by the absorbance at 280 nm. Full-wavelength spectra were obtained with a 0.5-nm step size and represent the average of 2 scans. The signal was converted to mean molar ellipticity (θ), using the equation: θ (in deg cm2 dmol−1) = millidegrees/[path length in millimeters × molar protein concentration × number of residues].
Analytical Ultracentrifugation
A sedimentation velocity study of MGP2-FL was conducted using the absorption optics of a Beckman Optima XL-I analytical ultracentrifuge with samples loaded into 2-sector cell assemblies run in the AN-60Ti rotor. Boundary movement was followed at 280 nm during centrifugation at 58 000 rpm (271 273g) and 20°C in buffer containing 10 mM sodium acetate, 150 mM NaCl with 0.5% DPC, or 1% bicelles at pH 4. D2O was used to density match the DPC or bicelles present in the buffer [22–24]. Between 60 and 70 scans were collected over the course of the sedimentation runs, of which a subset, beginning with those in which a clear plateau is evident between the meniscus and the boundary, was selected for time-derivative analysis, using DCDT+, version 2.4.2, by John Philo [25, 26]. The FL was analyzed at a concentration of 70 µM, and the corresponding buffer was used to blank each sample. Values of the buffer density and viscosity were calculated from the composition (including the D2O but neglecting the detergent), using Sedenterp, version 20120828 Beta (available at: http://sednterp.unh.edu/#). The partial specific volume of the FL was calculated from its sequence, also by using Sedenterp. The sedimentation parameters were corrected to standard conditions (20, w) using these values.
Liposomal Fusion Activity
To prepare large unilamellar vesicles with encapsulated fluorescent dye, lipid dispersions of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (4:1 molar ratio) were resuspended with HEPES buffer containing the fluorescent dye 8-aminonapthalene-1,3,6 trisulfonic acid (ANTS) and its quencher p-xylene-bis-pyridinium bromide (DPX). Ten cycles of freeze/thaw and 11 rounds of extrusion with a 100-nm membrane were performed to generate homogenous large unilamellar vesicles. Next, a 10-mL Sepharose CL-2B column was used to isolated liposomes with ANTS/DPX for the following assays. Lipid concentrations were determined using an organic phosphate assay [27]. An Infinite M1000 PRO plate reader was used to monitor liposomal content release induced by the FL constructs. Excitation was at 355 nm with an 8-nm slit width, and emission was monitored at 520 nm with a 12-nm slit width. Varying concentrations of FL (purified without detergent) were added to ANTS/DPX-encapsulated liposomes to test for their fusion activity. Buffer alone was used as baseline (0% content release), while buffer with 0.3% Triton was used as 100% content release.
Nuclear Magnetic Resonance
Isotopically labeled MGP2-FL was expressed in M9 minimum medium and purified as described above. Purified protein samples were reconstituted into 0.5% DPC micelles or 15% DMPC/DHPC bicelles (q = 0.33) and concentrated to 200 µM for NMR experiments. The NMR experiments were recorded on a Bruker Avance III 600 MHz spectrometer, using a Cryo-TCI probe, or on a Varian Inova 600 MHz spectrometer, using a Cryo-HXY probe. 15N-HSQC, HNCA, HNcoCA, HNCACB, HNCO, and HNcaCO experiments were recorded on uniformly 13C, 15N–enriched MARV GP2 FL samples in bicelles for backbone resonance assignments. All 3D experiments were recorded with nonuniform sampling and processed with MddNMR [28] and NMRPipe [29]. Referencing was made with respect to 4,4-dimethyl-4-silapentane-1-sulfonic acid.
RESULTS AND DISCUSSION
Protein Expression, Purification, and Reconstitution
The predicted MARV GP2 FL includes residues 508–551 of glycoprotein, as well as a single cysteine (C512) that is thought to disulfide bond to C557 contained in the N-heptad repeat region (NHR; Figure 1) [8]. We therefore produced a construct (MGP2-FL) that consists of residues 508–561, to allow disulfide bond formation between these 2 cysteine residues, presumably important for structure and mechanism of this region. MGP2-FL could be expressed in good yield from E. coli, and formation of the disulfide bond was confirmed by the Ellman test (data not shown). We found that MGP2-FL could be reconstituted effectively in membrane-like environments such as DPC micelles and DMPC/DHPC bicelles.
CD Studies
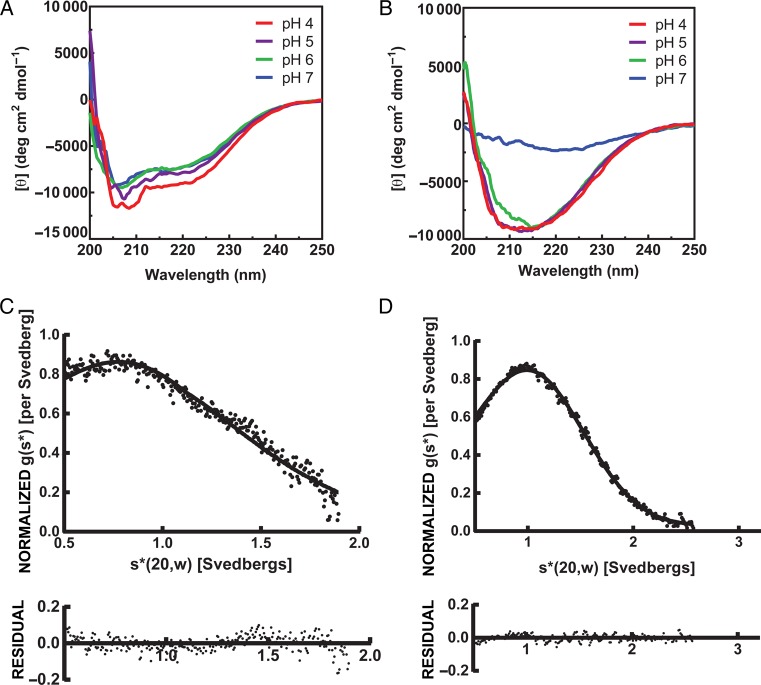
To explore whether the MGP2-FL structure exhibits a pH dependence, we recorded CD spectra under a variety of buffer conditions in both DPC micelles and lipid bicelles (Figure 2A and 2B). We found that MGP2-FL undergoes pH-dependent conformational changes in both DPC micelles and lipid bicelles, with the conformational change being more pronounced in bicelles. The overall spectra were consistent with an α-helical structure in DPC micelles, with slightly weakening intensity upon increasing pH. In lipid bicelles at pH 4–6, the spectra are consistent with mixed structure, showing a broad minimum at approximately 220 nm, but at pH 7, the spectra are featureless, suggesting a lack of defined structure under these conditions. These data are somewhat consistent with findings on the EBOV GP2 FL [21], in that the structure is pH dependent. However, EBOV GP2 FL shows a partial α-helix CD signature across this pH range in DPC micelles but with differing absolute and relative intensities at 208 nm and 222 nm, a change that is more pronounced than we observed here in DPC micelles with MGP2-FL. Furthermore, the EBOV GP2 FL CD spectra were consistent with an α-helical structure in small unilamellar vesicles at low pH. The solution NMR structures of EBOV GP2 FL at both pH 7 and 5.5 contained α-helical segments, although the overall topology differed between these 2 conditions [21]. Here, it appears the MARV GP2 FL is not as strongly α-helical and that a very pronounced change in conformational preference (potentially unfolding) occurs at pH >6.
Figure 2.
Circular dichroism and analytical ultracentrifugation of the Marburg virus GP2 fusion loop (MGP2-FL). A and B, Circular dichroism of (40 µM) MGP2-FL in n-dodecylphosphocholine (DPC) micelles (A) or 1,2-dimyristoyl-sn-glycero-3-phosphocholine/1,2-dihexanoyl-sn-glycero-3-phosphocholine (DMPC/DHPC) bicelles (B) at several pH conditions. C and D, Velocity analytical ultracentrifugation of (70 µM) MGP2-FL in DPC micelles (C) or DMPC/DHPC bicelles (D) at pH 4.
NMR Analysis
To gain residue-specific information about MGP2-FL, we produced it in isotopically labeled form for study by heteronuclear NMR. Since the corresponding EBOV FL is structurally somewhat malleable, we first examined the influence of the lipid environment on the MGP2-FL by comparing 1H15N-HSQC fingerprints of the protein in DPC detergent micelles and in DMPC/DHPC bicelles at pH 4. While a number of the amide cross-peaks were generally similar in the 2 environments (Figure 3A), there were also quite a few significant differences in chemical shift positions. In DPC, the protein also showed a much wider range of cross-peak intensities, with approximately 5–10 of the amide cross-peaks being barely visible in DPC (eg, L526, W532, F535, and L559; Figure 3A), indicative of some dynamic conformational exchange. Given these differences, we chose to use bicelles for more-extensive structural characterization. In bicelles (and also in DPC micelles; data not shown), the location of numerous cross-peaks differed at pH 4 and 7, confirming CD observations that structural preferences vary as a function of pH (Figure 3B). The backbone 1H, 13C, and 15N resonances of the MGP2-FL at pH 4 were assigned by standard triple-resonance NMR methods. These chemical shifts were used to estimate the secondary structure propensities for each residue, using the program SSP [30]. Comparison the MGP2-FL secondary structural predictions in lipid bicelles with those based on previously reported EBOV GP2 FL chemical shift assignments in DPC micelles indicates several significant differences (Figure 3C), with the MGP2-FL showing considerably more helical character for residues 524–532 (MARV numbering) and even more-substantial differences for residues 534–536 that form the tip of the loop in EBOV GP2 FL structure.
Figure 3.
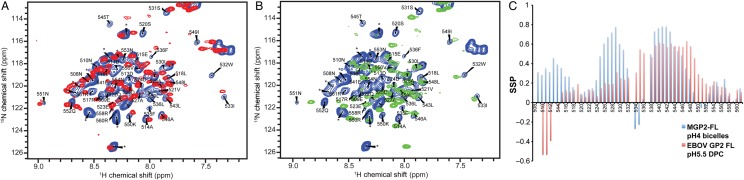
1H15N-HSQC spectra of the Marburg virus GP2 fusion loop (MGP2-FL) and secondary structure propensity (SSP)–based comparison with the Ebola virus (EBOV) GP2 FL. A, Overlay of HSQC spectra of MGP2-FL at pH 4 in 1,2-dimyristoyl-sn-glycero-3-phosphocholine/1,2-dihexanoyl-sn-glycero-3-phosphocholine (DMPC/DHPC) bicelles (blue) and n-dodecylphosphocholine (DPC) micelles (red). B, Overlay of HSQC spectra of MGP2-FL in DMPC/DHPC bicelles at pH 4 (blue) and pH 7 (green). Peaks marked with asterisks belong to the polyhistidine tag region in the construct. C, Comparison of the SSP [30] of MGP2-FL in DMPC/DHPC bicelles at pH 4 (blue) and EBOV GP2 FL in DPC at pH 5.5 (red). SSP values of EBOV GP2 FL were calculated using reported chemical shifts in previous work by Tamm et al [21]. Abbreviation: ppm, parts per million.
Analytical Ultracentrifugation
The EBOV glycoprotein prefusion structure, as well as both the EBOV and MARV GP2 postfusion structures, are trimeric outside of the context of membranes [5, 13, 15]. The molecular weights obtained from S20,w/D20,w values for MGP2-FL peptide are 7.15 kDa (68% confidence interval [CI], 6.79–7.53 kDa) and 14.52 kDa (68% CI, 13.96–15.07 kDa) for the peptide in DPC micelles and DMPC/DHPC bicelles, respectively (Figure 2C and 2D). These values are comparable to the calculated monomer mass of 9.38 kDa. Since MGP2-FL is a small, elongated peptide, deviation of the molecular weight determined by sedimentation velocity from the peptide mass is expected. Uncertainty in the partial specific volume in DPC micelles and bicelles also contributes to uncertainty in the measured molecular weight. However, both values are close to that of the monomer, confirming that the peptide has little propensity to trimerize under the experimental conditions analyzed.
In the EBOV prefusion glycoprotein structure, the GP2 FL adopts a hairpin-like structure, stabilized by interactions with each of the GP1 subunits with no direct interactions with other GP2 FL segments [5]. Therefore, it is reasonable to expect that, outside of the context of the prefusion spike, the FL itself would adopt a primarily monomeric conformation. In the postfusion GP2 structures of both EBOV and MARV, the NHR segment participates in interhelical trimer interactions in the region surrounding the C557 (MARV numbering), but in both crystallized constructs, the Cys was mutated to an alternative residue [13, 15]. Although MGP2-FL does contain portions of the NHR, these segments do not promote trimer formation in this context.
Pore-Forming Activity
The fusion peptide regions from a number of viral glycoproteins exhibit lipid perturbing activity. We explored the capacity of MGP2-FL to form pores in POPC:POPG vesicles, using an ANTS/DPX content release assay (Figure 4A). pH-dependent pore-forming activity was observed, with high activity at pH 4 and low activity at pH 7, both of which were dose dependent on MGP2-FL. These experiments were performed in the presence of excess Zn2+ at pH 4, which chelates His tags and prevents their protonation. Therefore, there is no contribution by protonation of the His tags to this pH-dependent behavior. These data are somewhat consistent with lipid-mixing experiments with the EBOV GP2 FL, which also shows pH-dependent activity. However, we note that it appears that the MGP2-FL has higher general activity than the EBOV GP2 FL because, in our hands, the MGP2-FL induces approximately 5% content release at protein to lipid ratios of 0.00027 (or a protein to liposome ratio of approximately 22), whereas 5% lipid mixing was observed for EBOV GP2 FL under similar conditions with similar liposomes [21]. We suggest that pore-forming activity is a higher threshold for membrane lytic or fusion activity since pore formation requires disruption of a full lipid bilayer, whereas lipid mixing requires only perturbation of the outer bilayer of ≥2 liposomes.
Figure 4.
Liposome content release assay and characterization of mutants. A, Release of 8-aminonapthalene-1,3,6 trisulfonic acid and p-xylene-bis-pyridinium bromide contents of wild-type Marburg virus GP2 fusion loop, as well as alanine mutants, at pH 4 and pH 7. B, Circular dichroism spectra of alanine mutant proteins at several pH conditions in 1,2-dimyristoyl-sn-glycero-3-phosphocholine/1,2-dihexanoyl-sn-glycero-3-phosphocholine bicelles at protein concentration of 40 µM.
Mutational Analysis of Core Residues
The EBOV GP2 FL undergoes a pH-dependent conformational change in DPC micelles; at low pH (5.5), a number of hydrophobic residues are exposed at the tip of the fist that is hypothesized to represent a fusion-competent conformation [9]. It is postulated that core interactions between L529, F535, and I544 are required to support this conformation, and mutation of L529 and I544 to alanine (each as a single mutation or as the double mutant) resulted in entry defects for GP-containing virus-like particles. The corresponding positions in MARV are similar at 2 of these positions (L530 and F536), but T545 (MARV) contains a hydroxyl group rather than being purely aliphatic, as is found at the aligning position in EBOV (I544) (Figure 1B). To explore whether similar interactions might be important for the MARV FL, we generated several single alanine point mutants—L530A, S531A, L543A, T545A, and F536A—and assessed their structural properties by CD and their capacity to induce leakage from liposomes (Figure 4). The L530A, T545A, and F536A mutations were intended to determine whether the same core residues in the EBOV GP2 FL play a role for MARV GP2. When considered in the context of the EBOV GP2 FL (pH 5.5) structure, there is the potential for a hydrogen bonding interaction between S531 and T545, and therefore the S531A mutation was used to explore this possibility. Also, since T545 contains some polarity, the neighboring aliphatic residue (L543) is a possible candidate for inclusion in the hydrophobic core, and therefore this residue was also mutated.
All mutants were found to have similar pH-dependent CD signatures relative to the wild-type protein, with a broad minimum at approximately 220 nm at pH <6 and featureless spectra at pH 7 in lipid bicelles. In addition, all mutants exhibited a pH-dependent liposome leakage activity similar to that of wild type, with high activity at pH 4 but low activity at pH 7. At pH 4, aliphatic mutants L530A, L543A, and F536A all showed similar levels of activity to the wild-type MGP2-FL, but S531A and T545A were slightly attenuated, showing approximately 50%–60% leakage at higher protein concentrations. Nonetheless, these differences were relatively subtle and overall the structural and lipid-perturbing behavior of all mutants was similar to wild-type MGP2-FL, thus suggesting specific side chain–side chain contacts among these positions do not contribute substantially to fusion active conformations.
Conclusions and Implications for Viral Membrane Fusion
Here, we have demonstrated that the MARV GP2 FL exhibits pH-dependent structure and pore-forming activity. While general aspects of these features resemble EBOV GP2 FL, there are clear differences. First, the CD and NMR chemical shift comparisons indicate that MARV GP2 FL adopts a distinct structure from EBOV GP2 FL at low pH and that the nature of the pH-dependent transition is sharper. The MARV GP2 FL undergoes a dramatic change between pH 6 and 7 in lipid bicelles, likely from a mixed structure to less structured, as opposed to the gradual conformational change across pH 4–7 that was reported with EBOV GP2 FL [21]. Also, the EBOV GP2 FL contains substantial α-helical character throughout this pH range, whereas the CD indicates a mixed structure for MARV GP2 FL at the lower pHs. The mutational data clearly indicate that L530, F536, and T545 of MARV GP2 FL are not as critical for structure or function as the corresponding residues in EBOV GP2 FL, which are proposed to form a core that supports the hydrophobic fist fusion-active conformation. Neighboring residues such as S531 and L543 also do not appear to play a strong role in stabilizing fusion-active conformations. Together, these results suggest that the fusion-active conformation of the MARV GP2 FL is distinct from that of EBOV GP2 FL but that there is convergence on the general pH-dependent activity. Higher-resolution structural studies will provide deeper insight into mechanism of the MARV GP2 FL relative to EBOV GP2 FL.
Functional data supporting a fusogenic role for the EBOV GP2 FL and, here, the MARV GP2 FL are strong, but the precise role of these segments in the membrane fusion cascade remains unclear. The lipid mixing activity of EBOV GP2 FL was modest, 5% under optimal conditions, suggesting that this segment may require other aspects for creation of a full fusion pore. However, here we have shown that pore-forming activity of MARV GP2 FL is strong under some conditions and this activity may be significant for membrane fusion in this case. The pH-dependent fusion activity is consistent with a model whereby the most active fusogenic conformations of GP2 are not deployed until late in the endosomal maturation pathway. For example, we have shown that the stability of the 6-helix bundle of the GP2 ectodomains from both EBOV and MARV are highly sensitive to pH [15, 31, 32]. Formation of the 6-helix bundle, which is postulated to draw the host and cell membranes into proximity, is also highly promoted under conditions of the matured endosome. Whether the fusogenic, or lipid-perturbing activities, of the FL participate at the stage of the extended intermediate or after collapse of the 6-helix bundle remains to be determined. In other systems, it has been proposed that membrane active segments of fusion subunits, such as the FL, participate in the transition from the hemifusion intermediate to the full fusion pore [33–35]. Nevertheless, the results reported here confirm a likely role for the FL during the membrane fusion cascade and highlight the requirement for this segment for viral entry.
Notes
Financial support. This work was supported by the National Institutes of Health (grants AI090249 [to J. R. L.] and GM072085 [to M. E. G.] and award 1S10OD016305).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol 2008; 15:690–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem 2001; 70:777–810. [DOI] [PubMed] [Google Scholar]
- 3.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins, multiple variations on a common theme. Crit Rev Biochem Mol Biol 2008; 43:189–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuhn JH, Becker S, Ebihara H, et al. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch Virol 2010; 155:2083–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 2008; 454:177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bale S, Dias JM, Fusco ML, et al. Structural basis for differential neutralization of ebolaviruses. Viruses 2012; 4:447–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 2005; 308:1643–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JE, Saphire EO. Neutralizing ebolavirus: structural insights into the envelope glycoprotein and antibodies targeted against it. Curr Opin Struct Biol 2009; 19:408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregory SM, Larsson P, Nelson EA, Kasson PM, White JM, Tamm LK. Ebolavirus entry requires a compact hydrophobic fist at the tip of the fusion loop. J Virol 2014; 88:6636–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller EH, Chandran K. Filovirus entry into cells - new insights. Curr Opin Virol 2012; 2:206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharyya S, Mulherkar N, Chandran K. Endocytic pathways involved in filovirus entry: advances, implications and future directions. Viruses 2012; 4:3647–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carette JE, Raaben M, Wong AC, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 2011; 477:340–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weissenhorn W, Carfí A, Lee K-H, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell 1998; 2:605–16. [DOI] [PubMed] [Google Scholar]
- 14.Malashkevich VN, Schneider BJ, McNally ML, Milhollen MA, Pang JX, Kim PS. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc Natl Acad Sci U S A 1999; 96:2662–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koellhoffer JF, Malashkevich VN, Harrison JS, et al. Crystal structure of the Marburg virus GP2 core domain in its post-fusion conformation. Biochemistry 2012; 51:7665–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller EH, Harrison JS, Radoshitzky SR, et al. Inhibition of Ebola Virus Entry by a C-peptide Targeted to Endosomes. J Biol Chem 2011; 286:15854–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins CD, Koellhoffer JF, Chandran K, Lai JR. C-peptide inhibitors of Ebola virus glycoprotein-mediated cell entry: Effects of conjugation to cholesterol and side chain-side chain crosslinking. Bioorg Med Chem Lett 2013; 23:5356–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basu A, Li B, Mills DM, et al. Identification of a small-molecule entry inhibitor for filoviruses. J Virol 2011; 85:3106–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Côté M, Misasi J, Ren T, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 2011; 477:344–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saphire EO. An update on the use of antibodies against the filoviruses. Immunotherapy 2013; 5:1221–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory SM, Harada E, Liang B, Delos SE, White JM, Tamm LK. Structure and function of the complete internal fusion loop from Ebolavirus glycoprotein 2. Proc Natl Acad Sci U S A 2011; 108:11211–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korendovych IV, Senes A, Kim YH, et al. De novo design and molecular assembly of a transmembrane diporphyrin-binding protein complex. J Am Chem Soc 2010; 132:15516–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanford C, Nozaki Y, Reynolds JA, Makino S. Molecular characterization of proteins in detergent solutions. Biochemistry 1974; 13:2369–76. [DOI] [PubMed] [Google Scholar]
- 24.Tanford C, Reynolds JA. Characterization of membrane proteins in detergent solutions. Biochim Biophys Acta 1976; 457:133–70. [DOI] [PubMed] [Google Scholar]
- 25.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem 2006; 354:238–46. [DOI] [PubMed] [Google Scholar]
- 26.Stafford WF. Boundary analysis in sedimentation transport experiments: A procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal Biochem 1992; 203:295–301. [DOI] [PubMed] [Google Scholar]
- 27.Ames BN. Assay of inorganic phosphate, total phosphate and phosphatases. Meth Enzym 1966; 8:115–8. [Google Scholar]
- 28.Kazimierczuk K, Orekhov VY. Accelerated NMR spectroscopy by using compressed sensing. Angew Chem-Int Edit 2011; 50:5556–9. [DOI] [PubMed] [Google Scholar]
- 29.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a multidimensional spectral processing system based on unix pipes. J Biomol NMR 1995; 6:277–93. [DOI] [PubMed] [Google Scholar]
- 30.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: Implications for fibrillation. Prot Sci 2006; 15:2795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison JS, Koellhoffer JF, Chandran K, Lai JR. Marburg virus glycoprotein GP2: pH-dependent stability of the ectodomain α-helical bundle. Biochemistry 2012; 51:2515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison JS, Higgins CD, Chandran K, Lai JR. Designed protein mimics of the Ebola virus glycoprotein GP2 -helical bundle: stability and pH effects. Prot Sci 2011; 20:1587–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reuven EM, Dadon Y, Viard M, Manukovsky N, Blumenthal R, Shai Y. HIV-1 gp41 Transmembrane Domain Interacts with the Fusion Peptide: Implication in Lipid Mixing and Inhibition of Virus-Cell Fusion. Biochemistry 2012; 51:2867–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen T, Pevsner-Fischer M, Cohen N, Cohen IR, Shai Y. Characterization of the interacting domain of the HIV-1 fusion peptide with the transmembrane domain of the T-cell receptor. Biochemistry 2008; 47:4826–33. [DOI] [PubMed] [Google Scholar]
- 35.Buzon V, Natrajan G, Schibli D, Campelo F, Kozlov MM, Weissenhorn W. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Path 2010; 6:e1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]