Abstract
Background. Marburg virus (MARV) is an emerging zoonotic pathogen that causes hemorrhagic fever. MARV VP24 (mVP24) protein interacts with the host cell protein Kelch-like-ECH-associated protein 1 (Keap1). Keap1 interacts with and promotes the degradation of IκB kinase β (IKKβ), a component of the IκB kinase (IKK) complex that regulates nuclear factor-κB (NF-κB) activity. We studied whether mVP24 could relieve Keap1 repression of the NF-κB pathway.
Methods. Coimmunoprecipitation assays were used to examine the interaction between Keap1 and IKKβ in the presence of wild-type mVP24 and mutants of mVP24 defective for binding to Keap1. Western blotting was used to determine levels of IKKβ expression in the presence of Keap1 and mVP24. NF-κB promoter-luciferase assays were used to determine the effect of mVP24 on Keap1-induced repression of activity.
Results. Expression of wild-type mVP24 disrupted the interaction of IKKβ and Keap1, whereas weakly interacting and noninteracting mVP24 mutants did not disrupt the interaction between Keap1 and IKKβ. The interaction of mVP24 with Keap1 enhanced IKKβ levels in the presence of Keap1. The interaction of mVP24 with Keap1 also relieved Keap1 repression of NF-κB reporter activity.
Conclusions. mVP24 can relieve Keap1 repression of the NF-κB pathway through its interaction with Keap1.
Keywords: Filovirus, Marburg virus, VP24, Keap1, NF-κB pathway
Marburg virus (MARV) and Ebola virus (EBOV), members of the family Filoviridae, are emerging zoonotic pathogens that cause severe disease in humans. Filoviruses encode a multifunctional VP24 protein that plays an important role in the formation of viral nucleocapsids, release of infectious virus particles, and modulation of viral RNA synthesis [1–7]. EBOV VP24 also inhibits the interferon signaling pathway through interaction with select karyopherin α proteins, preventing nuclear accumulation of tyrosine-phosphorylated signal transducer and activator of transcription 1 [8–11]. MARV VP24 (mVP24), however, does not inhibit interferon signaling or detectably interact with karyopherin α proteins [12]. Rather, mVP24 interacts with the cellular protein Kelch-like ECH-associated protein 1 (Keap1) [13, 14]. This interaction leads to the activation of a cytoprotective antioxidant response by disrupting the interaction of Keap1 with the antioxidant transcription factor nuclear factor (erythroid-derived 2)–like 2 (Nrf2) [13, 14].
Keap1, an E3 ubiquitin ligase, plays a role in the regulation of several cellular pathways through the interaction of its Kelch domain with cellular proteins, including Nrf2 and IκB kinase β (IKKβ) [15–18]. IKKβ, a serine/threonine kinase, is a member of the IκB kinase (IKK) complex. When the IKK complex is activated by a variety of stimuli, it targets the NF-κB inhibitory protein IκB for proteasomal degradation, causing the release of NF-κB transcription factors [19]. The interaction of Keap1 with IKKβ occurs through an ETGE motif in IKKβ, which is similar to the Keap1 binding motif of Nrf2 (ie, DXETGE) [15, 16, 20]. Keap1 targets IKKβ for degradation, leading to suppression of tumor necrosis factor α (TNF-α)-induced NF-κB reporter activity [15, 16].
As mVP24 can disrupt the interaction of Nrf2 and Keap1, the present study sought to determine whether mVP24 could disrupt the interaction of Keap1 and IKKβ, relieving Keap1 suppression on the NF-κB pathway. We demonstrate that the interaction between mVP24 and Keap1 disrupts the interaction of IKKβ with Keap1, stabilizing IKKβ expression in the presence of Keap1. The interaction of mVP24 and Keap1 also relieves Keap1 repression of NF-κB–dependent gene expression induced by TNF-α. These data suggest that mVP24 relieves the negative regulation of the NF-κB pathway mediated by Keap1.
METHODS
Coimmunoprecipitation Assays
Twenty-four hours after transfection with the indicated plasmids, HEK293T cells were lysed in NP-40 lysis buffer (50 mM Tris [pH 7.5], 280 mM NaCl, 0.5% Nonidet P-40, 0.2 mM ethylenediaminetetraacetic acid, 2 mM ethylene glycol tetraacetic acid, 10% glycerol, and protease inhibitor [cOmplete; Roche]). Anti-HA beads (Sigma-Aldrich) were incubated with lysates for 1 hour at 4°C, washed 5 times in NP-40 lysis buffer, and eluted by boiling for 5 minutes in sample buffer.
Cells
HEK293T cells were maintained in Dulbecco's minimal essential medium supplemented with 10% fetal bovine serum and cultured at 37°C and 5% CO2.
Plasmids
pRL-Flag-IKKβ was a kind gift from Dr Adrian Ting (Icahn School of Medicine at Mount Sinai). pCAGGS-HA-Keap1, Flag-Keap1, Flag-mVP24, and mVP24 mutants have previously been described [13]. The NF-κB firefly luciferase plasmid used contains 5 repeats of a canonical NF-κB promoter.
Antibodies and Cytokines
Monoclonal mouse anti-Flag M2, polyclonal rabbit anti-Flag, monoclonal mouse anti-HA, and polyclonal rabbit anti-HA antibodies were purchased from Sigma Aldrich. Recombinant human TNF-α was purchased from PeproTech.
Western Blotting
Lysates were run on 10% acrylamide sodium dodecyl sulfate–polyacrylamide gels (Lonza) and transferred to polyvinylidene difluoride membranes, which were probed with anti-FLAG and anti-HA antibodies and developed using Western Lightning ECL (Perkin-Elmer). Relative IKKβ levels were analyzed by ImageJ, normalizing IKKβ levels to the β-tubulin loading control.
NF-κB Reporter Assay
HEK293T cells were transfected with a NF-κB promoter firefly luciferase plasmid, a constitutively active Renilla luciferase plasmid (pRL-TK; Promega), and the indicated expression plasmids. Forty-eight hours after transfection, cells were treated with TNF-α (10 ng/mL) for 6 hours, after which a dual luciferase assay (Promega) was performed. Firefly luciferase values were normalized to Renilla luciferase values. Error bars represent the mean and standard error of the mean of triplicate samples. Statistical significance was assessed by 1-way analysis of variance and the Student t test.
RESULTS
mVP24 Disrupts IKKβ Binding to Keap1
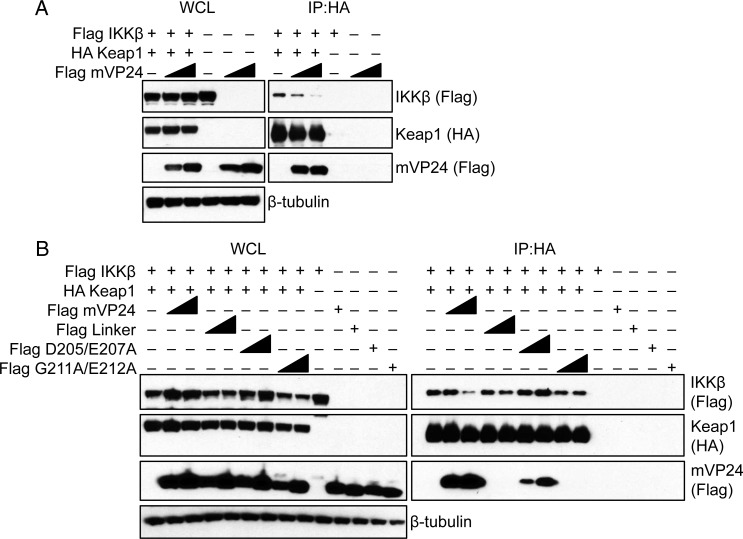
Previous studies identified Keap1 as an interacting partner of IKKβ [15, 16]. This interaction occurs through an ETGE motif on IKKβ and the Kelch domain of Keap1. Similarly, mVP24 has been shown to interact with the Kelch domain of Keap1 through a motif that also contains acidic residues upstream of a GE (ie, DIEPCCGE) [13]. To determine whether mVP24 can disrupt the interaction of Keap1 and IKKβ, we used a coimmunoprecipitation assay, expressing HA-Keap1, Flag-IKKβ, and increasing concentrations of Flag-mVP24 in HEK293T cells as indicated in Figure 1A. Twenty-four hours after transfection, cells were lysed and HA-Keap1 was immunoprecipitated. As expected, Flag-IKKβ coprecipitated with HA-Keap1 in the absence of Flag-mVP24 expression (Figure 1A). Upon expression of Flag-mVP24, a reduction in the level of IKKβ that coprecipitated with Keap1 was detected, with the reduction corresponding to the amount of mVP24 expressed (Figure 1A). To determine whether the interaction between mVP24 and Keap1 was required for the disruption of IKKβ and Keap1, previously described VP24 mutants with decreased or absent Keap1 binding were assessed [13]. Specifically, we tested the mVP24 mutants Linker and G211A/E212A, which no longer interact with Keap1, and D205A/E207A, which has decreased interaction with Keap1. Upon immunoprecipitation of HA-Keap1, the interaction with IKKβ was disrupted by wild-type mVP24, while the weakly interacting and noninteracting mVP24 mutants did not disrupt the IKKβ interaction with Keap1 (Figure 1B). This indicates that interaction of mVP24 with Keap1 is required to disrupt the Keap1:IKKβ interaction.
Figure 1.
Marburg virus VP24 (mVP24) disrupts IκB kinase β (IKKβ) binding to Kelch-like-ECH-associated protein 1 (Keap1). A, Coimmunoprecipitations (coIPs) with HA antibody were performed on lysates of HEK293T cells cotransfected with plasmids for HA Keap1, wild-type Flag mVP24, and Flag IKKβ. The coIP was performed 5 times with similar results; representative Western blots are shown for Flag and HA. B, HA Keap1, Flag IKKβ, and wild-type Flag mVP24 or mutants were analyzed by coIP as describe in panel A. The coIP was performed twice; representative Western blots are shown. Abbreviations: IP, immunoprecipitation; WCL, whole-cell lysate.
mVP24 Prevents Keap1-Targeted Degradation of IKKβ
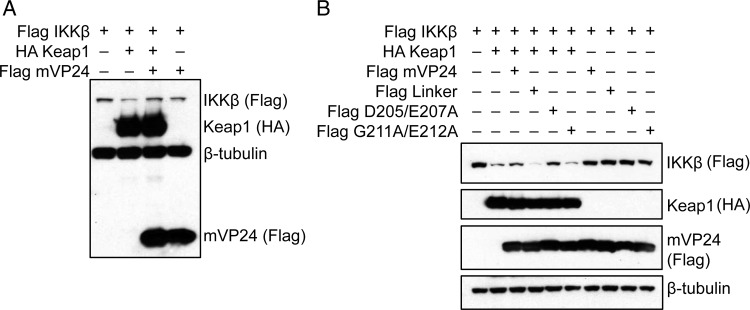
The interaction between Keap1 and IKKβ was previously demonstrated to result in the degradation of IKKβ [15, 16]. Because mVP24 disrupts the interaction of Keap1 and IKKβ, we asked whether mVP24 could prevent this loss of IKKβ. HEK293T cells were transfected with HA-Keap1, Flag-IKKβ, and Flag-mVP24 as indicated in Figure 2A. Twenty-four hours after transfection, cells were lysed and the levels of IKKβ were examined by Western blot. Coexpression of Keap1 and IKKβ resulted in a decrease in IKKβ as expected, with relative IKKβ expression levels decreasing to one-third of the level detected in the absence of Keap1 (Figure 2A). Expression of mVP24 with Keap1 and IKKβ prevented the loss of IKKβ, with IKKβ levels remaining at approximately two-thirds (68%) of those of the IKKβ-only control; coexpression of IKKβ and mVP24 in the absence of Keap1 had little impact on IKKβ expression, supporting a model whereby mVP24 prevents the Keap1-mediated loss of IKKβ (Figure 2A). We also determined the effect of the mVP24 mutants on Keap1-directed degradation of IKKβ. Interestingly, both wild-type mVP24 and the weakly interacting D205A/E207A mutant partially protected IKKβ expression in the presence of Keap1. The IKKβ level decreased in the presence of Keap1 to 20% of the level in the IKKβ-only control and increased to 34% and 43% of the IKKβ-only control level in the presence of mVP24 and D205A/E207A, respectively (Figure 2B). However, the noninteracting Linker and G211A/E212A mutants did not protect IKKβ from Keap1 degradation (Figure 2B). This suggests that the weakly interacting D205A/E207A mutant is able to stabilize IKKβ expression levels but that this does not occur exclusively through disruption of the interaction between Keap1 and IKKβ (Figure 1B). These data indicate that the interaction of mVP24 with Keap1 inhibits the Keap1-targeted decrease in IKKβ expression.
Figure 2.
Marburg virus VP24 (mVP24) prevents Kelch-like-ECH-associated protein 1 (Keap1)-targeted degradation of IκB kinase β (IKKβ). A and B, HEK293T cells were transfected with the indicated HA and Flag-tagged protein expression plasmids. Twenty-four hours after transfection, expression levels were analyzed by Western blot for HA and Flag. Western blots were performed at least 3 times; representative Western blots are shown.
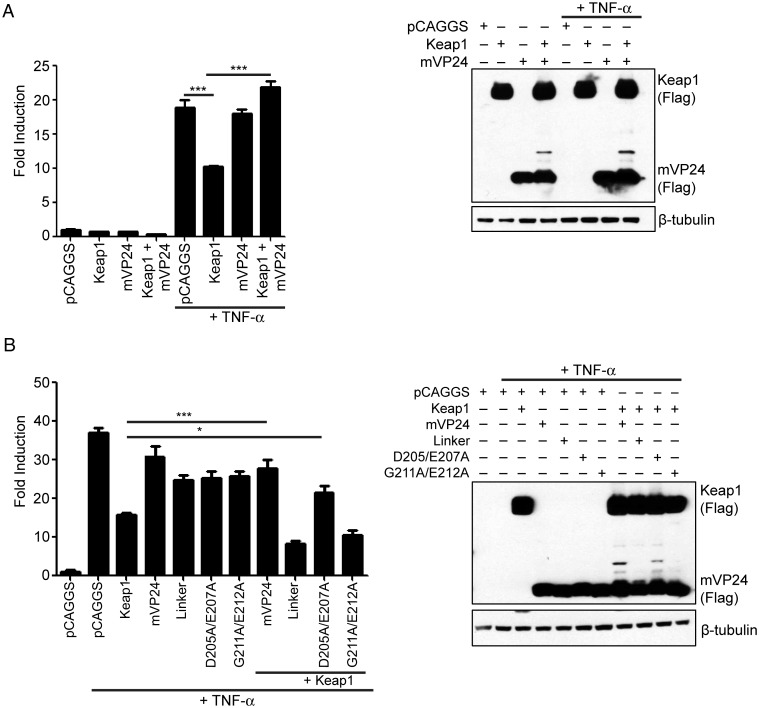
mVP24 Relieves Keap1 Repression of TNF-α–Stimulated NF-κB Activation
Keap1 has been shown to inhibit TNF-α–stimulated NF-κB activation [15, 16]. Because mVP24 prevents the degradation of IKKβ, we tested whether mVP24 expression could relieve the repression of Keap1 on NF-κB activation. HEK293T cells were transfected with an NF-κB promoter firefly luciferase plasmid, a constitutively active Renilla luciferase plasmid, and expression plasmids for Keap1 and mVP24, as indicated. Forty-eight hours after transfection, cells were treated with TNF-α (10 ng/mL) for 6 hours. As expected, Keap1 repressed the activation of the NF-κB reporter in the presence of TNF-α, and this repression was relieved when mVP24 was coexpressed (Figure 3A). To determine whether the relief of Keap1 repression requires the interaction of mVP24 and Keap1, we used our mVP24 Keap1-binding mutants in the same assay. Expression of the weakly interacting D205A/E207A mutant was able to relieve repression of Keap1 on the NF-κB reporter, although to a lesser extent than wild-type mVP24 (Figure 3B). The noninteracting Linker and G211A/E212A mutants did not restore activation (Figure 3B). These data suggest that the interaction between Keap1 and mVP24 is required to relieve Keap1 repression of NF-κB activity.
Figure 3.
Marburg virus VP24 (mVP24) relieves Kelch-like-ECH-associated protein 1 (Keap1) repression of tumor necrosis factor α (TNF-α)–stimulated nuclear factor-κB (NF-κB) activation. A and B, HEK293T cells were transfected with the NF-κB luciferase reporter plasmid, a constitutively expressed Renilla luciferase plasmid and pCAGGS, Flag Keap1, and Flag mVP24 or mutants as indicated. Forty-eight hours after transfection, cells were treated with 10 ng/mL TNF-α for 6 hours, after which luciferase activity was assayed. Western blots performed with anti-Flag antibody and β-tubulin as a loading control are on the right. Values represent the mean and standard error of the mean of triplicate samples, and statistical significance was assessed by a 1-way analysis of variance. *P < .05 and ***P < .001.
DISCUSSION
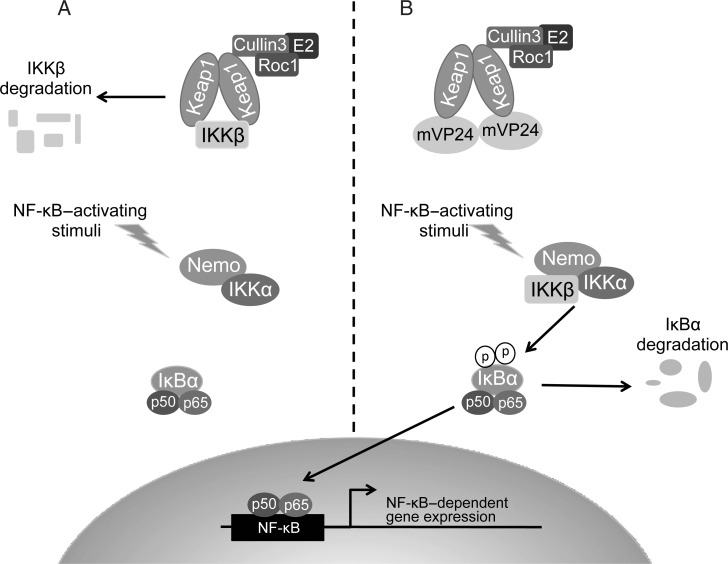
The IKK complex is a point of convergence between varied upstream signals and downstream activation of NF-κB transcription factors [19]. Activation of the NF-κB pathway leads to the expression of >100 target genes having roles in apoptosis, stress responses, and immune responses [21]. Here we demonstrate that the interaction between mVP24 and Keap1 relieves Keap1 repression of the NF-κB pathway by disrupting the interaction of IKKβ and Keap1, preventing the subsequent degradation of IKKβ (Figure 4). Therefore, while mVP24 does not directly activate the NF-κB pathway, it makes the NF-κB pathway more easily activated in the presence of stimuli.
Figure 4.
Working model. A, Kelch-like-ECH-associated protein 1 (Keap1) interacts with IκB kinase β (IKKβ), targeting it for degradation. The degradation of IKKβ prevents the cells from being able to respond to nuclear factor-κB (NF-κB)–activating stimuli. B, Marburg virus VP24 (mVP24) interacts with Keap1, disrupting the interaction of Keap1 and IKKβ. This frees IKKβ to interact with the IKK complex, allowing it to respond to NF-κB–activating stimuli.
Keap1 targets IKKβ, suppressing its expression through an interaction of the Keap1 Kelch domain and the ETGE motif in IKKβ. We show that wild-type mVP24 can stabilize IKKβ expression through disruption of the interaction of IKKβ and Keap1. Given that mVP24 D205A/E207A is unable to disrupt the interaction of IKKβ and Keap1, it was interesting to find that this mutant can stabilize IKKβ expression in the presence of Keap1. The mechanism of interaction between Nrf2 and Keap1 suggests a possible explanation for this observation. The interaction between Nrf2 and Keap1 is thought to use a hinge-latch mechanism [22]. In this model, 2 motifs in Nrf2, a high-affinity ETGE motif and a lower affinity DLG motif, bind the Keap1 homodimer [22]. Nrf2 is targeted for proteasomal degradation when bound to Keap1 through these 2 motifs. Disruption of Keap1 interaction with the weaker DLG motif, which can occur owing to modifications of Keap1 cysteine residues, stabilizes Nrf2 [22]. IKKβ also contains a DLG motif in addition to an ETGE motif [15]. While the DLG motif has been shown to be dispensable for interaction of IKKβ with Keap1, it is possible that it functions similarly to the DLG motif of Nrf2 and is required for targeted degradation of IKKβ. Therefore, it may be that the interaction of the weaker binding mVP24 D205A/E207A mutant with Keap1 is sufficient to disrupt the interaction of the IKKβ DLG motif with Keap1 without preventing the IKKβ ETGE motif with Keap1. This could result in stabilization of IKKβ expression while maintaining the interaction between Keap1 and IKKβ, thereby explaining the modest impact of mVP24 D205A/E207A on the interaction of Keap1 with IKKβ.
Expression of mVP24 stabilizes IKKβ, relieving Keap1-mediated negative regulation of the NF-κB pathway. This could result in the NF-κB pathway being more easily activated in MARV-infected cells. Activation of the NF-κB pathway could be beneficial to MARV by promoting cell survival and enhancing viral replication. Furthermore, the induction of cytokine expression by NF-κB activation could also recruit new target cells to be infected, leading to greater dissemination of the virus. Further study is required to understand the exact role of relieving Keap1 negative regulation on the NF-κB pathway during MARV infection.
Notes
Financial support. This work was supported by the National Institutes of Health (grants R01AI059536, U19AI109945 [C. F. B., principal investigator], and U19AI109664 [C. F. B., principal investigator] to C. F. B.).
Potential conflict of interest. Both authors: No reported conflicts.
Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Mateo M, Carbonnelle C, Martinez MJ, et al. Knockdown of Ebola virus VP24 impairs viral nucleocapsid assembly and prevents virus replication. J Infect Dis 2011; 204(suppl 3):S892–6. [DOI] [PubMed] [Google Scholar]
- 2.Bamberg S, Kolesnikova L, Moller P, Klenk HD, Becker S. VP24 of Marburg virus influences formation of infectious particles. J Virol 2005; 79:13421–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bharat TA, Riches JD, Kolesnikova L, et al. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol 2011; 9:e1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wenigenrath J, Kolesnikova L, Hoenen T, Mittler E, Becker S. Establishment and application of an infectious virus-like particle system for Marburg virus. J Gen Virol 2010; 91:1325–34. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y, Xu L, Sun Y, Nabel GJ. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol Cell 2002; 10:307–16. [DOI] [PubMed] [Google Scholar]
- 6.Noda T, Ebihara H, Muramoto Y, et al. Assembly and budding of Ebolavirus. PLoS Pathog 2006; 2:e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe S, Noda T, Halfmann P, Jasenosky L, Kawaoka Y. Ebola virus (EBOV) VP24 inhibits transcription and replication of the EBOV genome. J Infect Dis 2007; 196(suppl 2):S284–90. [DOI] [PubMed] [Google Scholar]
- 8.Mateo M, Reid SP, Leung LW, Basler CF, Volchkov VE. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J Virol 2010; 84:1169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid SP, Leung LW, Hartman AL, et al. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J Virol 2006; 80:5156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J Virol 2007; 81:13469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu W, Edwards MR, Borek DM, et al. Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host and Microbe 2014; 16:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valmas C, Grosch MN, Schumann M, et al. Marburg virus evades interferon responses by a mechanism distinct from ebola virus. PLoS Pathog 2010; 6:e1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards MR, Johnson B, Mire CE, et al. The Marburg virus VP24 protein interacts with Keap1 to activate the cytoprotective antioxidant response pathway. Cell Rep 2014; 6:1017–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Page A, Volchkova VA, Reid SP, et al. Marburgvirus hijacks nrf2-dependent pathway by targeting nrf2-negative regulator keap1. Cell Rep 2014; 6:1026–36. [DOI] [PubMed] [Google Scholar]
- 15.Kim JE, You DJ, Lee C, Ahn C, Seong JY, Hwang JI. Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation. Cell Signal 2010; 22:1645–54. [DOI] [PubMed] [Google Scholar]
- 16.Lee DF, Kuo HP, Liu M, et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell 2009; 36:131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh K, Wakabayashi N, Katoh Y, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 1999; 13:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem 2003; 278:21592–600. [DOI] [PubMed] [Google Scholar]
- 19.Scheidereit C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene 2006; 25:6685–705. [DOI] [PubMed] [Google Scholar]
- 20.Lo SC, Hannink M. PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex. J Biol Chem 2006; 281:37893–903. [DOI] [PubMed] [Google Scholar]
- 21.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol 2011; 12:695–708. [DOI] [PubMed] [Google Scholar]
- 22.Tong KI, Padmanabhan B, Kobayashi A, et al. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol 2007; 27:7511–21. [DOI] [PMC free article] [PubMed] [Google Scholar]