Abstract
Ebolaviruses are highly pathogenic in humans and nonhuman primates and pose a severe threat to public health. The interferon-induced transmembrane (IFITM) proteins can restrict entry of ebolaviruses, influenza A viruses, and other enveloped viruses. However, the breadth and mechanism of the antiviral activity of IFITM proteins are incompletely understood. Here, we employed ebolavirus glycoprotein–pseudotyped vectors and ebolavirus-like particles to address this question. We show that IFITM proteins inhibit the cellular entry of diverse ebolaviruses and demonstrate that type I interferon induces IFITM protein expression in macrophages, major viral targets. Moreover, we show that IFITM proteins block entry of influenza A viruses and ebolaviruses by different mechanisms and provide evidence that antibodies and IFITM proteins can synergistically inhibit cellular entry of ebolaviruses. These results provide insights into the role of IFITM proteins in infection by ebolaviruses and suggest a mechanism by which antibodies, though poorly neutralizing in vitro, might contribute to viral control in vivo.
Keywords: Ebola, glycoprotein, entry, IFITM, interferon
Ebolaviruses (EBOVs) are enveloped, negative-stranded RNA viruses within the Filoviridae family. The genus Ebolavirus contains 5 species, Zaire ebolavirus (single member: Ebola virus [EBOV]), Sudan ebolavirus (single member: Sudan virus [SUDV]), Bundibugyo ebolavirus (single member: Bundibugyo virus [BDBV]), Taï Forest ebolavirus (single member: Taï Forest virus [TAFV]), and Reston ebolavirus (single member: Reston virus [RESTV]) [1]. EBOV, SUDV, BDBV and TAFV are responsible for outbreaks of severe disease in sub-Saharan Africa, which are associated with high case fatality rates [2, 3]. In addition, an EBOV disease is currently ongoing in Western Africa [4] and is associated with 25 791 cases and 10 689 deaths (as of 15 April 2015) [5]. In contrast, RESTV is an Asian ebolavirus, which might be apathogenic in immunocompetent humans [6]. African [7] and Asian ebolaviruses [8] infect bats, which serve as a natural reservoir, and a related filovirus, Lloviu virus (LLOV; genus Cuevavirus), was identified in a European bat [9]. Thus, zoonotic transmission of ebolaviruses to humans can pose a significant threat to public health, and understanding virus–host cell interactions might yield insights into viral spread and pathogenesis and provide a basis for developing antiviral strategies.
The interferon (IFN) system is an integral component of the innate defenses against viral infection. The observation that ebolaviruses encode several IFN antagonists indicates that IFN-induced antiviral effector molecules can pose an effective blockade to viral spread [10]. The IFN-induced transmembrane proteins (IFITMs) IFITM1, IFITM2, and IFITM3 were shown to display antiviral activity against filoviruses and other viral pathogens [11–13]. Several studies suggest that IFITM proteins can block host cell entry of viruses by inhibiting the fusion of the viral membrane with an endosomal membrane. Thus, IFITM proteins inhibit influenza A virus (FLUAV) entry at the stage of hemifusion [14, 15], possibly by inducing the accumulation of endosomal cholesterol [16], although this mechanism is not undisputed [14, 17, 18]. Polymorphisms in the IFITM3 gene were shown by some studies [19, 20] but not all [21, 22] to be associated with increased susceptibility to severe influenza, indicating that IFITM3 poses a powerful block to FLUAV spread in infected humans. In contrast, it is presently unclear whether IFITM proteins inhibit entry of all African and Asian ebolaviruses as well as LLOV and whether they are expressed in major viral target cells. Finally, it remains to be determined whether IFITM proteins inhibit host cell entry of ebolaviruses and FLUAV by similar mechanisms. The present study addressed these questions, employing a vector system and novel EBOV-like reporter particles to analyze ebolavirus glycoprotein (GP)–driven entry into target cells and its inhibition by IFITM proteins.
MATERIAL AND METHODS
Cell Culture
Human embryonic kidney 293T cells were maintained in Dulbecco's minimal essential medium, supplemented with 10% fetal bovine serum (PAA), 100 U/mL penicillin, and 100 µg/mL streptomycin (Cytogen). RD cells stably expressing a C-terminal fragment of Gaussia luciferase (GLuc) were generated by selection of transfected cells in Dulbecco's minimal essential medium containing G418 at 1 mg/mL. Monocyte-derived macrophages (MDMs) were cultured in X-Vivo 10 medium (Lonza).
Production of MDMs
For the production of human MDMs, monocyte-enriched cells were isolated from thrombapheresis rings by Ficoll density gradient centrifugation. The amount of platelets in the preparations was reduced by centrifugation, and monocytes were collected by adhesion-mediated enrichment on plastic dishes followed by culture in monocyte adhesion medium (Roswell Park Memorial Institute 1640 medium supplemented with 7.5% human fibrin-depleted plasma and antibiotics). The next day, the cultures were washed with phosphate-buffered saline, and cells were detached and seeded in monocyte differentiation medium (X-Vivo 10 supplemented with 1% human fibrin-depleted plasma and antibiotics) and cultured for 6 days. Differentiation into MDMs was controlled by flow cytometric analysis of CD14 expression.
Plasmids
Plasmids encoding the GPs of vesicular stomatitis virus (VSV), Marburg virus (MARV; strain Musoke), murine leukemia virus (MLV), Lassa virus (LASV), Machupo virus (MACV; strain Carvallo), FLUAV (strain A/WSN/33; particles generated in cells expressing hemagglutinin [HA] and neuraminidase), EBOV (strain Mayinga), SUDV (strain Boniface), TAFV, RESTV, BDBV, and LLOV have been described elsewhere [17, 23, 24]. The retroviral vectors used for expression of IFITM proteins have also been described elsewhere [17]. The rhesus macaque IFITM homologues, IFITM1 (XM_001085444.2), IFITM3(1) (XM_001085567.2), and IFITM3(2) (XM_001085331.2) were amplified with polymerase chain reaction (PCR) from complementary DNAs and cloned into the pQCXIP vector. pQCXIP vectors encoding human and rhesus macaque IFITM proteins with a C-terminal myc tag were generated by PCR using myc-encoding primers.
The plasmid encoding the IFITM3-SVKS mutant was generated by PCR-based mutagenesis, as described elsewhere for IFITM1 [25]. To generate pQCXIP-CFP-IFITM1, the cyan fluorescent protein (CFP)-IFITM sequence was amplified from pSCFP3A-C1-IFITM1 and inserted into pQCXIP. pSCFP3A-C1-IFITM1 is based on pEGFP-C1, in which enhanced green fluorescent protein (EGFP) was replaced by super cyan fluorescent protein 3A (SCFP3A) [26], and IFITM1 was inserted via XhoI and EcoRI from pCAGGS-mod-IFITM1 [27]. pQCXIP-CFP-IFITM2 and 3 have been generated by subcloning IFITM genes from pQCXIP-IFITM plasmids via NotI and EcoRI into pQCXIP-CFP-IFITM1, thereby replacing IFITM1. Sequences encoding previously described N- and C-terminal fragments of human codon-optimized GLuc [28] were synthesized (Genscript). The fragment encoding the N-terminal portion of Gluc (GlucN), lacking a signal peptide, was fused to the C-terminus of EBOV VP40, lacking a stop codon, using overlap extension PCR, and the PCR product was inserted into plasmid pCAGGS. The fragment encoding GLucC was inserted into plasmid pcDNA3.1. The integrity of all PCR-amplified sequences was verified by automated sequence analysis.
Antibodies
Expression of IFITM proteins was detected using Western blot analyses with a monoclonal mouse anti-IFITM1 antibody (1:1000, Proteintech) and polyclonal rabbit anti-IFITM 2 antibody (1:2000, Proteintech), which is cross-reactive against IFITM3. IFITM proteins equipped with a C-terminal myc tag were detected using the monoclonal anti-myc antibody 9E10. Expression of β-actin was detected with mouse monoclonal anti–β-actin antibody (1:1000, Sigma). To detect STAT1 expression and phosphorylation, a polyclonal rabbit anti-STAT1α/β antibody and a polyclonal rabbit anti-phospho-STAT1 (Tyr701) antibody (both 1:1000; Cell Signaling) were used, respectively. Bound antibodies were visualized with horseradish peroxidase–conjugated secondary antibodies reactive against rabbit or mouse antibodies (Dianova, 1:5000–10 000). The antiserum used for neutralization experiments was produced by immunizing rabbits with an EBOV-GP1-Fc fusion protein.
Production of Retroviral Vectors
The production of vectors encoding IFITM proteins or vectors encoding luciferase and viral GPs under study was described elsewhere [17]. In brief, 293T cells were cotransfected with plasmids encoding MLV gag-pol, VSV-G, and pQCXIP-based or related vectors encoding either luciferase or IFITM proteins or chloramphenicol acetyltransferase (Cat) as a control. The culture medium was exchanged at 6 hours and supernatants harvested at 48 hours after transfection. Supernatants were sterile filtered, aliquoted, and stored at −80°C. Preparations of luciferase-encoding vectors were normalized for comparable transduction of 293T cells before usage in experiments.
Antiviral Activity of IFITM Proteins
To transiently transduce adherent cell lines, we seeded cells at a density of 104 cells per well in 96-well plates. The adherent cells were transduced with IFITM-encoding vectors by spinoculation [29] at 4000g for 30 minutes and incubated for 48 hours. Thereafter, the culture supernatants were replaced by 50 µL of fresh medium. Subsequently the cells were inoculated with 50 µl of luciferase-normalized vectors harboring the viral GP under study and incubated for 8 hours. Afterward, the supernatants were replaced with 150 µL of fresh culture medium, and luciferase activity in cell lysates was measured 72 hours after transduction using a commercially available kit (Promega; PJK). To analyze the effect of amphotericin B on the antiviral activity of IFITM proteins, we treated 293T cells with 10 µmol/L amphotericin B (Fisher Bioreagents) for 1 hour at 37°C before transduction with luciferase-encoding vectors.
IFN-Induced Expression of IFITM Proteins in Human MDMs
Differentiated human MDMs were stimulated for 24 hours with 1000–3000 U of human IFN-α2b, IFN-β, or IFN-γ (Antigenix) per milliliter or left untreated and harvested at different time points. As a control, IFITM expression in 293T cells transfected to express IFITM proteins was assessed.
Inhibition of GP-Mediated Transduction by Antibodies and IFITM Proteins
Pseudotypes bearing EBOV-GP, FLUAV-HA, or LASV-glycoprotein precursor (GPC) were incubated with a rabbit antiserum raised against EBOV-GP, the corresponding preimmune serum, or culture medium as a control, at a dilution of 1:500 for 30 minutes and 37°C. Subsequently, the pseudotypes were added to the cells, and the transduction efficiency determined as described above.
EBOV-like Particles
For production of EBOV-like particles, plasmids encoding the viral envelope protein and GLucN/VP40 were cotransfected into 293T cells employing TransIT-2020 (Mirus). Culture supernatants were harvested 48 hours after transfection and viruslike particles were concentrated by ultracentrifugation through a 20% sucrose cushion using a SW28 rotor. Target cells (RD/GLucC) were transiently transfected with plasmids encoding IFITM3 or Cat as control, using TransIT-2020, and then allowed to incubate for 24 hours. Subsequently, the cells were seeded into 96-well plates, incubated for 24 hours, and then spin-infected [29] for 1 hour at 4°C followed by incubation for 16 hours at 37°C. Finally, cells were harvested, and the GLuc activity in cell lysates was measured using a commercial kit (Pierce), according to the manufacturer's instructions.
Flow Cytometric Analysis of IFITM Expression
For flow cytometric analysis, 293T cells were transduced to express IFITM proteins N-terminally fused to CFP, harvested 48 hours after transduction, washed with phosphate-buffered saline, and fixed with 2% paraformaldehyde. Subsequently, CFP signals in transduced cells were determined by flow cytometry.
RESULTS
IFITM Proteins Inhibit Entry Driven by GPs From Viruses Representing All Species Within the Genus Ebolavirus
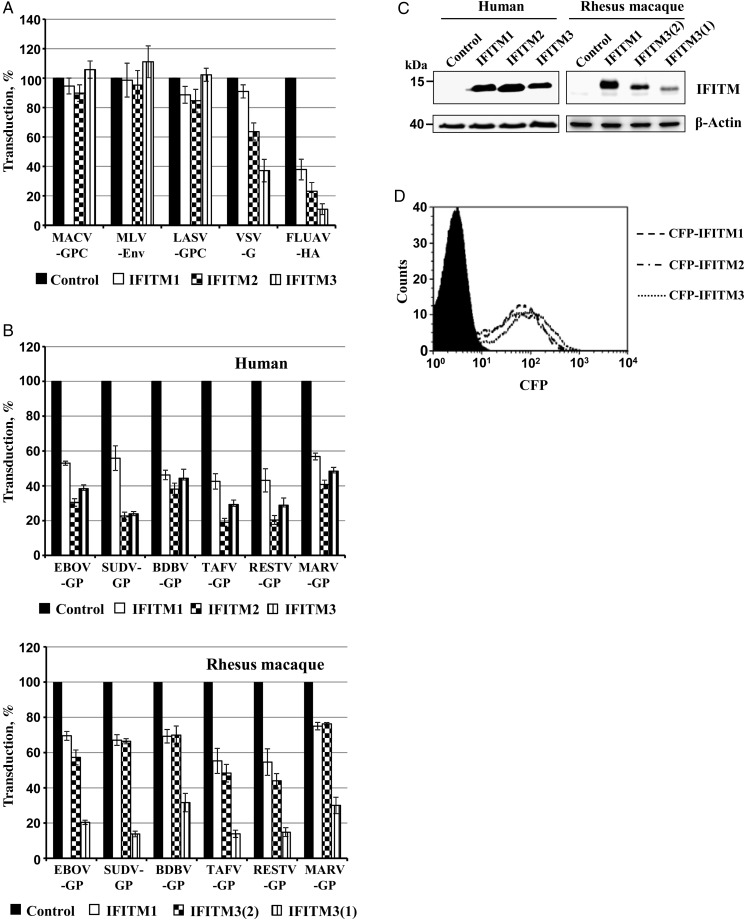
We first assessed whether susceptibility to inhibition by IFITM proteins is conserved between the GPs of viruses from all species within the genus Ebolavirus. For this, we employed retroviral vectors bearing GPs from ebolaviruses and analyzed transduction of 293T cells previously transduced to express IFITM proteins or Cat as negative controls, following an established protocol [17]. The 293T cell line was chosen because the cells can be readily transfected and are highly susceptible to transduction mediated by filovirus GPs.
Expression of IFITM proteins had no appreciable impact on transduction mediated by MLV-Env, LASV-GPC, and MACV-GPC and modestly reduced entry driven by VSV-G, whereas marked inhibition of FLUAV-HA–driven entry was observed (Figure 1A), in keeping with published data [11, 12, 17]. In contrast, entry driven by all EBOV-GPs tested and by MARV-GP was inhibited by IFITM proteins, with IFITM2 and IFITM3 displaying a somewhat higher antiviral activity than IFITM1 (Figure 1B, upper panel), despite comparable expression in transduced cells (Figure 1C and 1D).
Figure 1.
Interferon-induced transmembrane (IFITM) proteins inhibit entry driven by glycoproteins representing all viral species within the genus Ebolavirus. A, 293T cells, transduced to express human IFITM1, IFITM2, IFITM3, or chloramphenicol acetyltransferase (Cat), were subsequently transduced with vectors encoding luciferase and bearing the indicated viral entry proteins. Means are shown for 3 independent experiments performed with triplicate samples. Transduction of Cat-expressing cells (control) was set as 100%. Error bars indicate standard errors of the mean (SEMs). B, Experiment conducted as described in A but with examination of cells expressing human (top) or rhesus macaque (bottom) IFITM proteins. Means and SEMs are shown for 3 independent experiments performed with triplicate samples. Transduction of Cat-expressing cells (control) was set as 100%. C, Expression of myc-tagged IFITM proteins in transduced 293 T cells was analyzed by Western blot using anti-myc antibody. D, Expression of IFITM-cyan fluorescent protein fusion proteins in transduced 293T cells was analyzed by flow cytometry. Single representative experiments are shown in C and D and were confirmed in ≥1 separate experiment. Abbreviations: BDBV, Bundibugyo virus; EBOV, Ebola virus; FLUAV, influenza A virus; GP, glycoprotein; HA, hemagglutinin; LASV, Lassa virus; MACV, Machupo virus; MARV, Marburg virus; MLV, murine leukemia virus; RESTV, Reston virus; SUDV, Sudan virus; TAFV, Taï Forest virus; VSV, vesicular stomatitis virus.
The experimental infection of rhesus macaques with ebolaviruses and marburgviruses is an important animal model for ebolavirus disease in humans [30]. Therefore, we also determined whether IFITM proteins from rhesus macaques are expressed in transduced cells and inhibit GP-driven entry. The rhesus macaque genome does not encode IFITM2 [31] but encodes two variants of IFITM3, IFITM3(1) and IFITM3(2). Expression of rhesus macaque IFITM proteins was readily detectable, although expression of IFITM3(1) was reduced compared with that of IFITM3(2) (Figure 1C). The IFITM homologues of rhesus macaques displayed antiviral activity, with IFITM3(1) being most active (Figure 1B, lower panel), despite its reduced expression. Thus, IFITM proteins of human and rhesus macaque origin can block entry driven by GPs from diverse ebola- and marburgviruses in cell culture, suggesting that IFITM proteins could act as panfiloviral inhibitors in infected nonhuman primates and in humans.
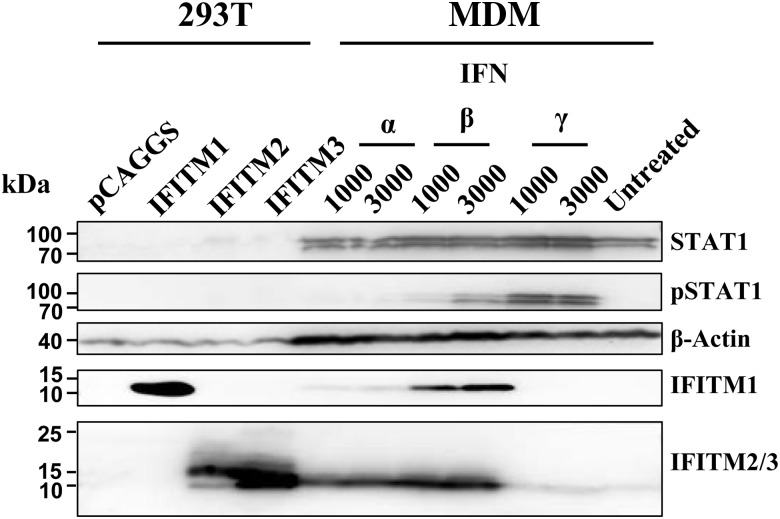
IFN Treatment Induces IFITM Expression in Macrophages
Macrophages are early and sustained targets of ebolavirus infection [32, 33]. Therefore, we assessed whether IFN treatment induces IFITM expression in human MDMs (Figure 2). Treatment of MDMs with IFN-α and IFN-β induced phosphorylation of STAT1 within 1 hour (not shown), and phosphorylated STAT1 was still detectable 24 hours after treatment with IFN-β. Expression of IFITM1 was modestly increased upon treatment with IFN-α, and a strong increase was observed upon treatment of MDMs with IFN-β, whereas both IFNs robustly induced expression of IFITM2/3 (note that an antibody reactive against both IFITM2 and IFITM3 was used, precluding discrimination between IFITM2 and IFITM3 signals). Finally, exposure of MDMs to IFN-γ induced marked STAT1 phosphorylation but had little effect on IFITM3 expression, suggesting that IFITM expression in human macrophages might be mainly induced by type I IFNs.
Figure 2.
Stimulation with type I interferon (IFN) induces IFN-induced transmembrane (IFITM) protein expression in human macrophages. Monocyte-derived macrophages were incubated with the indicated IFNs for 24 hours or left untreated and expression of STAT1, phosphorylated STAT1, β-actin, IFITM1, and IFITM2/3 was assessed by Western blot analysis. As controls, 293T cells were transfected with empty plasmid or expression plasmids for IFITM1–3. Similar results were obtained in 3 separate experiments. Abbreviation: MDM, monocyte-derived macrophages.
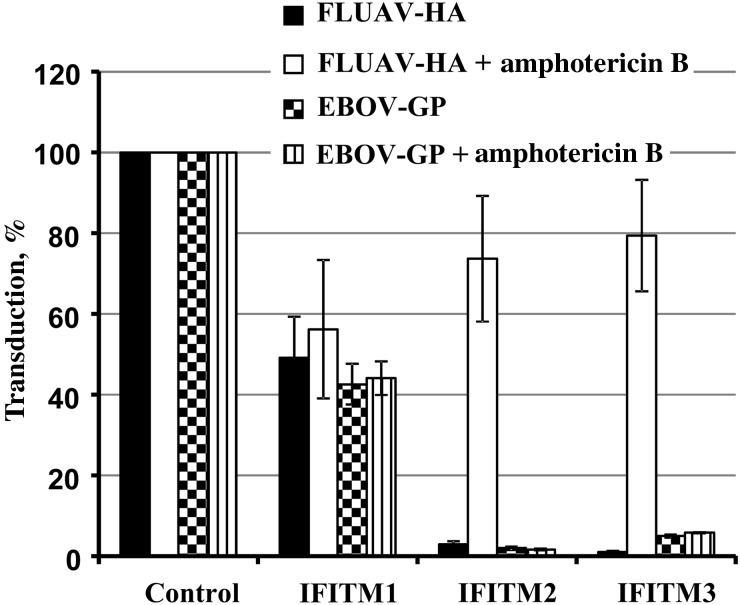
Amphotericin Rescues FLUAV-HA–Driven but Not EBOV-GP–Driven Entry From Inhibition by IFITM Proteins
Lin and colleagues [18] recently showed that inhibition of FLUAV-HA–driven host cell entry by IFITM3 is rescued by treatment of cells with the polyene macrolide antibiotic amphotericin B, a finding that has clinical implications and can be used as basis to study the mechanisms underlying the antiviral activity of IFITM proteins. We asked whether amphotericin B could also rescue EBOV-GP–driven host cell entry from inhibition by IFITM proteins. Amphotericin B had no effect on inhibition of FLUAV-HA–driven entry by IFITM1 but markedly increased entry into IFITM2- and IFITM3-expressing cells (Figure 3), as expected. In stark contrast, amphotericin B did not modulate EBOV-GP–driven entry (Figure 3), indicating that IFITM2 and IFITM3 inhibit FLUAV-HA– and EBOV-GP–mediated entry via different mechanisms.
Figure 3.
Amphotericin B does not rescue Ebola virus (EBOV) glycoprotein (GP)–driven entry from inhibition by interferon-induced transmembrane (IFITM) proteins. After 293T cells were transduced to express IFITM1, IFITM2, and IFITM3 or chloramphenicol acetyltransferase (control) and treated with 10 µmol/L amphotericin B for 1 hour, they were then transduced with luciferase-encoding murine leukemia virus particles bearing EBOV-GP or influenza A virus (FLUAV) hemagglutinin (HA). Means are shown for 3 independent experiments; error bars represent standard errors of the mean. Transduction of control cells was set as 100%.
Mutation of an SVKS Motif in IFITM3 Augments LLOV-GP–Driven Entry
LLOV was detected in dead Schreiber's bats (Miniopterus schreibersii) in Spain [9], and its potential transmission to humans might constitute a health threat. Therefore, we asked whether IFITM proteins block LLOV-GP–driven entry. Indeed, similar inhibition of EBOV- and LLOV-GP–driven transduction was observed upon expression of IFITM1, IFITM2, and IFITM3, indicating that the antiviral activity of IFITM proteins extends to LLOV (Figure 4A). These results were confirmed using a novel EBOV-like particle reporter system, which measures GLuc transcomplementation upon delivery of a VP40-fused GLuc fragment into cells expressing the complementary portion of GLuc (Figure 4B). This assay faithfully reproduces standard responses of authentic EBOV to entry inhibitors, such as ammonium chloride, CA074, and neutralizing antibodies (data not shown, K. L. and G. S.).
Figure 4.
Inhibition of Lloviu virus (LLOV) glycoprotein (GP)–driven entry by interferon-induced transmembrane (IFITM) proteins. A, 293T cells transduced to express human IFITM1, 2, 3 or chloramphenicol acetyltransferase (Cat) as a control were subsequently transduced with vectors bearing the indicated GPs and luciferase activity in cell lysates was determined. Means and standard errors of the mean (SEMs) are shown for 3 independent experiments performed with triplicate samples. Transduction of control cells was set as 100%. Abbreviations: FLUAV-HA, influenza A virus hemagglutinin; MLV-Env, murine leukemia virus . B, RD/Gaussia luciferase (GLucC) cells transduced to express human IFITM 3 or Cat as control were spin-infected with VP40/N-terminal portion of GLuc virus-like particles (VLPs) bearing the indicated GPs, and the luciferase activity in cell lysates was determined. Means and SEMs are shown for 2 experiments conducted with quintuplicate samples. Infection of control cells was set as 100%. Similar results were obtained when 293T cells transfected to express IFITM3 were analyzed. C, Western blot analysis of IFITM expression in transduced 293T cells. The expression of wild type-IFITM3 and an IFITM3 mutant in which the amino acids SVKS were mutated to alanine was analyzed. D, Experiment was conducted as described for A, but the SVKS-AAAA mutant was analyzed. Means and SEMs are shown for 3 independent experiments. Transduction of control cells was set as 100%. Abbreviation: MARV, Marburg virus. E, Experiment was carried as described for B, but the IFITM3 SVKS-AAAA mutant was included, and VLPs bearing no GP served as negative control. Results of a single experiment conducted with quintuplicate samples are shown and were confirmed in a separate experiment; error bars indicate standard deviations.
To identify determinants in IFITM3 required for its antifilovirus activity, we changed an SVKS motif located at the N-terminus of the conserved intracellular loop (amino acid residues 81–84) to AAAA, because this motif was previously shown to be important for the inhibition of other viruses by IFITM proteins [25, 34]. Mutation of SVKS in IFITM3, which had no appreciable effect on protein expression (Figure 4C), abrogated the blockade of EBOV-GP–driven entry and markedly augmented transduction mediated by LLOV-GP (Figure 4D). Similarly, cellular entry of EBOV-like particles bearing LLOV-GP was augmented by the SVKS mutant, and expression of this protein also increased EBOV-GP–dependent entry (Figure 4E). Thus, the SVKS motif is required for inhibition of EBOV-GP– and LLOV-GP–facilitated entry, and alteration of this motif can transform IFITM3 from an antiviral into a proviral factor.
IFITM3 Expression and Neutralizing Serum Synergistically Inhibit Viral Entry
Antibodies directed against the EBOV-GP can inhibit host cell entry and might contribute to control of viral spread in the infected host. We hypothesized that IFITM expression and antibodies directed against GP might synergistically inhibit GP-driven entry. To test this hypothesis, we assessed whether a poorly neutralizing antiserum raised in rabbits against EBOV-GP unfolds potent neutralizing activity when target cells are used that express IFITM proteins. The antiserum modestly inhibited EBOV-GP–driven entry into control cells (3-fold reduction) and had no appreciable effect on FLUAV-HA– and LASV-GPC–driven entry into these cells (Figure 5). EBOV-GP–mediated transduction of IFITM1- and IFITM2-expressing cells was markedly reduced by the serum, whereas the effect on FLUAV-HA– and LASV-GPC–driven entry was absent or modest, respectively. Furthermore, the roughly 10-fold inhibition of EBOV-GP–driven entry on IFITM3 expression alone was increased to 1000-fold when vectors were incubated with GP-specific antiserum, and similar effects were not observed for FLUAV-HA and LASV-GPC, although an 8–10-fold augmentation of inhibition efficiency was measured (Figure 5). Finally, preimmune serum did not modulate transduction efficiency, indicating that the effects observed were specific. Collectively, these results suggest that expression of IFITM3 and GP-specific antibodies might synergistically inhibit GP-driven entry.
Figure 5.
Synergistic inhibition of Ebola virus (EBOV) glycoprotein (GP)–driven entry by interferon-induced transmembrane (IFITM) proteins and GP-specific antiserum. 293T cells transduced to express IFITM1, IFITM2, and IFITM3 or chloramphenicol acetyltransferase as a control were transduced with vectors bearing the indicated GPs, which were preincubated with medium alone (control), 1:500 diluted rabbit preimmune serum, or 1:500 diluted rabbit anti–EBOV-GP antiserum for 30 minutes. The means and standard errors of the mean for 2 (Lassa virus [LASV]–GPC) or 4 (EBOV-GP and influenza A virus [FLUAV] hemagglutinin [HA]) experiments are shown.
DISCUSSION
The antiviral action of IFN-induced effector proteins can limit viral spread in the infected host, and understanding the mechanism underlying inhibition might help define novel targets for intervention. The present study demonstrated that IFITM proteins inhibit host cell entry driven by GPs representing all members of the Ebolavirus and Cuevavirus genus within the Filoviridae family. Interestingly, IFITM proteins might regulate a process that can either enhance or inhibit GP-driven entry, because an IFITM3 mutant markedly augmented LLOV-GP–dependent transduction. Moreover, evidence was obtained that IFITM proteins might inhibit FLUAV and EBOV entry via different mechanisms because the IFITM-mediated blockade of entry driven by FLUAV-HA but not EBOV-GP was rescued by amphotericin B treatment. Finally, our results suggest that neutralizing antibodies and IFITM proteins can synergistically inhibit EBOV-GP–facilitated entry.
Our study shows that IFITM proteins are expressed in macrophages upon exposure to type I IFNs, which are produced at high levels in the context of EBOV infection [35, 36]. Moreover, IFITM expression was reported to inhibit host cell entry of authentic EBOV and MARV [12], and our present findings demonstrate that entry of vectors pseudotyped with diverse filovirus GPs is also blocked. These observations indicate that IFITM proteins might constitute a panfiloviral defense in the infected host. This defense could slow down but not prevent viral spread and seems to be operative in humans and nonhuman primates, as evidenced by the blockade of filovirus-GP–driven entry by rhesus macaque IFITM proteins. Moreover, the panfiloviral inhibition suggests that all filovirus GPs, including LLOV-GP, depend on similar if not identical cellular factors and/or processes for host cell entry. This notion is in agreement with two recent reports [37, 38] indicating that the GPs of LLOV and EBOV use identical factors for attachment (C-type lectins) and entry (NPC1) and that entry driven by both GPs depends on GP proteolysis by endosomal cysteine proteases. The amount of GP incorporated into viral particles does not seem to modulate IFITM sensitivity. Thus, retroviral particles bearing low, intermediate, and high amounts of EBOV-GP and RESTV-GP exhibited comparable susceptibility to inhibition by IFITM proteins (Supplementary Figure 1), indicating that the block imposed by IFITM proteins might not be saturable by excess GP copies presented at the virion surface. In sum, IFITM proteins are comparably active against entry driven by all filoviral GPs, although subtle differences in inhibition efficiency might have remained undetected owing to the limited resolution of our assay, and it will be interesting to examine the contribution of these proteins to the control of viral spread in the host. IFITM knockout mice [19, 39] might be suitable tools for these endeavors.
Findings of a study by Amini-Bavil-Olyaee and colleagues [16] suggested that IFITM proteins might inhibit endosomal entry of enveloped viruses by inducing cholesterol accumulation in the endosomal membrane. However, subsequent work challenged this concept [14, 17, 18], and reduction of endosomal cholesterol levels did not rescue EBOV-GP–driven entry from blockade by IFITM proteins in the present study (not shown). Recent studies revealed that the antimycotic compound amphotericin B disables IFITM-dependent inhibition of FLUAV [18] and retrovirus entry [40] in cell culture, and amphotericin B treatment of mice was shown to abrogate control of FLUAV infection [18]. The underlying mechanism was not fully clarified, but a role for membrane fluidity was suggested. The present study shows that IFITMs might block FLUAV-HA- and EBOV-GP-mediated entry via different mechanisms, since EBOV-GP-driven entry was not rescued by amphotericin B treatment. Thus, further work is required to clarify how GP-driven entry is inhibited. These studies might be aided by the observation that mutation of an SVKS motif—required for the antiretroviral activity of IFITM1 [25] and the anti-dengue/FLUAV activity of IFITM3 [34] - potentiates LLOV-GP–driven entry, because it suggests that IFITM proteins might be able to positively and negatively regulate a cellular process critical for filovirus entry. This concept is in keeping with the finding that the human coronavirus OC43 can use IFITM proteins as cofactors for infectious viral entry into target cells [41].
The GP of ebolaviruses is the sole target for neutralizing antibodies, which can efficiently inhibit viral spread in the infected host when applied as postexposure prophylaxis [42]. We observed that IFITM3 and an otherwise poorly neutralizing antiserum directed against GP can synergistically inhibit EBOV-GP–driven entry. Although the responsible mechanism remains to be elucidated, this finding suggests that antibodies with unimpressive neutralizing capacity in standard cell culture systems, for instance Vero cells (which exhibit low endogenous IFITM expression [12]), might still unfold substantial antiviral activity in the host, where IFITM expression is induced by IFNs. In this context, a finding by Kajihara and colleagues [43] is noteworthy; they reported that antibodies unable to block cellular entry of MARV still displayed potent antiviral activity by blocking GP-promoted release of progeny particles from infected cells.
Collectively, IFITM proteins exhibit a broad activity against ebolaviruses and other filoviruses, and clarifying the underlying mechanism might yield valuable insights into viral spread and pathogenesis. In fact, a recent study suggested that blockade of viral entry by IFITM proteins might be a multifaceted process, because IFITM proteins were shown to be incorporated into the retroviral envelope thereby reducing infectivity of progeny virions [44].
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank H. Hofmann-Winkler for comments and support.
Financial support. This work was supported by the German Research Foundation (grant PO 716/8-1 to S. P.), the German Ministry for Research and Education (subproject within EBOCON [S. P.]), Leibniz Graduate School Emerging Infectious Diseases (S. P.), the Leibniz Foundation, and the National Institute of Allergy and Infectious Diseases (grant R21AI107165 to G. S.).
Potential conflicts of interest. All authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Kuhn JH, Becker S, Ebihara H et al. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch Virol 2010; 155:2083–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet 2011; 377:849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misasi J, Sullivan NJ. Camouflage and misdirection: the full-on assault of Ebola virus disease. Cell 2014; 159:477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO Ebola Response Team; Agua-Agum J, Ariyarajah A, Aylward B, et al. West African Ebola epidemic after one year—slowing but not yet under control. N Engl J Med 2015; 372:584–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. Ebola response roadmap—situation report. http://apps.who.int/ebola/current-situation/ebola-situation-report-15-april-2015.
- 6.Miranda ME, Miranda NL. Reston ebolavirus in humans and animals in the Philippines: a review. J Infect Dis 2011; 204(suppl 3):S757–60. [DOI] [PubMed] [Google Scholar]
- 7.Leroy EM, Kumulungui B, Pourrut X et al. Fruit bats as reservoirs of Ebola virus. Nature 2005; 438:575–6. [DOI] [PubMed] [Google Scholar]
- 8.Taniguchi S, Watanabe S, Masangkay JS et al. Reston ebolavirus antibodies in bats, the Philippines. Emerg Infect Dis 2011; 17:1559–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Negredo A, Palacios G, Vazquez-Moron S et al. Discovery of an ebolavirus-like filovirus in Europe. PLoS Pathog 2011; 7:e1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kühl A, Pöhlmann S. How Ebola virus counters the interferon system. Zoonoses Public Health 2012; 59(suppl 2):116–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brass AL, Huang IC, Benita Y et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 2009; 139:1243–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang IC, Bailey CC, Weyer JL et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog 2011; 7:e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perreira JM, Chin CR, Feeley EM, Brass AL. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol 2013; 425:4937–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog 2014; 10:e1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li K, Markosyan RM, Zheng YM et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog 2013; 9:e1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amini-Bavil-Olyaee S, Choi YJ, Lee JH et al. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 2013; 13:452–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wrensch F, Winkler M, Pöhlmann S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses 2014; 6:3683–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin TY, Chin CR, Everitt AR et al. Amphotericin B increases influenza A virus infection by preventing IFITM3-mediated restriction. Cell Rep 2013; 5:895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everitt AR, Clare S, Pertel T et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature 2012; 484:519–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YH, Zhao Y, Li N et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun 2013; 4:1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mills TC, Rautanen A, Elliott KS et al. IFITM3 and susceptibility to respiratory viral infections in the community. J Infect Dis 2014; 209:1028–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams DE, Wu WL, Grotefend CR et al. IFITM3 polymorphism rs12252-C restricts influenza A viruses. PLoS One 2014; 9:e110096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marzi A, Gramberg T, Simmons G et al. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J Virol 2004; 78:12090–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simmons G, Reeves JD, Grogan CC et al. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 2003; 305:115–23. [DOI] [PubMed] [Google Scholar]
- 25.Lu J, Pan Q, Rong L, He W, Liu SL, Liang C. The IFITM proteins inhibit HIV-1 infection. J Virol 2011; 85:2126–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kremers GJ, Goedhart J, van Munster EB, Gadella TW Jr. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Forster radius. Biochemistry 2006; 45:6570–80. [DOI] [PubMed] [Google Scholar]
- 27.Bertram S, Dijkman R, Habjan M et al. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J Virol 2013; 87:6150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat Methods 2006; 3:977–9. [DOI] [PubMed] [Google Scholar]
- 29.O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol 2000; 74:10074–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakayama E, Tomabechi D, Matsuno K et al. Antibody-dependent enhancement of Marburg virus infection. J Infect Dis 2011; 204(suppl 3):S978–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Liu J, Li M, Yang H, Zhang C. Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates. PLoS One 2012; 7:e49265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geisbert TW, Hensley LE, Larsen T et al. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol 2003; 163:2347–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez O, Leung LW, Basler CF. The role of antigen-presenting cells in filoviral hemorrhagic fever: gaps in current knowledge. Antiviral Res 2012; 93:416–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.John SP, Chin CR, Perreira JM et al. The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J Virol 2013; 87:7837–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutchinson KL, Rollin PE. Cytokine and chemokine expression in humans infected with Sudan Ebola virus. J Infect Dis 2007; 196(suppl 2):S357–63. [DOI] [PubMed] [Google Scholar]
- 36.Villinger F, Rollin PE, Brar SS et al. Markedly elevated levels of interferon (IFN)-gamma, IFN-alpha, interleukin (IL)-2, IL-10, and tumor necrosis factor-alpha associated with fatal Ebola virus infection. J Infect Dis 1999; 179(suppl 1):S188–91. [DOI] [PubMed] [Google Scholar]
- 37.Maruyama J, Miyamoto H, Kajihara M et al. Characterization of the envelope glycoprotein of a novel filovirus, Lloviu virus. J Virol 2014; 88:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng M, Ndungo E, Jangra RK et al. Cell entry by a novel European filovirus requires host endosomal cysteine proteases and Niemann-Pick C1. Virology 2014; 468–470:637–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey CC, Huang IC, Kam C, Farzan M. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog 2012; 8:e1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian J, Duff YL, Wang Y et al. Primate lentiviruses are differentially inhibited by interferon-induced transmembrane proteins. Virology 2015; 474:10–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao X, Guo F, Liu F et al. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc Natl Acad Sci U S A 2014; 111:6756–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saphire EO. An update on the use of antibodies against the filoviruses. Immunotherapy 2013; 5:1221–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kajihara M, Marzi A, Nakayama E et al. Inhibition of Marburg virus budding by nonneutralizing antibodies to the envelope glycoprotein. J Virol 2012; 86:13467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Compton AA, Bruel T, Porrot F et al. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe 2014; 16:736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.