Abstract
Background. Throughout the 2014–2015 Ebola outbreak in West Africa, major gaps were exposed in the availability of validated rapid diagnostic platforms, protective vaccines, and effective therapeutic agents. These gaps potentiated the development of prototype rapid lateral flow immunodiagnostic (LFI) assays that are true point-of-contact platforms, for the detection of active Ebola infections in small blood samples.
Methods. Recombinant Ebola and Marburg virus matrix VP40 and glycoprotein (GP) antigens were used to derive a panel of monoclonal and polyclonal antibodies. Antibodies were tested using a multivariate approach to identify antibody-antigen combinations suitable for enzyme-linked immunosorbent assay (ELISA) and LFI assay development.
Results. Polyclonal antibodies generated in goats were superior reagents for capture and detection of recombinant VP40 in test sample matrices. These antibodies were optimized for use in antigen-capture ELISA and LFI assay platforms. Prototype immunoglobulin M (IgM)/immunoglobulin G (IgG) ELISAs were similarly developed that specifically detect Ebola virus–specific antibodies in the serum of experimentally infected nonhuman primates and in blood samples obtained from patients with Ebola from Sierra Leone.
Conclusions. The prototype recombinant Ebola LFI assays developed in these studies have sensitivities that are useful for clinical diagnosis of acute ebolavirus infections. The antigen-capture and IgM/IgG ELISAs provide additional confirmatory assay platforms for detecting VP40 and other ebolavirus-specific immunoglobulins.
Keywords: Ebola, Ebola virus, filovirus, lateral flow immunodiagnostic, point-of-care testing, ELISA
Viral hemorrhagic fevers (VHFs) are serious, often fatal illnesses characterized by high fever, deregulation of the vascular system, and multiorgan failure in the acute phase of the disease. Members of the Filoviridae family of viruses are the causative agents of some of the most devastating VHFs known to occur in humans, with mortality rates as high as 90% during outbreaks [1]. Historically, filoviruses have caused relatively sporadic and geographically contained outbreaks in central and eastern Africa [2]. Casualty rates among significant outbreaks have ranged from 25% to 89% [3–5]. Beginning in March 2014, an outbreak of Ebola (also known as “Ebola hemorrhagic fever” and “Ebola virus disease”) swept through the West Africa nations of Guinea, Sierra Leone, and Liberia, with an estimated 25 515 cases and 10 572 deaths as of 8 April 2015 [6]. This outbreak was caused by a variant of Ebola virus (EBOV; formerly termed “Zaire ebolavirus”) [7, 8]. The West African Ebola outbreak has not been extinguished as of early April 2015; it is the longest, deadliest, costliest, and farthest reaching filovirus outbreak in recorded history, with recorded cases in Senegal, Nigeria, Spain, Mali, United Kingdom, and the United States [6].
At the onset of the 2014 Ebola outbreak, only polymerase chain reaction (PCR) assays were available and validated to diagnose EBOV infection in patient blood samples [9–11]. Although PCR platforms benefit from high sensitivity and specificity, they are not easily deployable and implementable in field settings and are not classifiable as point-of-care (POC) tests. True POC diagnostic tests should be fully portable, be independent of sustainable electricity, be able to rapidly generate sensitive and specific results, have extended shelf life at elevated temperatures, be affordable, and be easily manufactured. A critical need therefore exists to rapidly mobilize resources toward development of rapid and stand-alone diagnostic platforms that can test patients at the point of contact and thus avoid the need to collect samples for transport to testing locations [12]. This critical gap in rapid diagnostic assays for Ebola provide a motivation for development of effective, highly sensitive and specific, easy-to-use, adaptable, and cost-effective lateral flow immunodiagnostic (LFI) assays for public health laboratories, hospital-based clinical laboratories, and POC use. The potential use of filoviruses such as EBOV, Sudan virus (SUDV; formerly termed “Sudan Ebolavirus”), Bundibugyo virus (BDBV), and Marburg virus (MARV) as biological weapons further necessitates the development of rapid and accurate diagnostic assays for biodefense use [13].
A test that could detect filovirus antigens in a small blood sample would be suitable as a rapid diagnostic assay if the test had an appropriate sensitivity. In the early 1990s, Ksiazek et al developed an EBOV antigen-capture enzyme-linked immunosorbent assay (ELISA) that was evaluated initially by using tissue and serum samples from infected monkeys [14]. This ELISA was later evaluated by using samples collected from humans in the 1995 Ebola outbreak in Kikwit, Democratic Republic of the Congo, and exhibited an appropriate level of sensitivity early during disease progression [15], supporting a role for antigen-detection assays in outbreak settings. To our knowledge, no filovirus immunodiagnostic tests have progressed toward commercialization with the requisite level of clinical validation to support its clearance for use as a diagnostic assay in a regulatory jurisdiction. The ELISA and LFI assay prototypes have the ability to provide diagnostic standardization. In particular, LFI diagnostic assays are capable of meeting the rigorous and restrictive performance requirements of austere environments, while providing the antigen-detection sensitivities needed to implement treatment and containment plans in a timely manner.
The LFI assay is a rapid diagnostic test, a technology that provides simple, sensitive, and rapid detection of an analyte in biological fluids, such as whole blood or plasma, and is ideal for use in austere testing environments, where there may be a lack of laboratory infrastructure, such as reliable power, and no advanced biocontainment equipment. More importantly, the deployment of the rapid diagnostic test to hospitals and clinics throughout the region of Lassa fever endemicity permits rapid diagnosis of suspected cases, leading to expedited evacuation of affected individuals to centralized medical facilities capable of treating the disease, such as the Kenema Government Hospital (KGH) Lassa fever ward in eastern Sierra Leone, one of only 2 centers in the world equipped to diagnose, treat, and manage Lassa fever cases year round [16]. Further, widespread testing of patients with suspected Lassa fever helps contain possible outbreaks, owing to expedited follow up by outreach teams in remote settings, testing of possibly infected contacts, implementation of travel restrictions, and, ultimately, timely containment of outbreaks. Here, we describe the initial developmental efforts for an Ebola rapid diagnostic test.
METHODS
Human Subjects
Research involving human subjects at the KGH was approved by the Sierra Leone Ethics and Scientific Review Committee and the Tulane University Institutional Review Board, in accordance with National Institute of Allergy and Infectious Diseases, National Institutes of Health guidelines. Patients with suspected Lassa fever were enrolled after a preadmission evaluation to determine whether they met a previously described case definition for suspected Lassa fever [17]. Contacts of patients with suspected Lassa fever were also eligible for enrollment. All subjects enrolled in this study received a nonidentifying code number (G-series number). All patients enrolled through 25 May 2014, and whose sera was tested for filovirus-specific immunoglobulin M (IgM) and immunoglobulin G (IgG) provided informed consent and were enrolled as having suspected Lassa fever prior to the first diagnosis of an Ebola case in Sierra Leone. These subjects provided written informed consent for publication of their case details, as well as consent to test their samples for the presence of Lassa virus and other disease agents. An additional 49 subjects were enrolled in this study through June 2014 as having suspected Lassa fever, at which time testing for Lassa fever was temporarily suspended at the KGH Lassa fever ward, owing to the national emergency caused by the rapidly spreading Ebola outbreak into Sierra Leone. Human immunodeficiency virus and hepatitis C virus testing was not performed. Because of the Ebola outbreak, a waiver of written consent was granted for testing samples obtained from patients who enrolled after 25 May, all of which were originally collected for EBOV testing. All samples excess was aliquoted and stored. Approval from ethics committees was obtained to use this excess sample store for further testing. At no time during this study were the experimental platforms used to diagnose Ebola in patients suspected of having the disease.
Only KGH staff were involved in the administration of health care to patients in the KGH Lassa ward. All medical decisions were at the sole discretion of the attending physician in the KGH Lassa ward. Small blood volumes (typically 5 mL) for serum separation were collected from study subjects into vacutainer tubes by experienced phlebotomists and processed as previously described [5, 18, 19].
Generation of Recombinant Filovirus Antigens and Monoclonal and Polyclonal Antibodies
Recombinant filovirus VP40 antigens from EBOV, SUDV, and MARV have been generated and tested as target antigens for detection of antifilovirus antibodies [20]. Additionally, EBOV glycoprotein (GP) [21] was used in ELISA as a capture antigen for detection of circulating GP-specific IgG in the blood of infected and convalescent human subjects. These antigens were also used to generate polyclonal and monoclonal antibodies for assay development. Hybridoma clones were derived from mice immunized with EBOV and SUDV VP40 proteins and characterized by isotyping, ELISA complementation analysis, and Western blot to ascertain their independence. Filovirus-specific polyclonal goat antibodies were generated at ProSci (Poway, California) with purified antigens, using a 13-week immunization and blood specimen collection protocol, with monthly extensions. All animal protocols were approved by ProSci's institutional animal care and use committee. Filovirus-specific IgG was purified from hyperimmune goat sera by affinity purification on antigen-coupled chromatography resins. The EBOV nucleoprotein (NP)– and GP-specific monoclonal antibodies KZ51 [22] and KZ52 [23], respectively, were used as controls for assay development.
Prototype Filovirus IgG/IgM Capture ELISA
Purified antigens were used to develop an IgG/IgM capture ELISA. EBOV and SUDV VP40 antigens (75% identical in sequence) were coated separately onto microtiter plates and tested for reactivity with serum from a rhesus macaque infected with EBOV. The prototype IgG/IgM capture ELISA uses microwell plates coated with a mixture of recombinant EBOV and SUDV GP and VP40 antigens (TSRI, La Jolla, California). Controls include a human serum negative control and a reference consisting of EBOV and SUDV GP and VP40 antigen-specific peroxidase bioconjugates lyophilized in human serum. Reference, negative control, and patient serum samples diluted 1:100 were transferred to microwell plates and analyzed essentially as previously described for LASV IgM/IgG ELISA. Positive cutoffs were OD450 values of ≥0.320 for IgG ELISA and ≥0.200 for IgM ELISA.
Prototype Filovirus Antigen-Capture ELISA
Antibodies were initially tested using a multivariate approach based on checkerboard titration of each antibody against every other antibody to identify suitable antibody pairings. Each antibody was tested for both capture (ie, by coating onto the microtiter plate) and detection (ie, horseradish-peroxidase [HRP] conjugation) capability, and testing was performed over a range of plate coating concentrations and HRP-conjugate dilutions. The antigen-capture ELISA prototype uses microwell plates coated with EBOV-specific and SUDV-specific antibodies. Controls included a human serum negative control and a reference consisting of recombinant EBOV and SUDV antigens (TSRI, La Jolla, California) in lyophilized human serum. Diluted reference, controls, and patient serum or plasma diluted 1:9 were analyzed in a manner similar to that previously described for the Lassa virus antigen-capture ELISA [18, 19, 24] but with use of peroxidase-labeled EBOV NP–specific and EBOV VP40–specific caprine polyclonal antibodies as a secondary reagent. The cutoff for positive results was a concentration of 0.01 µg/mL or an OD450 of ≥0.120.
Prototype Filovirus Antigen-Capture LFI Assay
The ReEBOV antigen rapid tests (Corgenix, Broomfield, Colorado) have been developed using goat polyclonal antibodies specific for filovirus antigens. The immunochromatographic dipstick design is similar to that for the previously described ReLASV rapid diagnostic test [18, 19, 24]. The test controls comprise normal human serum as the negative control and recombinant filoviral antigen spiked into normal human serum as the positive control. Visual interpretation of results is performed after a 15–25-minute incubation time.
RESULTS
Filovirus Antigens and Antibodies
Select examples of recombinant filovirus protein expression in mammalian and bacterial cells are provided in Figure 1. These antigens were used to generate polyclonal and monoclonal antibodies for assay development. Demonstration that the monoclonal and polyclonal antibodies produced were capable of binding the appropriate filovirus antigens was achieved in several assays, including ELISA, immunoprecipitation, and Western blotting. As an example of the specificity, the goat polyclonal antibodies were obtained using Escherichia coli–produced recombinant VP40 and EBOV virus-like particles containing VP40, NP, and GP antigens (Figure 2). The panel of reagents assembled for development of prototype antigen and antibody-capture assay platforms in these studies is listed in Supplementary Table 1.
Figure 1.
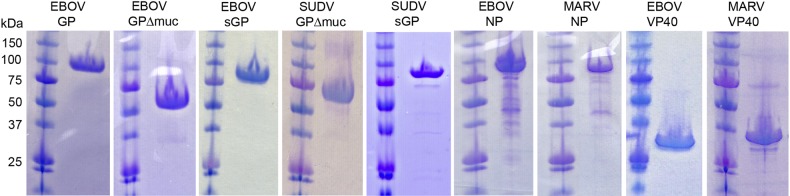
Recombinant filovirus protein expression in mammalian and bacterial cells. From left, Ebola virus (EBOV) glycoprotein (GP; mucin-containing), EBOV GP (mucin-deleted), EBOV secreted GP (sGP), Sudan virus (SUDV) GP (mucin-deleted), SUDV sGP, EBOV nucleoprotein (NP), Marburg virus (MARV) NP, EBOV VP40, and MARV VP40. GPs were expressed in either human 293T cells or in recombinant insect cell systems; those shown here were purified from 293T in multimilligram quantities for crystallization. NP and VP40 antigens were produced in Escherichia coli.
Figure 2.
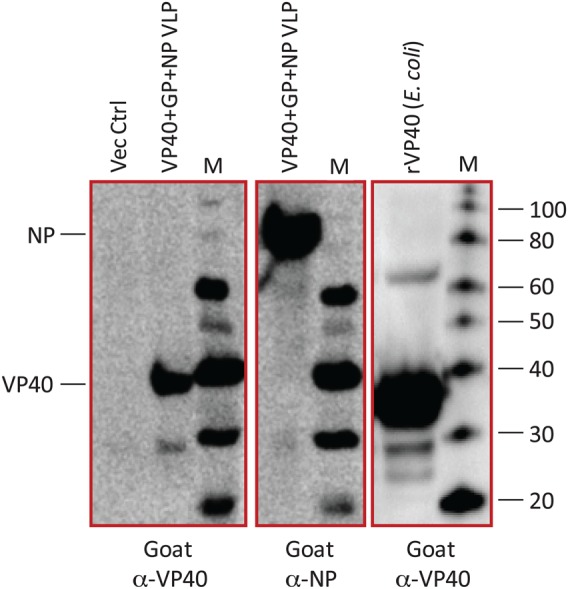
Escherichia coli–produced recombinant VP40 and Ebola virus (EBOV) virus-like particles (VLPs) precipitated from supernatants of HEK-293T/17 cell cultures transfected with EBOV VP40, EBOV nucleoprotein (NP), and EBOV glycoprotein (GP) mammalian expression constructs were resolved on nonreducing Bis-Tris/MES sodium dodecyl sulfate polyacrylamide gels. Proteins were transferred to nitrocellulose membranes and probed with goat anti-VP40 or goat anti-NP polyclonal antibodies (VHFC) and a rabbit anti-goat immunoglobulin G (H + L)-HRP (KPL). Blots were imaged with LumiGlo chemiluminescent substrate (KPL) in a GE ImageQuant LAS4000 imager.
Prototype ELISA for Detection of Filovirus-Specific Immunoglobulins
The matrix protein VP40 is the most abundantly expressed protein in the filoviral particle and infected cell [20, 25]. Evidence of unconventional secretion of VP40 in EBOV-infected cell supernatants and in the serum of experimentally infected animals has also been reported [26]. Virion-associated and soluble VP40 can therefore augment levels of this antigen available for sensitive diagnosis of EBOV infection in test samples. These studies therefore focused on the detection of VP40 antigen as a marker of active EBOV infection. Additionally, EBOV GP-specific circulating IgG was detected in sera from patients with acute infection and from convalescent patients, using ELISA with immobilized GP antigens, to measure the humoral response to Ebola infection.
EBOV VP40–specific and SUDV VP40–specific IgGs were captured and detected over a large dilution range with convalescent EBOV rhesus macaque serum, using ELISA with both immobilized antigens (Figure 3). In this format, EBOV-specific and SUDV-specific IgGs were detected over a dilution range of approximately 2 logs. An IgM response was not detected against either EBOV or SUDV VP40, presumably because the serum sample was collected at a time during convalescence, when an IgM response was no longer detectable (Figure 3).
Figure 3.
Detection of Ebola virus (EBOV) VP40–specific and Sudan virus (SUDV) VP40–specific immunoglobulin G (IgG) from rhesus macaque serum. Microplates were coated with either EBOV or SUDV VP40 and then overlaid with serial dilutions of serum from a rhesus macaque infected with EBOV. IgG contained in the serum was captured over a dilution range of approximately 2 logs, using either EBOV (black) or SUDV (red) VP40. No immunoglobulin M (IgM) was captured by EBOV (blue) or SUDV (green) over the same range because the IgM response had subsided at the time of serum collection.
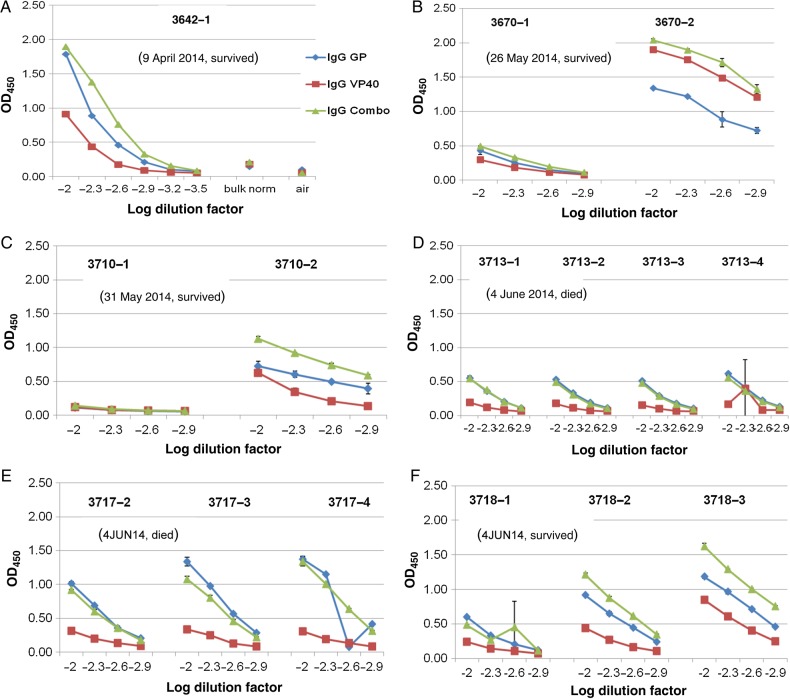
Sera from 6 patients with suspected Lassa fever who presented to the KGH between 9 April 2014 and 4 June 2014 and had at least 2 sequential blood specimens available were analyzed by ELISA for levels of EBOV VP40 and/or EBOV GP–specific IgGs throughout the time course (Figure 4). A dilution range between log2 and log2.9 were generated for 5 patients. As shown in Figure 4A, patient 3642 presented with a significant IgG titer against EBOV VP40 and GP. This patient presented to the KGH Lassa fever ward on 9 April 2014, nearly 2.5 months before the onset of the 2014 outbreak in Sierra Leone. The patient was from Magbema, a village in Kailahun District. The patient tested negative for Lassa virus by the LFI assay and antigen-capture ELISA and positive for LASV-specific IgM and IgG (data not shown). The patient was eventually discharged on 21 April 2014.
Figure 4.
Detection of Ebola virus (EBOV)–specific immunoglobulin G (IgG) from patients with suspected Lassa fever presenting to the Kenema Government Hospital Lassa fever ward between 9 April 9 2014 (G3642) and 4 June 2014 (G3718). Microplates were coated with either EBOV VP40 or glycoprotein (GP) or with a combination of both antigens and then were overlaid with serial dilutions of serum from patients with suspected Lassa fever. Polyclonal anti-human IgG–horseradish peroxidase was used as a detection reagent. Blood specimens obtained sequentially from most patients during hospitalization permitted analysis of seroconversion and increasing IgG titers to EBOV antigens. A single blood specimen was available from patient G3642. Sera were analyzed over a titration range, as permitted by available sample volumes. Patient G3670, who survived Ebola, showed a clear rise in titer between the first and second blood specimen collected, although precise end point titers could not be determined because the samples were too small for analysis. Patients G3710 and G3718, who also survived EBOV infection, registered a moderate increase in IgG titers to both EBOV antigens. Patient G3717, who died from Ebola, registered a substantial titer in the first blood specimen, which did not increase significantly over time. Patient G3713, who also died, did not register a significant titer to EBOV antigens throughout the interval during which samples were collected. IgG reactivity to VP40 (blue), GP (red), and the combination of both antigens (green) from duplicate sample analysis is plotted with standard deviations.
The first Ebola cases reported in Sierra Leone in 2014 were from Kailahun District, which borders the southeast region of Guinea, where the current outbreak is thought to have initiated. Patient 3670 (Figure 4B), from the city of Koindu, Kailahun District, presented with low levels of IgG against EBOV VP40 and GP. Following 5 days of hospitalization, EBOV VP40–specific and EBOV GP–specific IgG titers increased dramatically in this patient. This pattern was similarly observed in patient 3710 (Figure 4C) and patient 3718 (Figure 4F). In contrast, patient 3713 presented with a marginally positive IgG titer to EBOV GP and a negative titer to EBOV VP40, which remained relatively unchanged until death, after an additional period of hospitalization (Figure 4D). Similarly, a slow-rising IgG titer against EBOV GP was recorded in patient 3717, the other subject who died of Ebola, but with a very low and unchanged titer to EBOV VP40 antigen (Figure 4E).
Prototype ELISAs for Detection of Filovirus Antigens
Four candidate pairings were derived for EBOV or SUDV antigen detection, termed “pairs 1–4,” and 1 candidate pairing was derived for MARV antigen detection, termed “pair M1” (Supplementary Table 2). Interestingly, all of the candidate pairings used antibodies made against VP40, suggesting that the VP40 structure contains multiple antigenic sites suitable for detection. These antibody pairings were further optimized by testing coat concentrations and HRP-conjugate dilutions to obtain conditions that favored sensitivity and signal. For EBOV and SUDV, optimized pairings were then tested by ELISA over a dose response of either purified EBOV VP40 or SUDV VP40 spiked into a normal human serum control matrix. We observed that pair 1 and pair 4 each had the ability to detect both EBOV VP40 antigen (Figure 5A) and SUDV VP40 antigen (Figure 5B) with varying affinities and sensitivities (Table 1). Pair 1, pair 2, and pair 3 detected EBOV VP40 with near identical affinity and sensitivity, while pair 4 detected EBOV VP40 with reduced sensitivity. By contrast, pair 4 was better able to detect SUDV VP40 than was pair 3, which has a very high sensitivity for EBOV VP40. Variability among the pairs was likely the natural result of the particular epitopes on VP40 recognized by each antibody and viral species–specific amino acid differences contained in those sites.
Figure 5.
Prototype Ebola virus (EBOV) and Sudan virus (SUDV) antigen-capture enzyme-linked immunosorbent assay (ELISAs) detect EBOV and SUDV VP40. Microplates coated with capture antibodies were challenged with serial dilutions of either EBOV VP40 or SUDV VP40. A, Detection of EBOV VP40 with pair 1 (black), pair 2 (blue), or pair 3 (red) had nearly identical curves; EBOV VP40 detected with pair 4 (green) had a slightly reduced sensitivity. B, Detection of SUDV VP40 with pair 1 (black) showed greater variability, detection with pair 2 (blue) and pair 4 (green) had similar sensitivity, and detection with pair 3 (red) had the lowest sensitivity. C, Prototype Marburg virus (MARV) antigen-capture ELISA detected MARV VP40. Microplates coated with goat-α-MARV antibodies were challenged with serial dilutions of MARV VP40. Detection of captured MARV VP40 with the same self-pairing goat-α-MARV antibody (pair M1; black) showed dose-dependent detection, which lacked an upper plateau over the chosen dilution range.
Table 1.
Performance Characteristics of an Enzyme-Linked Immunosorbent Assay for Detection of Filovirus Antigen
| Antibody Pair | Designation | LOB, OD | LOD, ng/mL |
LOQ, ng/mL |
||
|---|---|---|---|---|---|---|
| EBOV VP40 | SUDV VP40 | EBOV VP40 | SUDV VP40 | |||
| Goat anti-Sudan VP40-HRP | Pair 1 | 0.237 | 10 | 3 | 30 | 100 |
| Goat anti-Zaire VP40-HRP | Pair 2 | 0.204 | 3 | 3 | … | … |
| Goat anti-Zaire VP40-HRP | Pair 3 | 0.269 | 10 | 100 | … | … |
| Goat anti-Sudan VP40-HRP | Pair 4 | 0.205 | 10 | 30 | … | … |
| MARV VP40 | MARV VP40 | |||||
| Goat anti-Marburg VP40-HRP | Pair M1 | 0.145 | 1 | 30 | ||
Abbreviations: EBOV, Ebola virus; HRP, horseradish-peroxidase; LOB, limit of blank; LOD, limit of detection; LOQ, limit of quantitation; MARV, Marburg virus; SUDV, Sudan virus.
We also generated an anti-MARV VP40 polyclonal antibody and analyzed it by ELISA (Figure 5C). We observed that this antibody was capable of self-pairing (likely because of the dimeric nature of native MARV VP40), and therefore we optimized this pairing and tested its ability to detect purified MARV VP40 spiked into a normal human serum control matrix. We observed that this pairing (pair M1) produced a dose response over the dilution range, although the response lacked an upper plateau (Table 1).
Interoperator and interday replicates (data not shown) of lead pairings (pair 1 for EBOV and SUDV and pair M1 for MARV) allowed us to calculate the corresponding limit of blank, limit of detection, and limit of quantitation (Table 1). These data suggest that the prototype antigen-detection ELISAs exhibited sensitivities that are useful for clinical diagnosis.
Prototype LFI Assays for Detection of Filovirus Antigens
Optimizing antibody pairings for use in ELISA does not necessarily predict equivalent performance in an LFI assay. Significant differences exist between the 2 platforms that require separate optimization strategies. To this end, we performed colloidal gold screening experiments and identified 1 monoclonal antibody (raised against EBOV VP40) that, qualitatively, retained its sensitivity and selectivity after colloidal gold conjugation. Likewise, the MARV VP40 polyclonal antibody retained some of its performance characteristics when conjugated to colloidal gold. These screening experiments allowed us to identify 3 antibody pairings to test in the LFI assay: 2 EBOV/SUDV pairings (pairs 5 and 6) and 1 MARV pair (pair M2; Supplementary Table 2). Once optimized, LFI strips were constructed and tested using purified antigen spiked into a normal human serum control matrix. LFI strips constructed with pair 5 exhibit a visual dose response at the test line when challenged with serial dilutions (300–0.1 µg/mL) of EBOV VP40 (Figure 6A). Pair 5 was also able to detect SUDV VP40 over the same range, although with reduced sensitivity (Figure 6B), indicating a selectivity of this pairing for EBOV VP40. LFI strips constructed with pair M2 exhibited a visual dose response at the test line when challenged with serial dilutions (300–0.1 µg/mL) of MARV VP40 (Figure 6C), albeit with low top-end sensitivity.
Figure 6.
Prototype Ebola virus (EBOV)/Sudan virus (SUDV) antigen-capture lateral flow immunodiagnostic (LFI) assays detect EBOV and SUDV VP40. LFI strips prepared with pair ES5 were challenged with a serial dilution (300-0.1 µg/mL) of either EBOV VP40 antigen (A) or SUDV VP40 antigen (B). Pair ES5 detected both antigens in a dose-dependent manner, with greater qualitative sensitivity observed for EBOV VP40 antigen. C, Prototype Marburg virus (MARV) antigen-capture LFI assay detects MARV VP40. LFI strips prepared with pair M2 were challenged with a serial dilution of MARV VP40 antigen. Pair M2 detected the antigen in a dose-dependent manner. Control (C) and test (T) lines on each set of strips are marked. Abbreviation: BCN, bulk control normal.
DISCUSSION
We have assembled a filovirus VP40 and GP antigen and antibody library from EBOV, SUDV, and MARV. Collectively, these tools provide a library of critical reagents for development of testing platforms that have furthered our understanding of how to rapidly diagnose active and convalescent filoviral infections. In this effort, we have generated and characterized 4 antigens and 10 antibodies with varying affinities and developed both ELISA and LFI assay prototypes with detection limits that are suitable for clinical diagnosis of filoviral infections.
The ongoing outbreak of Ebola in the West Africa enabled validation studies of a filoviral LFI assay and ELISA, using large panels of sera from patients with acute infection and those who were convalescent presenting to the KGH Lassa fever ward for treatment. The existing clinical research project (the Lassa Fever Program) facilitated our accelerated Ebola immunodiagnostics development program. Analytical performance testing using spiked recombinant antigens demonstrated that the current ELISA pairings for EBOV and SUDV exhibited sensitivities for detecting viral antigens in a blood or plasma sample that were similar to those we observed with Lassa virus rapid test prototypes. Extending the filovirus diagnostics program to include identification of additional antibody pairings for EBOV, SUDV, MARV, and BDBV will be useful for other regions in Middle Africa and Asia where outbreaks of filoviruses have occurred in the past.
The filovirus diagnostic platform can also be useful for epidemiological studies of filoviruses. In a recent study, we reported the detection of VP40-specific IgGs in a panel of archived and recently collected sera from patients with suspected Lassa fever, using prototype ELISAs [27]. This study and others [28–30] have established a pattern of filovirus seropositivity in West African populations that was quiescent until the recent outbreak and that could have resulted from the introduction of a new and highly virulent strain or from the adaptation of a previously circulating strain with enhanced virulence in humans. These observations will require additional studies to determine whether a filovirus antigenically related to EBOV may have circulated undetected in the local human population for months or longer prior to the 2014 outbreak.
A key feature of the LFI prototypes is their simplicity and relatively low cost, making them attractive for widespread use in a variety of clinical and field settings, with a focus on austere environments. These tests provide the ease of use necessary under field conditions and for personnel that may have varying levels of training. LFI diagnostic tests are robust within a wide range of temperature and stability conditions, making them well suited for transport, storage, and use in tropical regions. In addition, LFI technology uses low sample volumes, allowing for diagnosis using unprocessed whole blood specimens obtained from a finger stick. Further, the sensitivity of LFI assays in many cases meets or exceeds that of comparative protein-based assays, providing improved diagnostic power without the laboratory resource and training constraints associated with high-level technologies, as evidenced by previous work with the Lassa virus ELISA used during clinical research in Sierra Leone [18, 19, 24, 27]. Taken together, these characteristics suggest that the LFI diagnostic format can contribute to monoplexed and multiplexed pathogen detection for EBOV, other filoviruses, Lassa virus, and other pathogens, thereby providing a panel approach to the rapid triage of febrile patients on the basis of the contagion and the potential for disease transmission.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank members of the Viral Hemorrhagic Fever Consortium (Autoimmune Technologies, the Broad Institute, Corgenix, Harvard University, the Scripps Research Institute, Tulane University, the University of California–San Diego, the University of Texas Medical Branch, Vybion, Zalgen Labs, the Irrua Specialist Teaching Hospital Lassa Fever Program, and the KGH); the World Health Organization Mano River Union Lassa Fever Network and the Sierra Leone Ministry of Health and Sanitation, without which this work could not have been conducted; and the patients with serious febrile illnesses who presented to KGH, as well as their families, without whose cooperation this study would not have been possible.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) (grants AI067188, AI067927, AI070530, AI081982, AI082119, AI082805 AI088843, AI104216, AI104621, AI115754, and HHSN272200900049C), the Skaggs Institute for Chemical Biology, and the Burroughs Wellcome Fund (Investigators in the Pathogenesis of Infectious Disease award to E. O. S.).
Potential conflicts of interest. The Viral Hemorrhagic Fever Consortium (available at: http://www.vhfc.org) is a partnership of academic and industry scientists who are developing diagnostic tests, therapeutic agents, and vaccines for Lassa fever, Ebola, and other severe diseases. Tulane University and its various academic and industry partners have filed US and foreign patent applications on behalf of the consortium for several of these technologies. If commercial products are developed, consortium members may receive royalties or profits. This does not alter our adherence to all policies of the NIH and The Journal of Infectious Diseases on sharing data and materials.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Agua-Agum J, Ariyarajah A, Aylward B et al. . West African Ebola epidemic after one year--slowing but not yet under control. N Engl J Med 2015; 372:584–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Ebola outbreaks 2000–2014. http://www.cdc.gov/vhf/ebola/resources/outbreaks.html. Accessed 8 April 2015.
- 3.Kortepeter MG, Smith PW, Hewlett A, Cieslak TJ. Caring for patients with Ebola: a challenge in any care facility. Ann Intern Med 2015; 162:68–9. [DOI] [PubMed] [Google Scholar]
- 4.Fowler RA, Fletcher T, Fischer WA II et al. . Caring for critically ill patients with Ebola virus disease. Perspectives from West Africa. Am J Respir Crit Care Med 2014; 190:733–7. [DOI] [PubMed] [Google Scholar]
- 5.Schieffelin JS, Shaffer JG, Goba A et al. . Clinical illness and outcomes in patients with Ebola in Sierra Leone. N Engl J Med 2014; 371:2092–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. Ebola situation report—8 April 2015. http://apps.who.int/ebola/current-situation/ebola-situation-report-8-april-2015. Accessed 8 April 2015.
- 7.Baize S, Pannetier D, Oestereich L et al. . Emergence of Zaire Ebola virus disease in Guinea - preliminary report. N Engl J Med 2014; 371:1418–25. [DOI] [PubMed] [Google Scholar]
- 8.Gire SK, Goba A, Andersen KG et al. . Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 2014; 345:1369–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. Emergency Use Assessment and Listing (EUAL) procedure for Ebola virus disease (IVDs). http://www.who.int/diagnostics_laboratory/procurement/purchasing/en/. Accessed 8 April 2015.
- 10.Food and Drug Administration. Emergency use authorization. Available at: http://www.fda.gov/EmergencyPreparedness/Counterterrorism/MedicalCountermeasures/MCMLegalRegulatoryandPolicyFramework/ucm182568.htm - current. Accessed 8 April 2015.
- 11.Reusken C, Niedrig M, Pas S et al. . Identification of essential outstanding questions for an adequate European laboratory response to Ebolavirus Zaire West Africa 2014. J Clin Virol 2015; 62:124–34. [DOI] [PubMed] [Google Scholar]
- 12.Dhillon RS, Srikrishna D, Garry RF, Chowell G. Ebola control: rapid diagnostic testing. Lancet Infect Dis 2015; 15:147–8. [DOI] [PubMed] [Google Scholar]
- 13.Bray M. Defense against filoviruses used as biological weapons. Antiviral Res 2003; 57:53–60. [DOI] [PubMed] [Google Scholar]
- 14.Ksiazek TG, Rollin PE, Jahrling PB, Johnson E, Dalgard DW, Peters CJ. Enzyme immunosorbent assay for Ebola virus antigens in tissues of infected primates. J Clin Microbiol 1992; 30:947–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ksiazek TG, Rollin PE, Williams AJ et al. . Clinical virology of Ebola hemorrhagic fever (EHF): virus, virus antigen, and IgG and IgM antibody findings among EHF patients in Kikwit, Democratic Republic of the Congo, 1995. J Infect Dis 1999; 179(suppl 1):S177–87. [DOI] [PubMed] [Google Scholar]
- 16.Shaffer JG, Grant DS, Schieffelin JS et al. . Lassa fever in post-conflict Sierra Leone. PLoS Negl Trop Dis 2014; 8:e2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan SH, Goba A, Chu M et al. . New opportunities for field research on the pathogenesis and treatment of Lassa fever. Antiviral Res 2008; 78:103–15. [DOI] [PubMed] [Google Scholar]
- 18.Branco LM, Grove JN, Boisen ML et al. . Emerging trends in Lassa fever: redefining the role of immunoglobulin M and inflammation in diagnosing acute infection. Virol J 2011; 8:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Branco LM, Boisen ML, Andersen KG et al. . Lassa hemorrhagic fever in a late term pregnancy from northern Sierra Leone with a positive maternal outcome: case report. Virol J 2011; 8:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bornholdt ZA, Noda T, Abelson DM et al. . Structural rearrangement of Ebola virus VP40 begets multiple functions in the virus life cycle. Cell 2013; 154:763–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 2008; 454:177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meissner F, Maruyama T, Frentsch M et al. . Detection of antibodies against the four subtypes of Ebola virus in sera from any species using a novel antibody-phage indicator assay. Virology 2002; 300:236–43. [DOI] [PubMed] [Google Scholar]
- 23.Parren PW, Geisbert TW, Maruyama T, Jahrling PB, Burton DR. Pre- and postexposure prophylaxis of Ebola virus infection in an animal model by passive transfer of a neutralizing human antibody. J Virol 2002; 76:6408–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grove JN, Branco LM, Boisen ML et al. . Capacity building permitting comprehensive monitoring of a severe case of Lassa hemorrhagic fever in Sierra Leone with a positive outcome: case report. Virol J 2011; 8:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geisbert TW, Jahrling PB. Differentiation of filoviruses by electron microscopy. Virus Res 1995; 39:129–50. [DOI] [PubMed] [Google Scholar]
- 26.Reynard O, Reid SP, Page A et al. . Unconventional secretion of Ebola virus matrix protein VP40. J Infect Dis 2011; 204(suppl 3):S833–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boisen ML, Schieffelin JS, Goba A et al. . Multiple circulating infections can mimic the early stages of viral hemorrhagic fevers and possible human exposure to filoviruses in Sierra Leone prior to the 2014 outbreak. Viral Immunol 2015; 28:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoepp RJ, Rossi CA, Khan SH, Goba A, Fair JN. Undiagnosed acute viral febrile illnesses, sierra leone. Emerg Infect Dis 2014; 20:1176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becker S, Feldmann H, Will C, Slenczka W. Evidence for occurrence of filovirus antibodies in humans and imported monkeys: do subclinical filovirus infections occur worldwide? Med Microbiol Immunol 1992; 181:43–55. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez JP, Josse R, Johnson ED et al. . Antibody prevalence against haemorrhagic fever viruses in randomized representative Central African populations. Res Virol 1989; 140:319–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.