Abstract
The ongoing evolution of Ebolaviruses poses significant challenges to the development of immunodiagnostics for detecting emergent viral variants. There is a critical need for the discovery of monoclonal antibodies with distinct affinities and specificities for different Ebolaviruses. We developed an efficient technology for the rapid discovery of a plethora of antigen-specific monoclonal antibodies from immunized animals by mining the VH:VL paired antibody repertoire encoded by highly expanded B cells in the draining popliteal lymph node (PLN). This approach requires neither screening nor selection for antigen-binding. Specifically we show that mouse immunization with Ebola VLPs gives rise to a highly polarized antibody repertoire in CD138+ antibody-secreting cells within the PLN. All highly expanded antibody clones (7/7 distinct clones/animal) were expressed recombinantly, and shown to recognize the VLPs used for immunization. Using this approach we obtained diverse panels of antibodies including: (i) antibodies with high affinity towards GP; (ii) antibodies which bound Ebola VLP Kissidougou-C15, the strain circulating in the recent West African outbreak; (iii) non-GP binding antibodies that recognize wild type Sudan or Bundibugyo viruses that have 39% and 37% sequence divergence from Ebola virus, respectively and (iv) antibodies to the Reston virus GP for which no antibodies have been reported.
Ebolaviruses are negative-sense RNA filamentous viruses that cause very high morbidity and mortality1. Host cell entry is mediated first by the attachment of the heavily glycosylated glycoprotein (GP) on the viral envelope to the host cell encoded T-cell immunoglobulin and mucin domain 1 (TIM-1)2. Following cathepsin cleavage in the lysosome, GP mediates cellular entry by binding the host cell encoded Niemann-Pick C1 (NPC1)3. Five antigenically distinct ebolaviruses exhibiting 35–45% genome sequence divergence have been discovered4: Ebola virus (abbreviated as EBOV, formerly designated as Zaire ebolavirus); Sudan virus (SUDV); Bundibugyo virus (BDBV); Reston virus (RESTV, for which no zoonotic infections have been reported to date)5; and Taï Forest virus (TAFV, one incident of human infection)6. The recent EBOV outbreak in West Africa, centered in Guinea, Sierra Leone, and Liberia with isolated outbreaks in Nigeria and Mali, was the largest ever with a mortality rate estimated at 70% of recorded definitive clinical outcomes (http://www.cdc.gov/vhf/ebola/outbreaks/2014-west-africa/index.html)7. Phylogenetic comparison of isolates from the recent outbreak8 with 20 Ebolavirus genomes from earlier outbreaks suggested that the 2014 West African virus likely spread from central Africa within the past decade, having diverged from a common ancestor around 20049. The five Ebolavirus species have varying rates of molecular evolution, with the highest of 8.21 × 10−4 nucleotide substitutions/site/year for Reston virus10. The ongoing evolution of Ebolaviruses poses significant challenges to the development of immunodiagnostics. Specifically, there is a critical need for the discovery of panels of monoclonal antibodies with distinct affinities and specificities for different Ebolaviruses.
Antibodies to EBOV and SUDV have been produced from hybridomas11,12; by in vitro screening of synthetic Fab libraries13,14, and from human immune antibody libraries constructed from infected individuals15. However additional monoclonal antibodies to Ebolaviruses are urgently needed both for diagnostic purposes and as therapeutics16,17. Specifically, the generation of diagnostic antibodies to Ebolaviruses is complicated by the structural complexity of the GP, which is heavily glycosylated in a host cell-specific manner18,19 and subjected to proteolytic cleavage during entry20, as well as by the sequence diversity of the Ebolaviruses. Finally, characterization of useful antibodies to Ebolaviruses is limited by the safety concerns associated with handling the live virus.
Antibody discovery has relied either on the immortalization21 or cloning of antibodies isolated from individual B cells obtained from an antigen-challenged host22,23,24,25,26,27 or, alternatively, on the in vitro isolation from combinatorial libraries using a variety of screening techniques28. The current collection of antibody technologies is predicated on the isolation of clones that display high antigen binding. However, animal immunization induces the stimulation and expansion of a highly diverse population of B cells encoding an antibody repertoire with a wide range of antibody affinities22,23. Antibodies with low affinity nonetheless may exhibit other highly desirable properties, including broad cross-reactivity or heteroclitic specificity, i.e. stronger binding reaction to a different antigen other than the one used for immunization24,25,29. Unfortunately, there is no straightforward way to identify such interesting antibodies. For example, while the isolation of antibodies that bind to multiple antigens (e.g. to different flu hemagglutinins) or that neutralize rapidly evolving pathogens such as HIV-1 or flu has been accomplished by B cell cloning, the process typically requires the screening of many thousands of B cells and therefore is very laborious and expensive26,27,30,31.
In order to satisfy the need for a wider variety of antibodies to Ebolaviruses, we developed a novel approach to comprehensively mine the full suite of antibody diversity, shaped by in vivo selective mechanisms and generated within the boundary of reactive secondary lymphoid tissues in immunized animals. We reasoned that antibodies encoded by antigen-stimulated B cells that had undergone the greatest degree of expansion within the confinement of a secondary lymphoid organ are most likely to display desirable antigen recognition properties including heteroclite recognition of diverse Ebolaviruses. Briefly, mice were first immunized in the footpad with Ebola virus-like particles (VLPs). Footpad immunization triggers a strong and highly-focused immune reaction in the PLN, especially for particles <40 nm such as VLPs32. Antigen experienced, CD138+ B cells (plasmablasts) from the PLN were isolated and the natively paired VH:VL repertoire encoded by these cells was determined by NextGen sequencing33. Antibodies corresponding to the highest frequency VH:VL pairs, and thus likely arising from the most clonally expanded and highly-transcribing CD138+ B cells within the PLN, were expressed recombinantly and their binding properties were characterized in detail. In each of two mice tested, 7/7 antibodies encoded by the highest frequency antigen-draining PLN plasmablast sequences recognized the antigen (Ebola VLPs) with several binding to recombinant GPs with up to nM affinities. Interestingly, even though animals had been immunized with EBOV VLPs, mining of the expanded native B cell repertoire within the PLN resulted in antibodies that also recognized SUDV and BDBV. In a separate experiment we also employed this technique to generate the first RESTV GP-specific antibodies. Thus, the antibodies reported here collectively constitute a panel of reagents for the detection of most Ebolaviruses.
Results
Immunization of the PLN yields antigen-specific antibodies
Figure 1 summarizes our approach for mining the antibody repertoire encoded by the most highly expanded, antigen-experienced B cells following antigen stimulation. Footpad immunization leads to a strong inflammatory response in the draining popliteal lymph node (Fig. 1a). Unlike lymph nodes that drain sites of frequent extracorporeal interaction (e.g. the oral cavity or lungs), the germinal centers of the popliteal lymph node are normally relatively unstimulated34. Footpad immunization results in a marked increase in cellularity in the ipsilateral popliteal lymph node relative to the unstimulated contralateral lymph node35,36, and a large fraction of the constituent antibody-secreting B cells were expected to be antigen-specific.
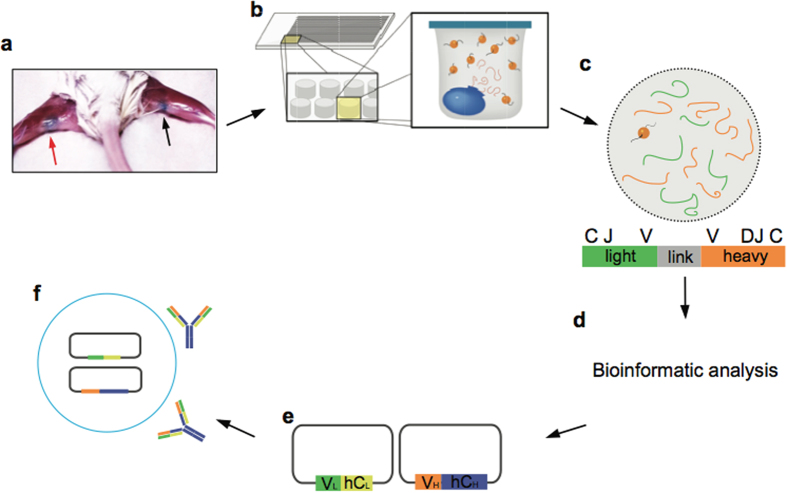
Figure 1. Isolation of antibodies by mining the paired VH:VL repertoire of draining popliteal lymph node (PLN) antibody-secreting B cells.
(a) Footpad immunization leads to a marked increase in cellularity within the ipsilateral popliteal lymph node relative to the contralateral lymph node (red and black arrows respectively). (b) PLN CD138+ cells isolated by magnetic sorting are deposited into 125 pL wells on PDMS slides that also contain poly(dT) beads. Cells are lysed in situ and mRNA is captured on the poly(dT) beads33. (c) The poly(dT) beads are emulsified and VH:VL amplicons are generated following reverse transcription and overlap extension PCR. (d) VH:VL amplicons are sequenced using Illumina 2 × 250 MiSeq and the highest frequency VH:VL pairs are identified via bioinformatics analysis. (e) Highest frequency VH (orange) and VL (green) genes are synthesized and cloned into IgH and IgL expression vectors containing human IgG1 (blue) and human kappa (yellow) constant regions, respectively. (f) Following co-transfection into Expi293 cells, recombinant IgG antibodies are expressed and purified.
Ebola VLPs were produced by co-transfection of HEK293FT cells with plasmids encoding the three major virus structural proteins: nucleoprotein (NP), VP40 and GP of the EBOV Mayinga strain and were purified by sucrose density centrifugation. Electron microscopy and gel staining confirmed that the VLPs displayed a morphology and consistency characteristic of EBOV virions (Supplementary Fig. 1). Three mice were immunized with Ebola VLPs in emulsified adjuvant in the left hind footpad, followed by boost immunization in the lateral hock to minimize pain and discomfort. An anti-VLP titer of >1:104 was observed after the second boost (Supplementary Fig. 2a) in all three mice, and two mice (denoted as ZM1 and ZM2) were chosen for further analysis. A final boost was administered and 6 days later the popliteal lymph nodes were extracted. As expected, the ispilateral PLN was observed to be hypertrophic (Supplementary Fig. 2b).
CD45R−CD19−CD138+ antibody secreting B cells were enriched by magnetic sorting and the paired VH:VL repertoire from single cells was determined following sequestration of the cells into 125 pL wells on PDMS slides33 (Fig. 1b). Approximately 1 × 105 CD45R−CD19−CD138+ plasmablasts were isolated by magnetic sorting, of which 2.5–3.5 × 104 cells were processed to create natively paired VH:VL amplicons. Linked VH:VL amplicons of approximately 850 bp were generated and then sequenced using Illumina MiSeq technology (Fig. 1c). High quality reads were clustered based on the CDRH3:CDRL3 sequences (Fig. 1d).
The PLN plasmablast IgG repertoire elicited by immunization with EBOV
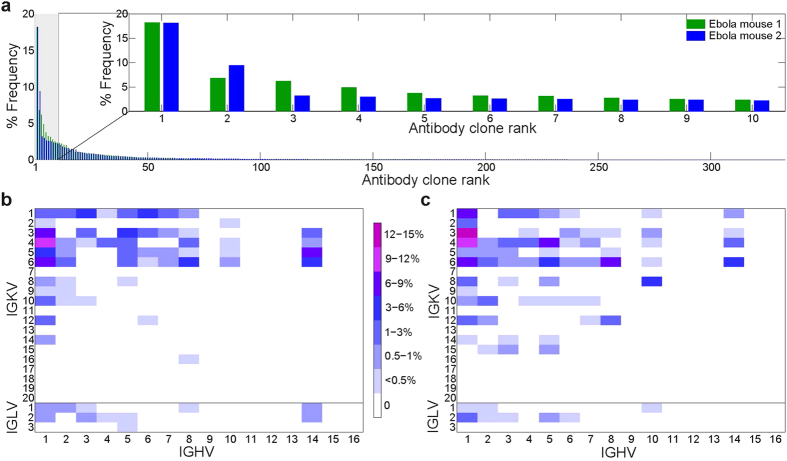
283 and 333 unique VH:VL pairs (represented by ≥2 sequence reads per pair each), comprising the PLN plasmablast repertoires were identified in mouse ZM1 and mouse ZM2, respectively. The repertoires from both Ebola VLPs immunized mice investigated were heavily skewed, with the top ten most abundant VH:VL pairs representing 53.9% and 48.4% of the total sequence counts in each mouse, respectively (Fig. 2a). We observed a strong bias in germline V-gene usage in immunized mice (Fig. 2b,c). Biased usage of IGHV subgroups 1–3 and 537,38, as well as IGKV1, 3, 4 and 6 have been previously observed in the repertoires of unimmunized mice38. Likewise, in Ebola VLPs immunized mice, IGHV1, IGHV5, as well as IGKV1, IGKV3, IGKV4, and IGKV6 heavy and light V genes were most strongly represented. Importantly, however, the IGHV8, IGHV14, and IGKV5 families were also strongly overrepresented in both mice immunized with EBOV VLPs. The enrichment of IGHV8 in the plasmablast repertoire from PLN of Ebola VLPs immunized animals is particularly noteworthy as this germline family is expressed at a very low level in mice37,38 and additionally it was shown to be utilized at very low frequencies in CD138+ lymphocytes from animals immunized with various other antigens22 (Supplementary Fig. 3a,b).
Figure 2. Characteristics of the PLN VH:VL repertoire in CD138+ antibody secreting cells.
(a) Polarization of VH:VL repertoire after immunization with EBOV VLPs. The frequency of each unique CDRH3:CDRL3 antibody clonotype is shown as a percentage of total sequencing read counts. CDRH3 sequencing reads having at least 96% identity at the nucleotide level were clustered and compiled, then analyzed for corresponding CDRL3 per pair. Sequences identified in <2 reads were excluded to minimize sequencing error33. Inset: frequency of the ten most frequently observed CDRH3:CDRL3 antibody clonotypes from each mouse. (b,c) VH:VL gene family usage of unique CDRH3:CDRL3 clonotypes in mouse ZM1 and ZM2, respectively.
The repertoire of Ebola VLPs immunized mice displayed an average CDRH3 length distribution similar to that observed in CD138+ repertoires previously reported22 (Supplementary Fig. 3c,d), although there was a slight skewing toward shorter CDRH3 lengths. Shorter CDRH3s are commonly found among antibodies that bind carbohydrates and thus the skewing observed here likewise may have reflected the elicitation of antibodies to the glycan component of the heavily glycosylated GP39. The plasmablast repertoire from EBOV VLPs immunized mice displayed a higher level of somatic hypermutation level in the framework 3 (FR3) heavy chain region compared to plasmablast repertoires reported for mice hyperimmunized with various other protein antigens22 (Supplementary Fig. 3e,f).
Construction and characterization of anti-EBOV VLP antibodies
The CDRH3 and CDRL3 amino acid sequences of the 7 highest frequency VH:VL clonotypes from each mouse, together with the respective V(D)J gene segments are listed in Table 1. Antibody genes for these 14 most prevalent VH:VL antibody clonotypes were synthesized and cloned as mouse V region-human constant domain chimeric antibodies, expressed in HEK293 cells, and characterized for antigen binding (Fig. 1e,f). KZ52, a very well characterized Ebola virus recognizing antibody isolated from a survivor of the 1995 Kikwit outbreak15,40, was expressed as a positive control. All 14 antibodies gave an ELISA signal above background on plates coated with the immunizing antigen, i.e. EBOV (Mayinga strain) VLPs (Fig. 3a). ELISA titer analysis revealed that the majority of the antibodies bound VLPs at a titer >1:1,000 (Supplementary Table 1). Interestingly, the ELISA signals did not correlate with the frequency of the respective antibodies in the PLN CD138+ repertoire.
Table 1. List of characterized EBOV antibodies sequenced from PLN CD138+ cells.
| Mouse | Rank (Name) | CDRH3 Sequence | CDRL3 Sequence | Gene Usage |
|---|---|---|---|---|
| 1 | 1 (ZM1.1) | ARSFAY | QQSNEDPYTF | IGHV1-IGHJ3:IGKV3-IGKJ2 |
| 2 (ZM1.2) | TGDGYYGFAY | FQGSHVPFT | IGHV6-IGHD2-IGHJ3:IGKV1-IGKJ4 | |
| 3 (ZM1.3) | ARGIGY | WQGTHFPFT | IGHV3-IGHJ3:IGKV1-IGKJ4 | |
| 4 (ZM1.4) | ARSTTATLDC | QQSDSWPTLT | IGHV14-IGHD1-IGHJ2:IGKV5-IGKJ5 | |
| 5 (ZM1.5) | ATISTATFPY | QQSDSWPTLT | IGHV1-IGHD1-IGHJ3:IGKV5-IGKJ5 | |
| 6 (ZM1.6) | ARRAMITTEGVDFDY | QQSRKVPWT | IGHV3-IGHD2-IGHJ2:IGKV3-IGKJ1 | |
| 7 (ZM1.7) | AREGYRYDWYFDV | QQRSSYPLT | IGHV1-IGHD2-IGHJ1:IGKV4-IGKJ5 | |
| 2 | 1 (ZM2.1) | TRSVSDY | WQGTHFPHT | IGHV1-IGHD2-IGHJ2:IGKV1-IGKJ5 |
| 2 (ZM2.2) | ARRTYRYDRFDY | QQWSSDPLT | IGHV1-IGHD2-IGHJ2:IGKV4-IGKJ5 | |
| 3 (ZM2.3) | TRRSNFPYYFDF | QQSIEDPFT | IGHV1-IGHD2-IGHJ2:IGKV3-IGKJ4 | |
| 4 (ZM2.4) | ARSELGATGFAY | QQGQSYPIFT | IGHV5-IGHD3-IGHJ3:IGKV15-IGKJ4 | |
| 5 (ZM2.5) | ARQKYGNYVLYWYFDV | QQWNSNPPT | IGHV5-IGHD2-IGHJ1:IGKV4-IGKJ4 | |
| 6 (ZM2.6) | TGMVTSY | LQHWNYPYT | IGHV6-IGHD2-IGHJ3:IGKV6-IGKJ2 | |
| 7 (ZM2.7) | VREGLGSYFDY | QQYYNYPRT | IGHV10-IGHD5-IGHJ2:IGKV8-IGKJ1 |
For each antibody, CDRH3:CDRL3 clonotypes and their V(D)J gene assignment are provided.
Figure 3. Functional characterization of IgG antibodies isolated via mining of the PLN CD138+ B cell repertoire.
(a) Binding to EBOV VLPs for antibodies encoded by the seven most frequently observed CDRH3:CDRL3 clonotypes from the sequenced PLN CD138+ B cell repertoires of each mouse. (b) Binding to purified EBOV recombinant GP for select antibodies as determined by ELISA. Binding to BSA as a control is shown in correspondingly colored dashed lines. Error bars represent the standard error of the mean for three technical replicates.
5/14 antibodies recognized the recombinant uncleaved form of the EBOV Mayinga strain GP (Fig. 3b). One antibody bound to GP with a single-digit nM equilibrium dissociation constant (ZM1.3, KD = 7.7 nM), as determined by SPR analysis (Table 2, Supplementary Fig. 4), while two others (ZM1.1 and ZM2.1) exhibited KD values in the 10 nM range. Finally, 2/5 GP specific IgGs, ZM1.2 and ZM1.6, exhibited lower affinities (KD = 156 and 635 nM, respectively (Table 2, Supplementary Fig. 4)), consistent with the lower titer of those antibodies for Ebola VLPs.
Table 2. SPR Binding kinetics and equilibrium dissociation constants (KD) towards uncleaved EBOV GP.
| Antibody ID | kon (M−1s−1) | koff (s−1) | KD (nM) |
|---|---|---|---|
| ZM1.1 | (3.12 ± 0.55) × 104 | (1.16 ± 0.07) × 10−3 | 37.7 ± 0.55 |
| ZM1.2 | (1.17 ± 0.41) × 104 | (1.71 ± 0.12) × 10−3 | 156 ± 47.4 |
| ZM1.3 | (3.12 ± 0.53) × 104 | (2.39 ± 0.62) × 10−4 | 7.71 ± 1.77 |
| ZM1.6 | (3.95 ± 0.7) × 103 | (2.46 ± 0.12) × 10−3 | 635 ± 93.8 |
| ZM2.1 | (1.47 ± 0.23) × 104 | (2.97 ± 0.07) × 10−4 | 20.5 ± 3.44 |
Experiments were performed in three technical replicates and data were fit to a 1:1 Langmuir binding model.
The three highest affinity, EBOV GP-specific antibodies (ZM1.1, ZM1.3 and ZM2.1) were tested for binding to VLPs encoding GPs from Ebola strains isolated from two different outbreaks: 034-KS (Democratic Republic of Congo, 2008, NCBI Accession number HQ613402) and Kissidougou-C15 (Kissidougou, Guinea, 2014, NCBI Accession number KJ660346) (Supplementary Fig. 5). Both ZM1.3 and ZM2.1 showed higher binding to Ebola 034-KS and Kissidougou-C15 VLPs than the well-studied KZ52 antibody (KD for purified Mayinga GP = 1.55 nM) (Supplementary Fig. 6a,b). We observed that KZ52 failed to recognize VLPs containing a Mayinga GP N550K variant in which Arg 550 residue was replaced with Lys, a mutation observed in Marburg GP41 (Supplementary Fig. 6c), while ZM1.1, ZM1.3 and ZM2.1 were still able to bind the Mayinga GP N550K variant. This finding indicates that ZM1.1, ZM1.3 and ZM2.1 likely recognize a different epitope than KZ52. Interestingly, ZM1.3 displayed heteroclitic specificity in that it bound better to EBOV 034-KS and Kissidougou-C15 VLPs than to the immunization Mayinga strain VLPs (EC50 of 0.334 nM and 0.39 nM, respectively compared to 1.7 nM for Mayinga VLPs). These antibodies also bound to live wild type Ebola virus (Supplementary Fig. 6d), indicating that they should be useful for developing diagnostic assays against primary biological samples.
Finally, encouraged by the results detailed above, we used the strategy described in Fig. 1 to develop antibodies that recognized Reston Ebola virus (RESTV, Reston, 1996, NCBI Accession number AB050936), an Ebolavirus for which no anti-GP monoclonal antibodies are available. RESTV VLPs were generated, mice were immunized as above, and the repertoire encoded by PLN plasmablasts was determined. We identified three antibodies designated RM2.4, RM3.2, and RM3.3 (Supplementary Table 2) that bound to both RESTV VLPs as well as to RESTV recombinant GP (Fig. 4).
Figure 4. Functional characterization of IgG antibodies isolated via mining of RESTV immunized PLN CD138+ B cell repertoire.
Binding to RESTV recombinant GP for select antibodies as determined by ELISA. Curves were fitted using 4-parameter logistic non-linear regression. Error bars represent the standard error of the mean for three technical replicates.
Diagnostic utility of non-GP binding antibodies
While 9/14 anti-EBOV VLP antibodies did not show binding to the GP and thus presumably recognized other VLP proteins (NP, VP40) they nonetheless are of diagnostic utility. Specifically, in addition to binding to EBOV Mayinga VLPs, antibodies ZM1.4, ZM1.7, ZM2.2, ZM2.3, ZM2.5, and ZM2.6 displayed measurable binding to Bundibugyo Virus (BDBV, Bundibugyo, 2007, NCBI Accession number FJ217161), while ZM1.7, ZM2.2, ZM2.5, and ZM2.6 also showed binding activity to Sudan Virus (SUDV, Gulu, 2000, NCBI Accession number AY729654) (Representative ELISA data is shown in Fig. 5a–c, also see Supplementary Table 3). Thus, antibodies elicited by immunization with EBOV VLPs bound differentially to phylogenetically diverse variants.
Figure 5. Binding of cross-reactive antibodies isolated via mining of the PLN CD138+ B cell repertoire to wild type EBOV, BDBV and SUDV viruses.
ELISA assays using antibodies ZM1.7, ZM2.5, ZM2.6, and KZ52 for the detection of (a) wild type Ebola virus; (b) wild type Bundibugyo virus; and (c) wild type Sudan virus. Assays were performed in two technical replicates. Lines represent measurements fitted via 4-parameter logistic nonlinear regression for EC50.
Discussion
Here we report a facile and rapid approach for generating large panels of distinct monoclonal antibodies that, unlike existing antibody discovery platforms, does not rely on screening for antigen binding; and further describe application of this approach by developing panels of diagnostic antibodies for Ebola virus strains. We found that in multiple animals immunized with EBOV or RESTV VLPs, the repertoire of CD138+ plasmablasts in the draining PLN was dominated by highly expanded, antigen-specific, antibody sequences. We find that these highly expanded sequences include antibodies that display high affinity to GP, bind to live, wild-type virus and, somewhat surprisingly, display diverse specificities to VLPs from different Ebolaviruses. Among the antibodies isolated, six recognized live Bundibugyo or both Bundibugyo and Sudan viruses in addition to EBOV. These results demonstrate the power of mining the antibody repertoire of highly expanded B cells after immunization for discovery of antibodies with interesting properties.
The ability to isolate 7 (and possibly more) distinct antibodies with very different CDR3 sequences per animal as well as a much larger number of somatic variants whose sequences are also available in the VH:VL sequence database provides a rich source of antibodies for practical purposes. Undoubtedly, many PLN B cells encoding antigen-specific antibodies are likely to have been subject to more limited expansion and thus are present at a lower abundance within the repertoire. However, such medium or low abundance antibody sequences within the repertoire are present at comparable levels to those elicited by environmental stimuli and thus recognizing unrelated antigens. Therefore the low abundance antigen-specific antibody sequences in the repertoire cannot be identified directly without significant additional effort.
The method we have pioneered should prove particularly useful for the facile development of diagnostic (and possibly therapeutic) antibodies. Starting from antibody-secreting B cells, it took us only 3 weeks to produce and characterize a diverse set of antigen-specific antibodies with a variety of useful specificities. This method should be particularly valuable for assessing and combating fast spreading pandemics.
In case of rapidly spreading emerging diseases such as Ebola, rapid and robust diagnostics are critical for treatment and disease control. For Ebola in particular, validated PCR based assays suitable for field work in third world countries are not available. Highly sensitive antibody based immunodiagnostics are extremely important and easy to implement42. However, a dearth of monoclonal antibodies for emergent Ebolaviruses limits the ability to use antigen-capture technologies for viral identification of early-stage infections. The panel of antibodies identified and characterized here should be useful for diagnostic applications including discriminating SUDV, BDBV, and EBOV. By using multiplex immunoassay platforms such as the Luminex MagPlex® technology, it should be possible to multiplex up to 50 different antibodies with varying specificities for a broad range of epitopes, thus ensuring wide coverage of Ebolaviruses variants. Studies to incorporate the antibodies we have described here onto the Luminex MagPlex® diagnostic platform for field applications are on-going.
Methods
VLP Production and Characterization
VLPs were produced by co-transfection of HEK293FT (Invitrogen) cells with plasmids encoding the three major virus structural proteins, NP, VP40 and GP. All open reading frames for virus structural proteins were obtained from NCBI and were codon optimized for mammalian cell expression using Gene Designer (DNA 2.0), synthesized (Epoch Life Science) and inserted into either pcDNA3 (for Ebola GP) or pCAGGS (all other genes) mammalian expression plasmids. Sequences of the structural protein ORFs were verified and are available upon request to R. A. D. Cells were transfected with each of 5 μg NP, 5 μg VP40 and 1 μg GP encoding plasmids by the calcium chloride/BES transfection method. After 24 h cells were washed with DMEM, which was replaced with DMEM containing 10% (v/v) FBS. After an additional 24 h, the culture supernatant was collected and clarified by centrifugation at 3750 rpm for 30 min at 4 oC to remove cell debris. The supernatants were then overlaid onto a 5 mL 20% (w/v) sucrose cushion in 20 mM NaCl, 20 mM HEPES, pH 7.4 in an SW28 ultracentrifuge tube (Beckman). The VLPs were then pelleted by centrifugation at 28,000 rpm for 2 h at 4 oC in an SW28 rotor. To further purify VLPs, the pellet was resuspended in PBS and overlaid onto a 20 to 60% sucrose step gradient (5% increments) in 20 mM NaCl, 20 mM HEPES, pH 7.4 in a SW55 rotor tube (Beckman). The gradient was centrifuged at 38,000 rpm for 2 h at 4 oC after which an opaque band corresponding to VLPs was visible. Fractions were collected corresponding to the band as well as directly above and below it and analyzed by SDS-PAGE, staining for total protein with Krypton stain (Thermo Scientific) as well as immunoblotting. The middle fraction containing the peak of the VLPs was stored at −80°C until required. For immunoblotting, proteins were transferred to nitrocellulose membranes and stained using broadly reactive polyclonal antibodies against GP (gift from Dr. Andrew Hayhurst, Texas Biomedical Research Inst.), VP40 (gift from Dr. Ricardo Carrion, Texas Biomedical Research Inst.) or NP (IBT Bioservices). A specific monoclonal antibody against Zaire Ebolavirus GP was also used (4F3, IBT Bioservices). Appropriate secondary antibodies were purchased from LiCor Biosciences. Blots were imaged using a LiCor Odyssey SA imager. Electron microscopy of VLPs was performed at the University of Texas Health Sciences Center, San Antionio, Department of Pathology electron microscopy facility. VLPs were adhered to copper grids. The samples were fixed in glutaraldehyde and osmium tetroxide was used as contrast agent. Images were captured on a Philips 208S digital imaging electron transmission microscope.
Immunizations and Serum Titer Determination
The study was approved by the University of Texas Institutional Animal Care and Use Committee (AUP-2013–00015). All animal experiments were carried out in accordance with the approved protocol. VLPs in PBS pH 7.4 were emulsified in a 1:1 ratio with TiterMax Gold adjuvant (Sigma). For footpad immunizations, 20 μL (containing a total of 5 μg VLPs) antigen/adjuvant mixture was injected into the subcutaneous space of three BALB/c mice; for lateral hock injections, up to 50 μL was injected into the subcutaneous space just proximal to the lateral aspect of the ankle. Mice were immunized at footpad on day 0 for primary immunizations, and in the lateral hock on days 21, 35, and 77 for secondary immunizations. At days 10, 28, and 42 mice were bled for titration of the antigen-specific response. In order to determine serum antibody titer, mice were restrained in a tube restrainer, the tail wiped with isopropyl alcohol, and small incisions made with a fresh scalpel blade to nick the tail vein. 20–50 μL blood was obtained and allowed to coagulate at room temperature (RT) for 30 min, followed by centrifugation at 13,000 g for 15 min to pellet the clot. The serum was then used for ELISA assays. High binding ELISA plates (Corning) were coated overnight (O/N) at 4°C with 50 μL of 4 μg/mL Zaire Ebolavirus VLPs in PBS pH 7.4. Antigen solution was decanted and plates were then blocked at RT for 2 h in 2% milk (w/v) in PBS. Blocking solution was then decanted and plates were then incubated with 50 μL of serum diluted three-fold from 1:100 to 1:218,700 in 2% milk (w/v) in PBS for 1 h. Plates were then aspirated and washed 3 times with PBS containing 0.05% tween-20 (PBST), then incubated with 50 μL of 1:5000 diluted goat anti-mouse HRP secondary antibody (Jackson ImmunoResearch) for 1 h. Plates were washed 3 times with PBST, and incubated with 50 μL TMB-Ultra (Thermo Scientific) for 15 min. The reaction was quenched with 50 μL 2 M H2SO4 and absorbance was read at 450 nm on a Tecan M200 plate reader.
Tissue Collection, Cell Isolation, and Subtype Purification
After determination of significant titer for Ebolavirus VLPs (signal evident above background at dilution >1:10,000), mice were administered a final boost at day 77, and lymph nodes were collected 6 days later. For lymph nodes collection, mice were injected with 5–10 μL of 2% Evans Blue (Sigma) in PBS into the footpad. 30 min post-injection, mice were sacrificed by carbon dioxide asphyxiation followed by cervical dislocation. The skin and fur around the leg was removed to reveal the blue-stained popliteal lymph node (Fig. 1a), located just behind the knee. The lymph node was isolated and stored in PBS pH 7.4 supplemented with 0.1% (w/v) BSA, 2 mM EDTA in a 6-well plate (Corning). Lymph node was homogenized by mechanical disruption using two 18G needles. The cells were then passed through a 70 μm cell strainer (Corning), with additional disruption using the plunger from a 3 mL syringe to aid passage of single cells. Cells were then spun down at 500 g for 10 min in a swinging bucket rotor. The cell pellets were then resuspended in 2 mL red blood cell lysis buffer (155 mM NH4Cl, 12 mM NaHCO3, 0.1 mM EDTA) and incubated at room temperature for 3.5 min. The lysis reaction was quenched by adding 20 mL PBS buffer followed by centrifugation at 500 g for 10 min at RT. Cells were washed again with 5 mL PBS buffer and resuspended in a final volume of 1 mL buffer. Plasma cells were then isolated using the Miltenyi Plasma Cell Isolation kit (Miltenyi Biotec). Briefly, non-plasma cells were depleted by magnetic labeling of CD49b and CD45R followed by enrichment of magnetically-labeled CD138+ cells. CD45R is a pan-B cell marker expressed on naïve and activated B lymphocytes, but not on antibody-secreting cells. Conversely, CD138 is expressed on pre-B and immature B-lymphocytes in the bone marrow, lost upon emigration into secondary lymphoid tissues, and re-expressed upon differentiation into plasma cells.
Single Cell VH:VL Sequencing
Sorted cells were analyzed by single B cell VH:VL sequencing as previously described33. Briefly, single cells were isolated into 125 pL wells printed in PDMS along with poly(dT) conjugated magnetic beads. Cell lysis and capture of mRNA was performed in situ, and beads were collected and emulsified to serve as template for emulsion overlap extension RT-PCR. A follow-up nested PCR resulted in 850 bp amplicons containing linked genetic information for VH and VL genes. 850 bp amplicons were analyzed using the Illumina MiSeq 2 × 250 platform. VH and VL genes were amplified separately for full-length VH and VL analysis using the Illumina MiSeq platform as previously described33.
Sequence Analysis
Raw MiSeq data was analyzed as previously described33. Briefly, raw data were filtered for a minimum Phred quality score of 20 over 50% of nucleotides to ensure high read quality in the CDR3 regions of heavy and light genes. Sequence data were submitted to the IMGT information system for V-D-J germline gene mapping. Sequences were filtered for in-frame V-D-J junctions and VH:VL pairs were compiled by exact CDRH3:CDRL3 nucleotide match. CDRH3 junction nucleotide sequences were clustered to 96% identity and resulting clusters with > = 2 VH:VL reads were ranked by MiSeq read counts33. Due to read length limitations of current next-generation sequencing technology, the complete VH and VL genes were also sequenced and analyzed separately. Full-length VH and VL genes were filtered for a minimum Phred quality score of 20 over 50% of nucleotides and were compiled by CDRH3 and CDRL3 exact nucleotide match. Consensus sequences of VH and VL genes (i.e. from all reads passing quality filters and that contained exact matches to the CDRH3:CDRL3 pair of interest) were used for antibody gene synthesis, expression, and in vitro analysis33.
IgG Synthesis, Expression, and Purification
Consensus VH and VL genes were designed and purchased as gBlocks (Integrated DNA Technologies) and cloned into the pcDNA3.4 vector (Invitrogen) containing Oryctolagus cuniculus IgG leader peptide as fusions to human IgG1 and kappa constant regions, respectively. Sequences of both the heavy and the light chain for each antibody variant were confirmed by Sanger sequencing. Plasmids for each antibody variant were transfected into Expi293 cells (Invitrogen) at a 1:3 heavy:light ratio. After incubating at 37 °C with 8% CO2 at 125 rpm for 6 days, the supernatant containing secreted antibodies was collected by centrifugation at 500 g for 15 min at 25°C. Supernatant was passed over a column of 0.5 mL Protein A agarose resin (Thermo Scientific) three times to ensure efficient binding. After washing with 20 column volumes of PBS, antibodies were eluted with 3 mL 100 mM citric acid pH 3.0 and immediately neutralized with 500 μL 1 M Tris pH 8.0. Antibodies were buffer exchanged into PBS, pH 7.4 utilizing Amicon Ultra-30 centrifugal spin columns (Millipore) for storage and subsequent use.
ELISA
Costar 96-well ELISA plates (Corning) were coated with 50 μL of 4 μg/mL recombinant Ebola Glycoprotein (a gift from Dr. Erica O. Saphire, The Scripps Research Institute) or Ebola virus VLPs. The coated plates were incubated at 4 °C O/N, after which they were decanted and blocked with 2% milk in PBS for 2 h at RT. After blocking, 1:5 serially diluted antibodies were applied to the plates for 1 h, after which 1:5000 diluted donkey anti-human IgG HRP-conjuated secondary antibodies were applied (Jackson ImmunoResearch) for 1 h. For detection, 50 μL TMB-Ultra substrate was applied for 10 min before quenching with 50 μL 2 M H2SO4. Absorbance was measured at 450 nm using a Tecan M200 plate reader. Data were analyzed and fitted for EC50 using a 4-parameter logistic nonlinear regression model in the Prism software.
Surface Plasmon Resonance
Antibody affinity to recombinant Ebola GP protein was measured by surface plasmon resonance using a BIAcore 3000 biosenor (Biacore). In order to fit the responses to 1:1 Langmuir binding model for more accurate affinity determination, antibodies were immobilized on the CM5 sensor chip (GE Healthcare) using the amine coupling chemistry. All binding experiments were done in HBS-EP buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3.4 mM EDTA, and 0.005% P20 surfactant) (GE Healthcare). GP was injected in triplicates at concentrations 80, 100, 200, 300, 400, 500, and 600 nM with a flow rate of 60 μL/min for 2 min and a dissociation time of 10 min. Regeneration of the antibody was performed by a single injection of 100 mM citric acid, pH 3.0. The response generated by flowing GP over a bovine serum albumin (BSA) coupled surface was used as control and was consequently subtracted. All kinetic parameters were determined in BIAevaluation 3.0 software and were reported as the average of three technical replicates.
Wild type virus ELISA assays
All work with wild type virus was performed at BSL4 at Texas Biomedical Research Institute. All virus stocks were cultivated on Vero-E6 cells in DMEM with 2% FBS and antibiotics. When 80% of cells began showing a cytopathic effect, the culture supernatant containing virus was collected. Virus was purified as for VLPs by pelleting cell debris and then pelleting virus from the culture supernatants through 20% sucrose in 20 mM NaCl and 20 mM HEPES, pH 7.4. The virus pellets were resuspended in PBS and stored in aliquots at −80 °C until needed. Virus titers were determined by conventional plaque assay using Vero-E6 cells. For ELISA assays, an aliquot of virus was thawed and the equivalent of 106 PFU of virus was diluted 1:3 into RIPA buffer. This was then diluted 1:100 into 10 mM sodium phosphate buffer, pH 7.4. After coating O/N, plates were washed with PBS containing 0.1% Tween-20 and incubated with each antibody starting at 1:100 of a 1 mg/mL stock and then over serial 4-fold dilutions on the plates. The secondary antibody was anti-human IgG HRP conjugate from Pierce. TMB Ultra substrate (Life Technologies) was used to detect antibody binding on plates. All assays were performed at least in duplicate and repeated 3 times. ELISAs were analyzed and fitted for EC50 using a 4-parameter logistic nonlinear regression model in the Prism software.
Additional Information
How to cite this article: Wang, B. et al. Facile Discovery of a Diverse Panel of Anti-Ebola Virus Antibodies by Immune Repertoire Mining. Sci. Rep. 5, 13926; doi: 10.1038/srep13926 (2015).
Supplementary Material
Acknowledgments
We would like to thank C. Das for aid in antibody expression, Dr. G. Ippolito for insightful advice and critical reading of the manuscript, and Dr. C. Lee for help with SPR experiments. We also thank Dr. E. O. Saphire for providing recombinant Ebola virus GP. This study was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI096228 (A.L.A.) and by the Defense Threat Reduction Agency grant HDTRA1-12-C-0105 and HDTRA1-12-C-007. O.I.L. was funded by NIH CORE funding NIH 1K12GM102745.
Footnotes
Author Contributions B.W., C.A.K., O.I.L., S.M.C., I.V., A.L.A., R.A.D. and G.G. designed the experiments. B.W., C.A.K., O.I.L., B.J.D., S.A.K., E.L.J., J.J., A.B.R., A.N.R., J.R.B. and R.A.D. performed the experiments, B.W., C.A.K., O.I.L., B.J.D., R.A.D., A.D.E. and G.G. analyzed the data. B.W., C.A.K., O.I.L., I.V., R.A.D., A.D.E. and G.G. wrote the manuscript.
References
- Bowen E. T. W. et al. Viral Haemorrhagic Fever in Southern Sudan and Northern Zaire Preliminary Studies on the Aetiological Agent. Lancet 309, 571–573 (1977). [DOI] [PubMed] [Google Scholar]
- Kondratowicz A. S. et al. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. USA. 108, 8426–8431 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J. E. et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner J. S. et al. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 4, e1000212 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M. E. G. & Miranda N. L. J. Reston ebolavirus in humans and animals in the Philippines: a review. J. Infect. Dis. 204 Suppl, S757–S760 (2011). [DOI] [PubMed] [Google Scholar]
- Le Guenno B. et al. Isolation and partial characterisation of a new strain of Ebola virus. Lancet 345, 1271–1274 (1995). [DOI] [PubMed] [Google Scholar]
- Team W. E. R. Ebola Virus Disease in West Africa - The First 9 Months of the Epidemic and Forward Projections. N. Engl. J. Med. 371, 1481–1495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baize S. et al. Emergence of Zaire Ebola Virus Disease in Guinea. N. Engl. J. Med. 371, 1418–1425 (2014). [DOI] [PubMed] [Google Scholar]
- Gire S. K. et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345, 1369–1372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. A. et al. Molecular evolution of viruses of the family Filoviridae based on 97 whole-genome sequences. J. Virol. 87, 2608–2616 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias J. M. et al. A shared structural solution for neutralizing ebolaviruses. Nat. Struct. Mol. Biol. 18, 1424–1427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X. et al. Characterization of Zaire ebolavirus glycoprotein-specific monoclonal antibodies. Clin. Immunol. 141, 218–227 (2011). [DOI] [PubMed] [Google Scholar]
- Koellhoffer J. F. et al. Two synthetic antibodies that recognize and neutralize distinct proteolytic forms of the ebola virus envelope glycoprotein. Chembiochem 13, 2549–2457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. et al. Synthetic Antibodies with a Human Framework That Protect Mice from Lethal Sudan Ebolavirus Challenge. ACS Chem. Biol. 9, 2263–2273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T. et al. Ebola Virus Can Be Effectively Neutralized by Antibody Produced in Natural Human Infection. J. Virol. 73, 6024–6030 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren P. W. H. I., Geisbert T. W., Maruyama T., Jahrling P. B. & Burton D. R. Pre- and Postexposure Prophylaxis of Ebola Virus Infection in an Animal Model by Passive Transfer of a Neutralizing Human Antibody. J. Virol. 76, 6408–6412 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X. et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514, 47–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H., Nichol S. T., Klenk H-D., Peters C. J. & Sanchez A. Characterization of filoviruses based on differences in structure and antigenicity of the virion glycoprotein. Virology 199, 469–473 (1994). [DOI] [PubMed] [Google Scholar]
- Lin G. et al. Differential N-Linked Glycosylation of Human Immunodeficiency Virus and Ebola Virus Envelope Glycoproteins Modulates Interactions with DC-SIGN and DC-SIGNR. J. Virol. 77, 1337–1346 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood C. L. et al. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J. Virol. 84, 2972–2982 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler G. & Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497 (1975). [DOI] [PubMed] [Google Scholar]
- Reddy S. T. et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat. Biotechnol. 28, 965–969 (2010). [DOI] [PubMed] [Google Scholar]
- Saggy I. et al. Antibody isolation from immunized animals: comparison of phage display and antibody discovery via V gene repertoire mining. Protein Eng. Des. Sel. 25, 539–549 (2012). [DOI] [PubMed] [Google Scholar]
- Lane D. & Koprowski H. Molecular recognition and the future of monoclonal antibodies. Nature 296, 200–202 (1982). [DOI] [PubMed] [Google Scholar]
- Ghosh S. & Campbell A. M. Multispecific monoclonal antibodies. Immunol. Today 7, 217–222 (1986). [DOI] [PubMed] [Google Scholar]
- Wu X. et al. Rational Design of Envelope Identifies Broadly Neutralizing Human Monoclonal Antibodies to HIV-1. Science 329, 856–861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D. et al. A Neutralizing Antibody Selected from Plasma Cells That Binds to Group 1 and Group 2 Influenza A Hemagglutinins. Science 333, 850–856 (2011). [DOI] [PubMed] [Google Scholar]
- Bradbury A. R. M., Sidhu S., Dübel S. & McCafferty J. Beyond natural antibodies: the power of in vitro display technologies. Nat. Biotechnol. 29, 245–254 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Regenmortel M. H. V. Specificity, polyspecificity, and heterospecificity of antibody-antigen recognition. J. Mol. Recognit. 27, 627–39 (2014). [DOI] [PubMed] [Google Scholar]
- Walker L. M. et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L. M. et al. Broad and Potent Neutralizing Antibodies from an African Donor Reveal a New HIV-1 Vaccine Target. Science 326, 285–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S. T., Swartz M. A. & Hubbell J. A. Targeting dendritic cells with biomaterials: developing the next generation of vaccines. Trends Immunol. 27, 573–579 (2006). [DOI] [PubMed] [Google Scholar]
- DeKosky B. J. et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat. Biotechnol. 31, 166–169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamala T. Hock immunization: a humane alternative to mouse footpad injections. J. Immunol. Methods 328, 204–214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleichmann H. Studies on the Mechanism of Drug Sensitization: T-cell-Dependent Popliteal Lymph Node Reaction to Diphenylhydantoin. Clin. Immunol. Immunopathol. 18, 203–211 (1981). [DOI] [PubMed] [Google Scholar]
- Ravel G. & Descotes J. Popliteal lymph node assay: facts and perspectives. J. Appl. Toxicol. 25, 451–458 (2005). [DOI] [PubMed] [Google Scholar]
- Kaushik A. & Lim W. The primary antibody repertoire of normal, immunodeficient and autoimmune mice is characterized by differences in V gene expression. Res. Immunol. 147, 9–26 (1996). [DOI] [PubMed] [Google Scholar]
- Lu J. et al. IgG variable region and VH CDR3 diversity in unimmunized mice analyzed by massively parallel sequencing. Mol. Immunol. 57, 274–283 (2014). [DOI] [PubMed] [Google Scholar]
- Schoonbroodt S. et al. Engineering Antibody Heavy Chain CDR3 to Create a Phage Display Fab Library Rich in Antibodies That Bind Charged Carbohydrates. J. Immunol. 181, 6213–6221 (2008). [DOI] [PubMed] [Google Scholar]
- Lee J. E. et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454, 177–182 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy B. et al. Characterization of Marburg virus glycoprotein in viral entry. Virology 358, 79–88 (2007). [DOI] [PubMed] [Google Scholar]
- Towner J. S. et al. Rapid Diagnosis of Ebola Hemorrhagic Fever by Reverse Transcription-PCR in an Outbreak Setting and Assessment of Patient Viral Load as a Predictor of Outcome. J. Virol. 78, 4330–4341 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.