Abstract
Recent advances in the study of the CRISPR/Cas9 system have provided a precise and versatile approach for genome editing in various species. However, the applicability and efficiency of this method in large animal models, such as the goat, have not been extensively studied. Here, by co-injection of one-cell stage embryos with Cas9 mRNA and sgRNAs targeting two functional genes (MSTN and FGF5), we successfully produced gene-modified goats with either one or both genes disrupted. The targeting efficiency of MSTN and FGF5 in cultured primary fibroblasts was as high as 60%, while the efficiency of disrupting MSTN and FGF5 in 98 tested animals was 15% and 21% respectively, and 10% for double gene modifications. The on- and off-target mutations of the target genes in fibroblasts, as well as in somatic tissues and testis of founder and dead animals, were carefully analyzed. The results showed that simultaneous editing of several sites was achieved in large animals, demonstrating that the CRISPR/Cas9 system has the potential to become a robust and efficient gene engineering tool in farm animals, and therefore will be critically important and applicable for breeding.
Genome-editing technologies rely on the use of engineered nucleases to induce cellular DNA repair mechanisms and introduce programmable, site-specific genetic modifications in diverse systems1. These programmable endonucleases include zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and, most recently developed, the clustered regularly interspaced short palindromic repeats CRISPR-associated 9 (Cas9) system. The CRISPR/Cas9 system uses short, single-guide RNAs (sgRNA) that recognize the target DNA, then programmed the Cas9 towards targets that are complementary to the first 20 nucleotides of the sgRNA2. Compared with ZFNs and TALENs, the RNA-guided CRISPR/Cas9 system demonstrates its precious, versatile and robust merits for targeted genome editing in a variety of species, including model organisms, as well as crops and animals that are crucial to agriculture.
As one of the first domestic farm animals, the goat is one of the most important livestock species and provides a variety of products, including fiber, milk, meat, and hides. Furthermore, goats have also been used as a model in biomedical studies3,4,5. Although specific gene knockout strategies based on homologous recombination (HR) and somatic cell nuclear transfer (SCNT) have been established in goats6, precise gene modification of the goat genome is still challenging. In a previous study on disruption of four genes simultaneously in goat primary fibroblasts by the CRISPR/Cas9-mediated approach, only the myostatin (MSTN) knockout fibroblasts were achieved and resulted in live-born goats by SCNT7. However, an anti-biotic selection cassette is normally essential for isolating single-cell colonies from seeded donor cells, and reconstructed embryos have a low developmental potential, leading to a relatively low SCNT targeting efficiency (for example, 1–5% in pigs and >10% in cattle)8.
Co-injection of Cas9 mRNA and sgRNA into one-cell stage embryos has been demonstrated to be an efficient approach for the generation of genetically modified mice, rats, monkeys and pigs9,10,11,12, which encourages us to extend the application of this strategy to gene targeting in goats. In the present study, through co-injection of one-cell stage embryos of cashmere goats with sgRNAs of two functional genes (MSTN and FGF5) and Cas9 mRNA, targeted modifications of one or two genes were achieved at an efficiency around 26.5%. We also carefully analyzed the on- and off-target mutations of the targeted genes in the somatic tissues and gonads, providing comprehensive evidence for the efficiency and reliability of injection of zygotes with Cas9 mRNA and sgRNAs for the generation of gene-modified farm animals.
Results and Discussion
Design of sgRNAs
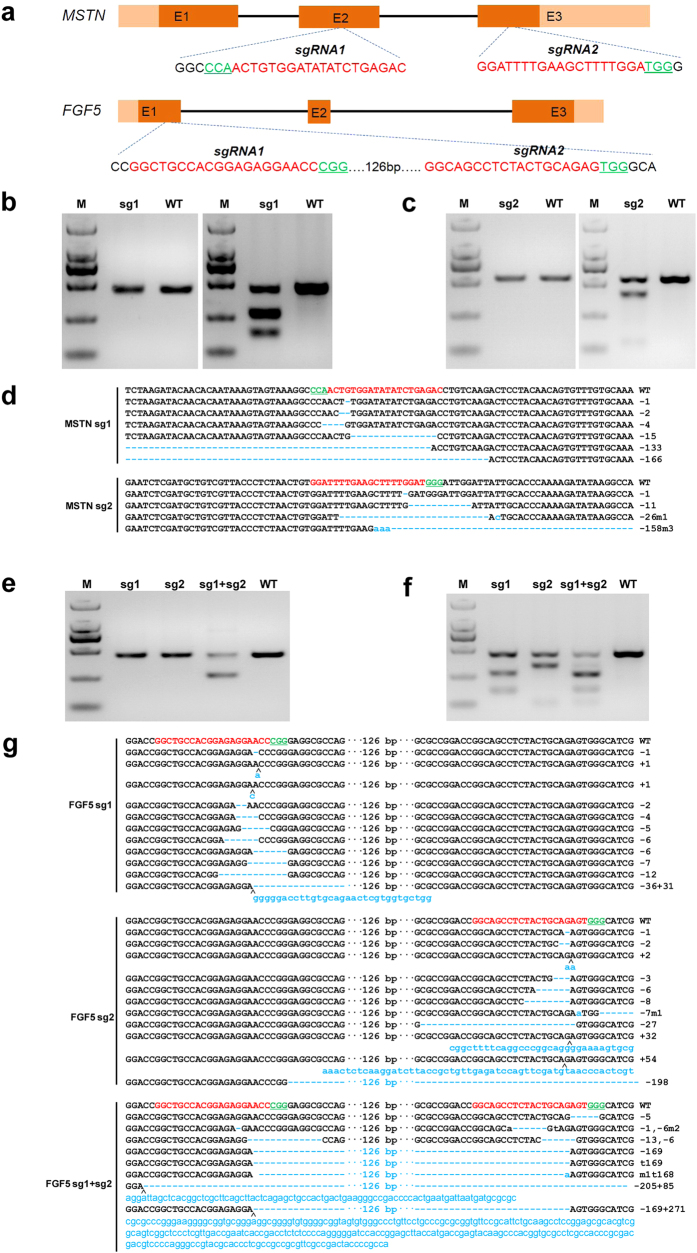
The Shannbei cashmere goat is a cultivated dual-purpose breed that provides both meat and fibers (fine cashmere). In an attempt to improve the performance of cashmere goats considering both meat and cashmere production purposes, two genes that are associated with muscle development (MSTN) and hair length (FGF5), were selected as target genes. MSTN is a secreted growth differentiation factor that inhibits muscle differentiation and growth. Function loss of MSTN is known to cause an increased muscle mass phenotype in several mammals, including mice, dogs, cattle and humans13,14,15,16. Fibroblast growth factor 5 (FGF5), a secreted signaling protein during the hair growth cycle, inhibits hair growth by blocking dermal papilla cell activation17, and is regarded as the causative gene underlying the angora phenotype (long hair coat) in mice18. Mutations in FGF5 underlie trichomegaly (excessively long eyelashes) in humans19, as well as being associated with hair length in other mammal species such as cats20, dogs21 and donkeys22. MSTN and FGF5 were therefore selected to generate gene-modified cashmere goats with both genes disrupted by the CRISPR/Cas9 system. To ensure the successful targeting, two sgRNAs independently targeting exons 2 and 3 of MSTN, and two sgRNAs targeting the first exon of FGF5, were selected as described23 (Fig. 1a).
Figure 1. Evaluation of sgRNA:Cas9-mediated modifications of MSTN and FGF5 in goat fibroblasts.
(a) Schematic diagram of MSTN and FGF5 partial protein coding region and the targeting loci of sgRNA:Cas9. sgRNAs targeting sites are presented in red. PAM sequences are highlighted in green and underlined. (b) Left panel, PCR products of the targeted exon 2 of MSTN from goat fibroblasts transfected with Cas9 and MSTN sgRNA1. Right panel, detection of sgRNA:Cas9-mediated on-target cleavage of MSTN by T7E1 cleavage assay. PCR products from left panel were subjected to T7E1 cleavage assay. M, marker; WT, wild type PCR product from fibroblasts that have not treated with CRISPR/Cas9. (c) Left panel, PCR products of the targeted exon 3 of MSTN from goat fibroblasts transfected with Cas9 and MSTN sgRNA2. Right panel, detection of sgRNA:Cas9-mediated on-target cleavage of MSTN by T7E1 cleavage assay. PCR products from left panel were subjected to T7E1 cleavage assay. (d) Sequences of modified MSTN alleles. Target sequences complementary to MSTN sgRNAs are in red text; the mutations are blue, lower case; insertions (+), deletions (−) or mutation (m) are shown to the right of each allele. (e) PCR products of the targeted exon 1 of FGF5 from goat fibroblasts transfected with Cas9 and FGF5 sgRNA1, FGF5 sgRNA2 and FGF5 sgRNA1 & 2, respectively. (f) Detection of sgRNA:Cas9-mediated on-target cleavage of FGF5 by T7E1 cleavage assay. PCR products from (e) were subjected to T7E1 cleavage assay. (g)Sequences of modified FGF5 alleles. Target sequences complementary to FGF5 sgRNAs are in red text; the mutations are blue, lower case; insertions (+), deletions (−), mutation (m) or turnover (t) i shown to the right of each allele.
Efficiency of the CRISPR/Cas9 System in Fibroblasts
Although microinjection of zygotes with Cas9 mRNA and sgRNA had not previously been tested in goats, the successful application of this system in embryos of other species12,24, suggested that this system could work in goat embryos. To verify this possibility, we overexpressed the Cas9 nuclease and sgRNAs targeting MSTN and FGF5 in goat fibroblasts isolated from goat foetus. Subsequently, genomic DNA was extracted from fibroblasts 72 h after transfection, and screened for the presence of site-specific gene modification by PCR amplification of the region around the targeted site (Fig. 1b, c and e) using a T7 endonuclease I (T7E1) cleavage assay. The T7E1 cleavage bands were visible in the targeted genes (Fig. 1b, c and f). The cleavage was characterized further by Sanger sequencing, which displayed overlapped peaks in the sequencing chromatographs (Fig. 1d,g), suggesting different genotypes from target modifications. Distinguishable indels were observed at the target sites of MSTN and FGF5 with a range of mutation sizes (Fig. 1d,g, Supplementary Table S1). These data demonstrated that the designed sgRNAs work efficiently with Cas9 on targeted genes in the cultured goat fibroblasts.
Generation of Gene-Modified Goats
To establish target modification in goats, a total of 926 early zygotes (one-cell stage) were surgically collected from 79 naturally mated ewes; ~11.72 zygotes were obtained from each donor. The Cas9 mRNA and sgRNA mixture targeting MSTN and FGF5 was injected into the cytoplasm of the embryos at the one-cell stage. After the injected embryos developed to the two-cell stage (~24 h, 37 °C), only 416 (48.25%) of the 862 injected embryos were transferred into 137 pseudopregnant mothers. On average, 3.04 injected embryos were transferred to each recipient (Table 1). Of the 137 recipient ewes, 64 pregnancies (46.7%) were established according to observation of the estrus cycles. After full-term gestation, which lasted around 150 days, 93 lambs from surrogates were successfully delivered, 14 of which died immediately after birth (Fig. 2a and Table 1).
Table 1. Summary of production of gene-modified goats via CRISPR-Cas9.
| Goats forzygotecollection | Collectedembryos | Cas9-sgRNA injectedembryos (one-cell stage) |
Recipientgoatsa | Newborns | Alive goats | Gene-modified goats | |
|---|---|---|---|---|---|---|---|
| Injectedembryos | Transferredembryos | ||||||
| 79 | 926 | 862 | 416 | 137 | 93 | 79 | 26 |
aFive surrogate ewes had aborted.
Figure 2. Detection of sgRNA:Cas9-mediated modifications of MSTN and FGF5 in lambs and tissues.
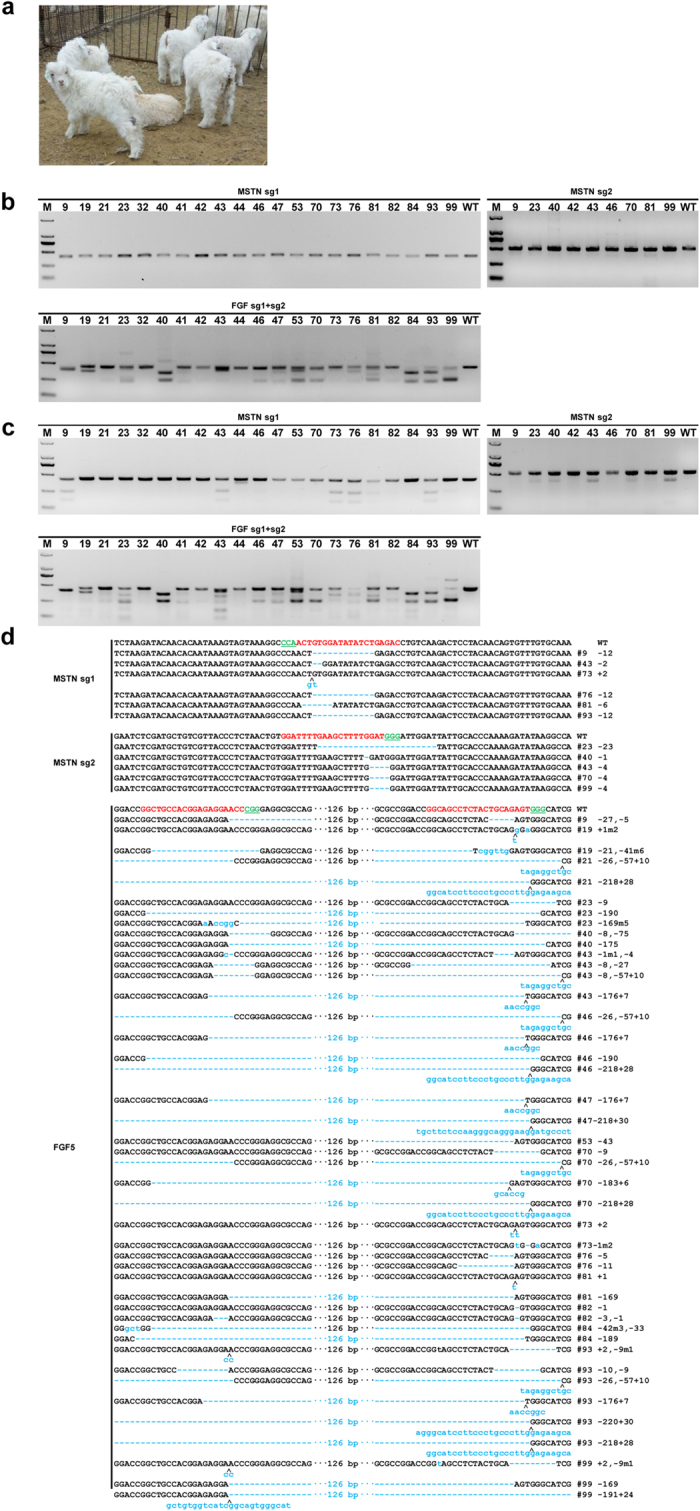
(a) Photographs of 30-day-old gene-modified lambs (Photo taken by X.W.). (b) PCR products of the targeted region of MSTN and FGF5 from founder goats co-microinjected with a mixture of Cas9 mRNA and sgRNAs. (c) Detection of sgRNA:Cas9-mediated on-target cleavage of MSTN and FGF5 by T7E1 cleavage assay. All PCR products from (b) were subjected to T7E1 cleavage assay. All the samples were digested by T7E1, suggesting that all founders carry MSTN and FGF5 mutations. (d) Sequencing results of modified MSTN and FGF5 loci detected in lambs.
Genomic DNA was isolated from aborted, dead and live lambs. In total, 98 individuals (including 93 delivered and 5 aborted) were used for genotyping. The sgRNA:Cas9-mediated genome modifications were first screened using the genomic DNA as described above. Genotyping was performed by PCR amplification, T7E1 assay, and TA-cloning sequencing. The additional bands were observed by PCR amplification of the target region in some of the infant goats (Fig. 2b), indicating that genomic modification occurred in these goats. Then, the PCR products of all the animals were subjected to the T7E1 cleavage assay (Fig. 2c). Impressively, the cleavage products for MSTN were observed in 15 infants (15.3%) and FGF5 in 21 infants (21.4%), with 10 lambs showing disruption of two genes simultaneously (Table 2). In total, 26 lambs (26/98, 26.5%) demonstrated the disruption of one or two genes, indicating efficient genomic modifications in the infant goats. As expected, different types of indels were observed, and further confirmed by Sanger sequencing (Fig. 2d).
Table 2. The efficacy of Cas9-mediated modifications in goat fibroblasts and tested individuals.
| Target gene | Fibroblasts | Tested individuals |
|---|---|---|
| MSTN | 15/26 (57.7%) | 15/98 (15.3%) |
| FGF5 | 11/18 (61.1%) | 21/98 (21.4%) |
| FGF5&MSTN | − | 10/98 (10.2%) |
Currently, there are two descriptions about generation of genetically modified goat and sheep via the CRISPR/Cas9 system. Ni et al. reported the application of the CRISPR/Cas9 system to disrupt four genes (MSTN, NUP, PrP, and BLG) in goat primary fibroblasts, and generated live-born goats via SCNT approach for the single MSTN gene. Only the MSTN biallelic mutations were used for SCNT; seven pregnancies were yielded from 21 transfers, and two live-born goats were eventually obtained7. Through co-injection of Cas9 mRNA and sgRNA targeting a single gene-MSTN, Han et al. described the successful generation of MSTN knockout sheep at a targeting efficiency of 5.7% (2/35)25. Taken together, our results support that direct microinjection of Cas9 mRNA and sgRNAs into one-cell stage embryos is efficient in goats, and could be applied to produce gene-modified animals.
Cas9-Mediated Genome Targeting Extensively Integrates into Different Tissues of Goats
To further evaluate the integration of the Cas9-mediated modifications into the derivatives of three germ layers, we performed extensive analysis of target mutagenesis in seven different somatic tissues (heart, liver, spleen, lung, kidney, skin, and muscle) and testis from the aborted and dead lambs. PCR amplification and T7E1 cleavage assay showed similar cleavage patterns among different tissue (Fig. 3a,b), suggesting that the Cas9-mediated mutations did occur in various goat tissues.
Figure 3. Detection of sgRNA:Cas9-mediated targeting in different tissues.
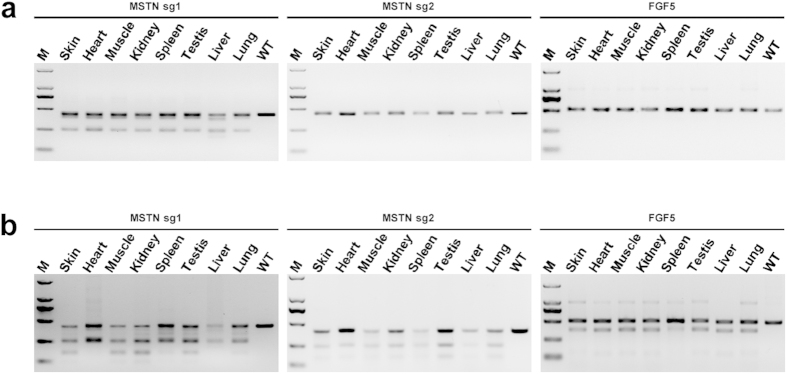
(a) Dead goat #D24 was chose for tissue distribution analysis of on –target mutations. PCR products of the targeted regions of MSTN and FGF5 from seven tissues. (b) Detection of sgRNA:Cas9-mediated on-target cleavage of MSTN and FGF5 by T7E1 cleavage assay. All PCR products from (a) were subjected to T7E1 cleavage assay. All the samples were digested by T7E1, suggesting that all the 7 tested tissues carry MSTN and FGF5 mutations.
Off-Target Detection
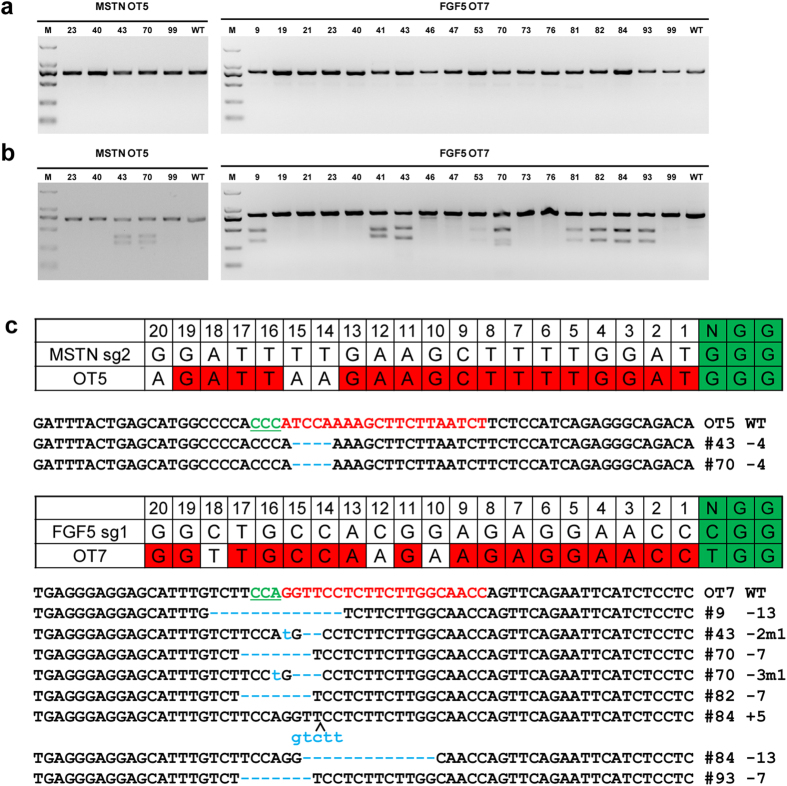
Previous studies have described that off-target mutagenesis occurred in Cas9 medicated human cell lines25, mice26 and zebrafish27, suggesting that mismatches between the sgRNA and target DNA exist due to Cas9-mediated DNA cleavage25. To verify whether off-target mutations occurred in these Cas9 mediated goats, we predicted putative off-target sites using SeqMap28 and assessed the off-target mutations in both fibroblasts and founder animals. A total of 13 (6 for MSTN and 7 for FGF5) most potential off-target sites were predicted across the goat genome (Supplementary Table S2). Out of the 13 off-target sites, two sites (OT5 and OT7) showed off-target modifications in goat fibroblast cells (Supplementary Table S3, Fig S1). The remaining 11 off-target sites were therefore excluded for further examination in the founder animals. Subsequently, the PCR products at OT5 and OT7 were subjected to the T7E1 cleavage assay using the DNA from founder animals, we found that mutations at these two sites indeed occurred in some of the founder animals (Fig. 4a,b). To further verify the off-target cleavage events, PCR products from MSTN-disrupted animals and FGF5-disrupted animals were subjected to Sanger sequencing, showing #43, #70 of MSTN mutated founders and #9, #43, #70, #82, #84, #93 of FGF5 mutated founders had off-target modification (Fig. 4c). Except #41 of FGF5, which had a TGG-------GGA/AGGAGGGGTGGGG SNP 113 bp before OT7, no mutations were found in #53 and #81 of FGF5 mutated founders, indicating the slight off-target efficiency. In addition, #46 and #47 of FGF5 showed false positive cut bands due to confirmed poly-T structure. Considering the off-target effects are site-dependent, we believe the off-target mutagenesis can be avoided by choosing a single or several sites.
Figure 4. Detection of the MSTN and FGF5 sgRNA:Cas9-mediated off-target cleavages in vivo.
(a) PCR products of the potential off-target sites of MSTN and FGF5 sgRNA:Cas9 from founder lambs. A total of 13 potential off-target sites most homologous to MSTN and FGF5 sgRNA were named OT1 to OT13. OT5 and OT7 were selected and PCR amplified from genomic DNA from founders. (b) Detection of sgRNA:Cas9-mediated off-target cleavage of MSTN and FGF5 by T7E1 cleavage assay. All PCR products from (a) were subjected to T7E1 cleavage assay. (c) Sequencing results of PCR products.
Taken together, we present here the successful generation of gene-modified goats targeting multiple genomic loci via the CRISPR/Cas9 approach. We further carefully analyzed the sgRNA:Cas9-mediated on- and off-target mutation in various somatic tissues, as well as testis, providing detailed information about generating gene-modified goats by injection of zygotes with Cas9 mRNA and sgRNAs.
Materials and Methods
Ethical statement
Eleven rams (2–3 years old, body weight: 30–50 kg) and 216 ewes (79 donors and 137 recipients, 2–3 years old, body weight: 24–40 kg) were used in this study. The animals were regularly maintained in the Shaanbei Cashmere Goat Farm of Yulin University. All the protocols involving the use of animals were in accordance with approved guidelines of the Animal Care and Use Committee of the Northwest A&F University (Approval ID: 2011ZX08008-002).
Design of Cas9/sgRNA
To construct the recombinant vector for the preparation of sgRNA by in vitro transcription, the two complementary DNA oligos shown in Supplementary Table S4 were annealed to be double-stranded, and then sub-cloned into the pUC57-T7-gRNA vector as described9. The constructed recombinant vector was completely linearized by DraI endonuclease and used as the template. sgRNAs were produced by in vitro transcription using the MEGAshortscript kit (Ambion) and purified using the MEGAClear kit (Ambion) according to the manufacturer’s instructions. Using the Cas9 mRNA in vitro transcription vector (Addgene No. 44758) as templates, Cas9 mRNA was produced and purified according to description9. The sequence of both the Cas9 and sgRNAs targeting MSTN and FGF5 are listed in Supplementary Table S5.
Cas9/sgRNA efficiency test in goat fetal fibroblasts
We established a fibroblast cell line from 40-day-old goat fetus trunk tissue. The fibroblasts went through five passages cultured in DMEM medium (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin streptomycin (Gibco) to achieve 80–90% confluency on the day of transfection. The transfection procedure was carried out using Lipofectamine 2000 Reagent (Invitrogen) according to the manufacturer’s instructions. Briefly, goat fibroblasts were transfected with MSTN sgRNA1 (0.8 ug), MSTN sgRNA2 (0.8 ug), FGF5 sgRNA1 (0.8 ug), FGF5 sgRNA2 (0.8 ug) and FGF5 sgRNA1 & 2 (0.4 ug/each), along with 0.8 μg of Cas9 plasmid by Lipofectamine 2000 in a 24-well culture plate. 24 h after transfection, 10 mg /mL of blasticidine S hydrochloride was added to the medium at a 1:1000 dilution for 24 h. Genomic DNA was extracted from fibroblasts 72 h after transfection using a saturated solution of phenol and chloroform, then the DNA was precipitated with alcohol and sodium acetate. Subsequently, a T7E1 cleavage assay was performed as described by Shen et al.9. Briefly, the targeted fragments were amplified by PrimerSTAR HS DNA polymerase (TaKaRa, DR010A) from the genomic DNA, then purified with a PCR cleanup kit (Axygen, AP-PCR-50). The primers for amplifying MSTN and FGF5 targeted fragments are listed in Supplementary Table S6. The purified PCR products were denatured and re-annealed in NEBuffer 2 (NEB) using a thermocycler (BioRad). The PCR products were digested with T7E1 (NEB, M0302L) for 30 min at 37 °C and then separated on a 2.5% agarose gel. The PCR products with mutations detected by the T7E1 cleavage assay were then sub-cloned into T vector (Takara). For each sample, the colonies were picked randomly and sequenced with the M13F (–47) primer: 5′-CGC CAG GGT TTT CCC AGT CAC GAC-3′).
Production of gene-modified goats via zygote injection with Cas9/sgRNA
Healthy ewes with regular estrus cycles were selected as donors for zygote collection. Zygotes were collected through surgical oviduct flushing from the donors by estrus synchronization and superovulation treatment as we previously described30,31. In brief, donors were treated with EAZI-BREED CIDR Sheep and Goat Device (CIDR, contain progesterone 300 mg) by inserting in vagina for 14 days, and the superovulation was performed 60 hours prior to CIDR Device removal. A total of 260 mg follicle stimulating hormone (FSH) (Folltropin®-V) was administered by intramuscular injection in 7 dosages, at 12 h intervals (the first dose was 70 mg and other doses were decrease progressively to 25 mg). And subsequently injected with 0.1 mg cloprostenol after 60 hours from FSH was administered. Estrous detection was carried out 12 h after CIDR withdrawal and mating were repeated at 8 h intervals.
Goat zygotes at the one-cell stage (around 14 h post-fertilization) were surgically collected and were immediately transferred into TCM199 medium (Gibco, NY, USA). Cas9 mRNA (2 ng/μL) and sgRNAs (5 ng/μL for each sgRNA) targeting MSTN and FGF5 were mixed and injected into the cytoplasm of fertilized oocytes using the FemtoJect system (Eppendorf, Hamburg, Germany). The injection pressure, injection time and compensatory pressure were 45 kpa, 0.1 s and 7 kpa, respectively. Microinjection was conducted in manipulation medium TCM199 on the heated platform of the Olympus micromanipulation system ON3. After injection, the zygotes were cultured in Quinn’s Advantage Cleavage Medium (Sage Biopharma, NJ, USA) for 24 h at 37 °C, and then transferred to Quinn’s Advantage Blastocyst Medium (Sage Biopharma, NJ, USA) at 37 °C, 5% concentration of carbon dioxide and saturated humidity conditions. The surrogate animals for transfer were determined according to their oestrus cycles. About three divisive embryos were transferred into the ampullary-isthmic junction of the oviduct of the surrogate ewes. Pregnancy was determined by observing the oestrus behaviors of surrogate ewes every ovulation circle.
T7E1 cleavage assay and sequencing
Samples including blood, heart, liver, spleen, lung, kidney, skin, testis, and muscle, were collected and digested in lysis buffer (0.4 M NaCl, 2 μM EDTA, 1% SDS, 10 μM Tris-HCl, and 100 μg/ml Proteinase K). The genomic DNA of the sample was extracted from the lysate by the phenol-chloroform protocol, and recovered by alcohol precipitation. A T7E1 cleavage assay was performed as described9.
Off-target assay
To determine the site-specific cleavage of the CRISPR-Cas9 system in vivo, the potential off-target loci were searched using an open tool, SeqMap29. The mismatch parameter for the target sequence was set as 5. ‘NGG’ and ‘NAG’ were chosen as the protospacer adjacent motifs (PAM). The sites with 7 bp conserved proximal to PAM and total mismatches <5, and sites with total mismatches <4, were chosen as potential off-target sites for subsequent testing. The selected potential off-target sites were initially PCR-amplified using genomic DNA from cultured goat fibroblasts. The PCR products were then subjected to a T7E1 cleavage assay. The potential off-target sites yielding the typical cleavage bands were considered as candidates and further evaluated in the founder animals by PCR amplification, T7E1 cleavage assay and TA sequencing. The information on the off-target loci and primer pairs used are listed in Supplementary Tables S7.
Additional Information
How to cite this article: Wang, X. et al. Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci. Rep. 5, 13878; doi: 10.1038/srep13878 (2015).
Supplementary Material
Acknowledgments
The authors would like to thank those who assisted with goat herd management, lamb care and technical work in the lab related to this project including: Chongqi Tian, Yong Mu, Sen Ma, Bibo Li and Yankun Liu, as well as the entire Huang Lab for their support on lab techniques. This work is supported by National Natural Science Foundation of China (31372279, 31402038, 31171377), and by grants from the Major Projects for New Varieties of Genetically Modified Organisms of China (2014ZX08008-002) and Provincial Key Projects of Shaanxi (2014KTDZ02-01).
Footnotes
Author Contributions Conceived and designed the experiments: X.W., H.Y., A.L., X.H., B.M., Y. J., W.Z., L.Q. and Y.C. Performed the experiments X.W., H.Y., A.L., X.H., B.M., H.Y., J.Z., H.Z., S.H., Z.D., L.S., Y.N., B.S., B.C., X.Z., Y.Y., Y.J., J.L., X.C., X.T. and Y.W. Analyzed the data: X.W., J.Z, Y.N., B.S., B.C., G.Z. and X.H. Wrote the manuscript: X.W., H.Y., A.L., J.Z., X.H. and Y.C. All authors read and approved the final manuscript.
References
- Gaj T., Gersbach C. A. & Barbas C. F. III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in Biotech. 31, 397–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Lander E. S. & Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon E. et al. Osteochondral regeneration using a novel aragonite-hyaluronate bi-phasic scaffold in a goat model. Knee Surg Sports Traumatol Arthrosc. 22, 1452–1464 (2013). [DOI] [PubMed] [Google Scholar]
- Walschot L. H. B., Aquarius R., Schreurs B. W., Verdonschot N. & Buma P. Osteoconduction of impacted porous titanium particles with a calcium-phosphate coating is comparable to osteoconduction of impacted allograft bone particles: In vivo study in a nonloaded goat model. J. Biomed Mater Res. 100B, 1483–1489 (2012). [DOI] [PubMed] [Google Scholar]
- Faisal S. M. et al. Evaluation of a Salmonella vectored vaccine expressing Mycobacterium avium subsp. paratuberculosis antigens against challenge in a goat model. PLoS ONE. 8, e70171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefer C. L. et al. Generation of Dwarf Goat (Capra hircus) Clones Following Nuclear Transfer with Transfected and Nontransfected Fetal Fibroblasts and In Vitro-Matured Oocytes. Biol Reprod. 64, 849–856 (2001). [DOI] [PubMed] [Google Scholar]
- Ni W. et al. Efficient gene knockout in goats using CRISPR/Cas9 system. PLoS ONE. 9, e106718 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrels W., Ivics Z. & Kues W. A. Precision genetic engineering in large mammals. Trends in Biotech. 30, 386–393 (2012). [DOI] [PubMed] [Google Scholar]
- Shen B. et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 23, 720–723 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. et al. Generating rats with conditional alleles using CRISPR/Cas9. Cell Res. 24, 122–125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y. et al. Generation of Gene-Modified Cynomolgus Monkey via Cas9/RNA-Mediated Gene Targeting in One-Cell Embryos. Cell. 156, 836–843 (2014). [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. Efficient generation of gene-modified pigs via injection of zygote with Cas9/sgRNA. Sci. Rep. 5, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron A. C. & Lee S.-J. Suppression of body fat accumulation in myostatin-deficient mice. J Clin Invest. 109, 595–601 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher D. S. et al. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 3, e79 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobet L. et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 17, 71–74 (1997). [DOI] [PubMed] [Google Scholar]
- Kim J., Petrella J. K., Cross J. M. & Bamman M. M. Load-mediated downregulation of myostatin mRNA is not sufficient to promote myofiber hypertrophy in humans: a cluster analysis. J Appl Physiol. 103, 1488–1495 (2007). [DOI] [PubMed] [Google Scholar]
- Ota Y. et al. Fibroblast Growth Factor 5 Inhibits Hair Growth by Blocking Dermal Papilla Cell Activation. Biochem Biophys Res Commun. 290, 169–176 (2002). [DOI] [PubMed] [Google Scholar]
- Hébert J. M., Rosenquist T., Götz J. & Martin G. R. FGF5 as a regulator of the hair growth cycle: Evidence from targeted and spontaneous mutations. Cell. 78, 1017–1025 (1994). [DOI] [PubMed] [Google Scholar]
- Higgins C. A. et al. FGF5 is a crucial regulator of hair length in humans. PNAS. 111, 10648–10653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehler J. S. et al. Four Independent Mutations in the Feline Fibroblast Growth Factor 5 Gene Determine the Long-Haired Phenotype in Domestic Cats. J Hered. 98, 555–566 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieu E. et al. Coat Variation in the Domestic Dog Is Governed by Variants in Three Genes. Science. 326, 150–153 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand R., Tiret L. & Abitbol M. Two recessive mutations in FGF5 are associated with the long-hair phenotype in donkeys. Genet Sel Evol. 46, 65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Esvelt K. M. & Church G. M. Cas9 as a versatile tool for engineering biology. Nat Meth. 10, 957–963 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D. & Joung J. K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotech. 32, 347–355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., et al. One-step generation of myostatin gene knockout sheep via the CRISPR/Cas9 system. Front Agr Sci Eng. 1, 2–5 (2014). [Google Scholar]
- Fu Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotech. 31, 822–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruscha A. et al. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 140, 4982–4987 (2013). [DOI] [PubMed] [Google Scholar]
- Jiang H. & Wong W. H. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 24, 2395–2396 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., et al. Studies on optimized method of synchronized estrus in goats. Southwest China J Agr Sci. 17, 664–667 (2004). [Google Scholar]
- Li J., et al. Study on the superovulatioin in Boer goat. Southwest China J Agr Sci. 4, 92–95 (2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.