Abstract
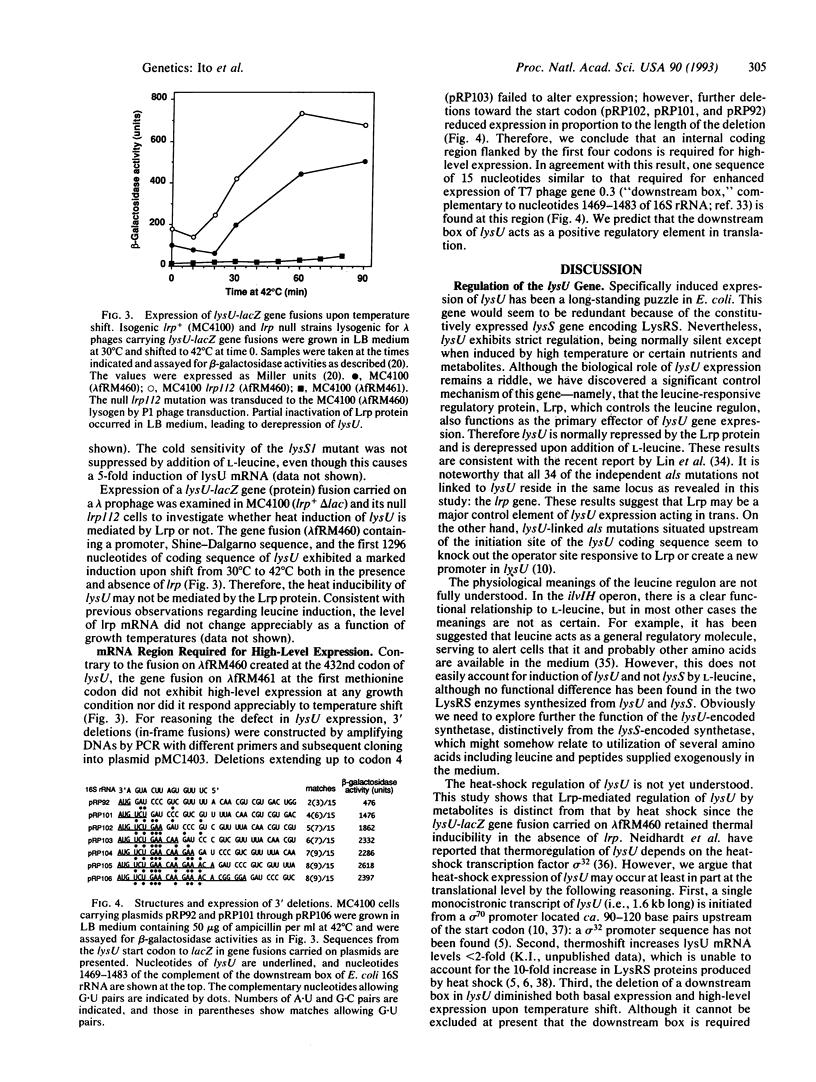
Lysyl-tRNA synthetases [L-lysine:tRNA(Lys) ligase (AMP-forming), EC 6.1.1.6] are synthesized from two distinct genes in Escherichia coli, lysS (constitutively) and lysU (inducibly), but neither the physiological significance nor the mechanism of differential regulation of these two genes is understood. We have constructed a null mutation of lysS that causes cold-sensitive lethality and then used this mutant to acquire and characterize several bypass mutations called als (abandonment of lysS). Cold-resistant survivors were isolated either spontaneously or by transposon-mediated disruption, and all caused derepression of lysU transcription. One class of als mutations is linked to lysU and presumably affects the cis regulatory element. Mutations of the other class map within the lrp gene, which encodes the leucine-responsive regulatory protein (Lrp). A lysU-lacZ gene fusion study revealed that lysU is susceptible to thermal regulation in the absence of lrp and that a small mRNA region immediately downstream of the initiation codon is required for potentially high-level expression. These results suggest that lysU is part of the leucine regulon and is both negatively controlled by Lrp and positively regulated by a potential translational enhancer sequence. This sequence is similar to that of the "downstream box" complementary to nucleotides 1469-1483 of 16S rRNA, which can be universally found in tRNA synthetase genes of E. coli.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Carter-Muenchau P., Wolf R. E., Jr Growth-rate-dependent regulation of 6-phosphogluconate dehydrogenase level mediated by an anti-Shine-Dalgarno sequence located within the Escherichia coli gnd structural gene. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1138–1142. doi: 10.1073/pnas.86.4.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Clark R. L., Neidhardt F. C. Roles of the two lysyl-tRNA synthetases of Escherichia coli: analysis of nucleotide sequences and mutant behavior. J Bacteriol. 1990 Jun;172(6):3237–3243. doi: 10.1128/jb.172.6.3237-3243.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich R. V., Hirshfield I. N. Mapping of the constitutive lysyl-tRNA synthetase gene of Escherichia coli K-12. J Bacteriol. 1987 Nov;169(11):5311–5313. doi: 10.1128/jb.169.11.5311-5313.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faxén M., Plumbridge J., Isaksson L. A. Codon choice and potential complementarity between mRNA downstream of the initiation codon and bases 1471-1480 in 16S ribosomal RNA affects expression of glnS. Nucleic Acids Res. 1991 Oct 11;19(19):5247–5251. doi: 10.1093/nar/19.19.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J., Newman E. B. Derivation of glycine from threonine in Escherichia coli K-12 mutants. J Bacteriol. 1975 Jun;122(3):810–817. doi: 10.1128/jb.122.3.810-817.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A., Casadaban M. J. Mini-mu bacteriophage with plasmid replicons for in vivo cloning and lac gene fusing. J Bacteriol. 1986 Oct;168(1):357–364. doi: 10.1128/jb.168.1.357-364.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshfield I. N., Bloch P. L., Van Bogelen R. A., Neidhardt F. C. Multiple forms of lysyl-transfer ribonucleic acid synthetase in Escherichia coli. J Bacteriol. 1981 Apr;146(1):345–351. doi: 10.1128/jb.146.1.345-351.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshfield I. N., Bukald N. E. Effect of alanine, leucine and fructose on lysyl-transfer ribonucleic acid ligase activity in a mutant of Escherichia coli K-12. J Bacteriol. 1973 Jan;113(1):167–177. doi: 10.1128/jb.113.1.167-177.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshfield I. N., Tenreiro R., Vanbogelen R. A., Neidhardt F. C. Escherichia coli K-12 lysyl-tRNA synthetase mutant with a novel reversion pattern. J Bacteriol. 1984 May;158(2):615–620. doi: 10.1128/jb.158.2.615-620.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshfield I. N., Yeh F. M., Sawyer L. E. Metabolites influence control of lysine transfer ribonucleic acid synthetase formation in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1364–1367. doi: 10.1073/pnas.72.4.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., Ito K., Nakamura Y. Differential regulation of two genes encoding lysyl-tRNA synthetases in Escherichia coli: lysU-constitutive mutations compensate for a lysS null mutation. Mol Microbiol. 1992 Jul;6(13):1739–1745. doi: 10.1111/j.1365-2958.1992.tb01346.x. [DOI] [PubMed] [Google Scholar]
- Kawakami K., Jönsson Y. H., Björk G. R., Ikeda H., Nakamura Y. Chromosomal location and structure of the operon encoding peptide-chain-release factor 2 of Escherichia coli. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5620–5624. doi: 10.1073/pnas.85.15.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Lin R., Ernsting B., Hirshfield I. N., Matthews R. G., Neidhardt F. C., Clark R. L., Newman E. B. The lrp gene product regulates expression of lysU in Escherichia coli K-12. J Bacteriol. 1992 May;174(9):2779–2784. doi: 10.1128/jb.174.9.2779-2784.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévêque F., Gazeau M., Fromant M., Blanquet S., Plateau P. Control of Escherichia coli lysyl-tRNA synthetase expression by anaerobiosis. J Bacteriol. 1991 Dec;173(24):7903–7910. doi: 10.1128/jb.173.24.7903-7910.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévêque F., Plateau P., Dessen P., Blanquet S. Homology of lysS and lysU, the two Escherichia coli genes encoding distinct lysyl-tRNA synthetase species. Nucleic Acids Res. 1990 Jan 25;18(2):305–312. doi: 10.1093/nar/18.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikuni O., Kawakami K., Nakamura Y. Sequence and functional analysis of mutations in the gene encoding peptide-chain-release factor 2 of Escherichia coli. Biochimie. 1991 Dec;73(12):1509–1516. doi: 10.1016/0300-9084(91)90185-4. [DOI] [PubMed] [Google Scholar]
- Nagai H., Yuzawa H., Yura T. Interplay of two cis-acting mRNA regions in translational control of sigma 32 synthesis during the heat shock response of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10515–10519. doi: 10.1073/pnas.88.23.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Masui Y., Inouye M. Use of a lac promoter-operator fragment as a transcriptional control switch for expression of the constitutive lpp gene in Escherichia coli. J Mol Appl Genet. 1982;1(4):289–299. [PubMed] [Google Scholar]
- Neidhardt F. C., VanBogelen R. A. Positive regulatory gene for temperature-controlled proteins in Escherichia coli. Biochem Biophys Res Commun. 1981 May 29;100(2):894–900. doi: 10.1016/s0006-291x(81)80257-4. [DOI] [PubMed] [Google Scholar]
- Oka A., Sugisaki H., Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981 Apr 5;147(2):217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- Platko J. V., Willins D. A., Calvo J. M. The ilvIH operon of Escherichia coli is positively regulated. J Bacteriol. 1990 Aug;172(8):4563–4570. doi: 10.1128/jb.172.8.4563-4570.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex J. H., Aronson B. D., Somerville R. L. The tdh and serA operons of Escherichia coli: mutational analysis of the regulatory elements of leucine-responsive genes. J Bacteriol. 1991 Oct;173(19):5944–5953. doi: 10.1128/jb.173.19.5944-5953.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricca E., Aker D. A., Calvo J. M. A protein that binds to the regulatory region of the Escherichia coli ilvIH operon. J Bacteriol. 1989 Mar;171(3):1658–1664. doi: 10.1128/jb.171.3.1658-1664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengart M. L., Fatscher H. P., Fuchs E. The initiation of translation in E. coli: apparent base pairing between the 16srRNA and downstream sequences of the mRNA. Nucleic Acids Res. 1990 Apr 11;18(7):1719–1723. doi: 10.1093/nar/18.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan L. R., D'Ari R., Newman E. B. The leucine regulon of Escherichia coli K-12: a mutation in rblA alters expression of L-leucine-dependent metabolic operons. J Bacteriol. 1990 Aug;172(8):4529–4535. doi: 10.1128/jb.172.8.4529-4535.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBogelen R. A., Vaughn V., Neidhardt F. C. Gene for heat-inducible lysyl-tRNA synthetase (lysU) maps near cadA in Escherichia coli. J Bacteriol. 1983 Feb;153(2):1066–1068. doi: 10.1128/jb.153.2.1066-1068.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Willins D. A., Ryan C. W., Platko J. V., Calvo J. M. Characterization of Lrp, and Escherichia coli regulatory protein that mediates a global response to leucine. J Biol Chem. 1991 Jun 15;266(17):10768–10774. [PubMed] [Google Scholar]