Abstract
All-atom simulation data are presented for ternary mixtures of palmitoyl sphingomyelin (PSM), cholesterol, and either palmitoyl oleoyl phosphatidyl choline or dioleoyl phosphatidyl choline (DOPC). For comparison, data for a mixture of dipalmitoyl phosphatidyl choline (DPPC), cholesterol, and DOPC are also presented. Compositions corresponding to the liquid-ordered phase, the liquid-disordered phase, and coexistence of the two phases are simulated for each mixture. Within the liquid-ordered phase, cholesterol is preferentially solvated by DOPC if it is available, but if DOPC is replaced by POPC, cholesterol is preferentially solvated by PSM. In the DPPC mixtures, cholesterol interacts preferentially with the saturated chains via its smooth face, whereas in the PSM mixtures, cholesterol interacts preferentially with PSM via its rough face. Interactions between cholesterol and PSM have a very particular character: hydrogen bonding between cholesterol and the amide of PSM rotates the tilt of the amide plane, which primes it for more robust hydrogen bonding with other PSM. Cholesterol-PSM hydrogen bonding also locally modifies the hexagonal packing of hydrocarbon chains in the liquid-ordered phase of PSM mixtures.
Introduction
It has been recognized since at least the early 1970s that the plasma membrane of eukaryotic cells is heterogeneous (1,2). In later experiments, lateral heterogeneity was incorporated into a model for functional domains in the plasma membrane, termed rafts, which are hypothesized to organize membrane components for signaling, trafficking, and other purposes (3). However, the raft hypothesis remains controversial, in part due to the difficulty of performing controlled experiments in live cells to probe organization on nanometer length scales. Nevertheless, experiments by Eggeling et al. (4) and Honigmann et al. (5,6) indicated nanoscale heterogeneity in live cells, and Sanchez et al. (7) demonstrated that these heterogeneities are characterized by tightly packed lipids (8), as may be induced by cholesterol (Chol).
Lipid bilayers containing Chol and one or more other lipids are frequently used as models for laterally heterogeneous cell membranes (9,10). The putative raft fraction of cell membranes is reported to be enriched in sphingolipids and Chol (11). At low concentrations, Chol increases the order of the hydrocarbon chains of fluid-phase, saturated-chain phospholipids, whereas at higher concentrations it induces the liquid-ordered (Lo) phase (12,13). Adding a third component with a lower melting temperature (Tm), such as dioleoyl phosphatidylcholine (DOPC), supports the coexistence of the Lo phase with the conventional disordered-chain fluid phase (here called Ld for liquid disordered). Phase diagrams and insights into the chain order for such mixtures have been obtained by various experimental methods, including NMR (14–17), neutron (18) and x-ray (19) diffraction, Förster resonance energy transfer (FRET) (20), and fluorescence microscopy (21). (For comprehensive reviews of binary and ternary phase diagrams, see Marsh (22,23).) However, the extent to which these systems serve as good models of cell membranes remains an open question.
The length scale of Lo domains in the two-phase region depends strongly on composition (18,24,25). DOPC/dipalmitoyl phosphatidylcholine (DPPC)/Chol, perhaps the best-studied ternary mixture, forms micron-scale domains that are observable by diffraction-limited fluorescence microscopy in giant unilamellar vesicles (GUVs) (26). Replacing DOPC by palmitoyloleoyl phosphatidylcholine (POPC) renders the composition more physiological and reduces the length scale of Lo domains to tens of nanometers (18,24). Feigenson and co-workers have described the former mixture as a type II mixture and the latter as a type I mixture (9).
The saturated-chain, high-Tm phospholipids that one usually finds in the outer leaflet of the mammalian plasma membrane are not glycerolipids like DPPC, but rather are sphingolipids. For example, palmitoyl sphingomyelin (PSM) is the most abundant sphingolipid in the Maden-Darby canine kidney cell line, comprising roughly 50% by mole (27). Like DPPC, PSM will also form an Lo phase in collaboration with Chol, and has a gel-to-fluid-phase transition temperature very similar to that of DPPC (∼41.4°C for PSM compared with 41.5°C for DPPC (28)). Unlike DPPC, PSM has an amide group in the backbone, and thus is well positioned to hydrogen bond with Chol. Accordingly, the Lo phases formed by saturated glycerolipids may well be different from those formed by sphingolipids, with consequences for the length scale of Lo domains and partitioning of integral membrane proteins between the two phases. Indeed, a mixture of POPC/PSM/Chol forms domains that are large enough to be observed by diffraction-limited fluorescence microscopy, in contrast to DPPC/POPC/Chol (21,28,29). (FRET experiments on brain sphingomyelin (SM), however, revealed nanodomains (25).) This is of particular interest in light of our recently published study regarding lateral heterogeneity within the Lo phase formed by DPPC/DOPC/Chol, in which simulations yielded regions of hexagonally ordered, mostly trans chains of DPPC separated by interstitial regions enriched in Chol and DOPC (30).
Coarse-grained simulations of ternary mixtures (31,32) and all-atom simulation data for mixtures of PSM with Chol (including ternary mixtures) have been reported previously (33–35). However, recent advances in force-field development for SMs (36) (driven by recent experimental data (37)) and the availability of specialized hardware that allows simulations of up to 10 μs (the Anton computer (38)) motivated us to revisit the problem. Here, we present all-atom simulation data for ternary mixtures of PSM/POPC/Chol, PSM/DOPC/Chol, and DPPC/DOPC/Chol. Compositions corresponding to the Lo and Ld phases, as well as the Lo/Ld coexistence regime, are studied for all cases. A detailed analysis of the lipid-lipid interactions in each phase for all three mixtures reveals significant differences between the DPPC and PSM mixtures. A molecular view of the interactions reveals a special role for hydrogen bonding between the amide of SM and Chol, such that the amide plane of PSM is primed for a particular hydrogen-bonding configuration. This in turn modifies the local hexagonal order in the Lo phase as compared with a mixture of DPPC/DOPC/Chol.
Materials and Methods
A total of nine different systems were prepared for three different ternary mixtures: PSM/DOPC/CHOL, PSM/POPC/CHOL, and DPPC/DOPC/CHOL. In each case, compositions were selected for the Lo and Ld phase from the endpoints of an experimentally determined tie-line as published by Veatch et al. (26) for the DOPC mixture, or from unpublished data for PSM mixtures (Gerald Feigenson, Cornell University, personal communication, 2014). Initial configurations were generated by CHARMM-GUI (39,40) and checked for ring penetration artifacts. A third system was simulated for each tie-line, in which roughly equal areas of each phase (after equilibration of the total areas) were combined into a single system to yield a model of the interfacial region. A summary of the nine systems is presented in Table 1.
Table 1.
List of Simulations
| Mixture | Composition | Phase | Duration (μs) | Temp (K) | Total Lipids | Area/Lipid (Å2) |
|---|---|---|---|---|---|---|
| PSM/POPC/Chol | 0.61/0.08/0.31 | Lo | 9.08 | 295 | 560 | 40.5(0.2) |
| PSM/POPC/Chol | 0.23/0.69/0.08 | Ld | 1.14 | 295 | 560 | 54.8(0.5) |
| PSM/POPC/Chol | 0.47/0.32/0.21 | Lo/Ld | 6.88 | 295 | 506 | N/A |
| PSM/DOPC/Chol | 0.64/0.03/0.33 | Lo | 0.95 | 295 | 560 | 40.4(0.2) |
| PSM/DOPC/Chol | 0.15/0.82/0.03 | Ld | 0.81 | 295 | 574 | 62.7(0.6) |
| PSM/DOPC/Chol | 0.43/0.38/0.19 | Lo/Ld | 4.54 | 295 | 452 | N/A |
| DPPC/DOPC/Chol | 0.55/0.15/0.30 | Lo | 5.05 | 298 | 512 | 40.9(0.2) |
| DPPC/DOPC/Chol | 0.29/0.6/0.11 | Ld | 2.02 | 298 | 400 | 57.1(0.8) |
| DPPC/DOPC/Chol | 0.37/0.36/0.27 | Lo/Ld | 9.43 | 298 | 380 | N/A |
The total number of lipids in each system ranged from 380 to 560, and the lipids were solvated with at least 30 TIP3P waters per lipid. The lipids were modeled with the CHARMM36 force field (36,41), and Chol was modeled with Pitman et al.’s (42) original Chol parameters. (The more recent update of Chol includes NBFIX corrections between atoms in the hydrocarbon region, resulting in a large number of NBFIX corrections, which degrades the performance of Anton.)
Each system was minimized, heated, and equilibrated by velocity rescaling, and then integrated under NPT conditions using a Langevin thermostat (0.5 ps−1 coupling frequency) and piston (200 fs period, 100 fs decay time) with NAMD v2.9 on local resources. The NPT simulations were performed for at least 40 ns, with a 2.0 fs timestep and hydrogen positions constrained by SHAKE with a tolerance of 10−5. Long-range electrostatics were computed using particle-mesh Ewald with a tolerance of 10−6, 4th-order spline interpolation, and a mesh size of 1.0 Å, and van der Waals potentials and forces were shifted smoothly to zero between 10 and 12 Å.
The systems were then transferred for production simulation to Anton, and input files from the NAMD restart files were generated using a python script available on the Anton wiki. On Anton, the multigrator was used to integrate the equations of motion, with the use of a Martyna-Tobias-Klein (43) barostat and Nosé-Hoover (44,45) temperature control. The long-range cutoff varied by simulation, but in every case was at least 11.5 Å. Electrostatics were computed by the Gaussian-split Ewald (46) method, with parameters chosen by the Anton preprocessing scripts.
Results
Lo phases contain a locally hexagonal substructure
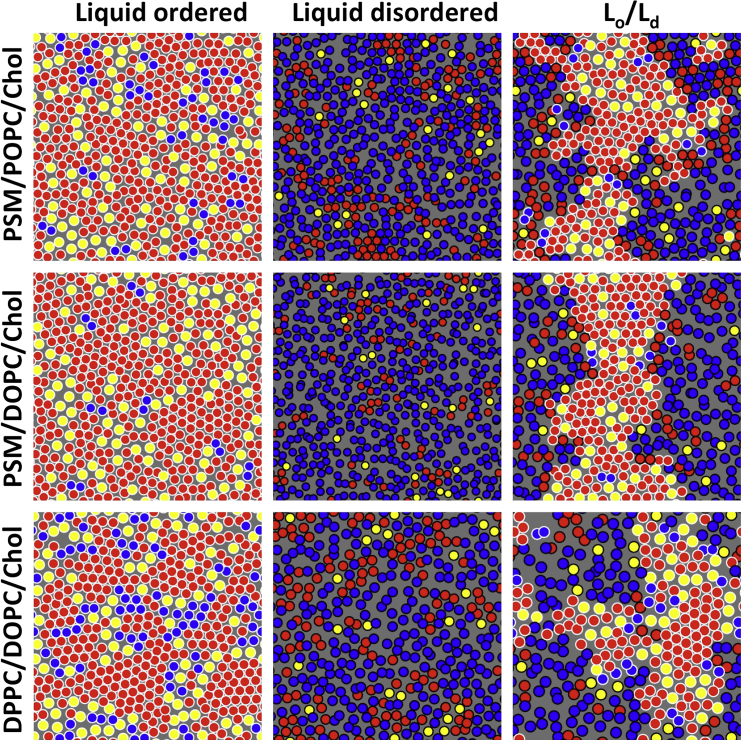
Images of the final configurations are shown in Fig. 1, and movies for the PSM/POPC/Chol systems are included in the Supporting Material. In the Lo/Ld coexistence systems, the state of each lipid is determined by a previously published hidden Markov model (30). The locally hexagonal substructure in the Lo phase, previously reported for a mixture of DPPC/DOPC/Chol (30), is readily apparent in the PSM Lo phase as well. The PSM Lo phases presented here contain a similar fraction of Chol but fewer unsaturated chains than the DPPC case (Table 1); nonetheless, the Chol is effective in generating defects that prevent the formation of the longer-range hexagonal order that would be observed in the gel. The tightly packed nature of the Lo phase leads to a slow, collective dynamics, which, likely due to hydrogen bonding of the amide PSM (discussed in detail below), is even slower in the PSM Lo phase (36).
Figure 1.
Lateral chain ordering in Lo and Ld phases. The final configurations of nine simulations are shown. A single leaflet is shown with the center of mass of hydrocarbon chains and Chol rendered as discs. A white border indicates an Lo phase lipid and a black border indicates an Ld phase lipid, with phases determined in the coexistence systems by the hidden Markov model. Red indicates PSM or DPPC, blue indicates DOPC or POPC, and yellow indicates Chol. Column order: Lo phases (left), Ld phases (middle), and coexistence systems (right). Row order: PSM/POPC/Chol (top), PSM/DOPC/Chol (middle), and DPPC/DOPC/Chol (bottom). The same configurations are rendered in space-filling representation in Fig. S1. To see this figure in color, go online.
Comparison of lipid interactions in ternary mixtures
A central question in efforts to understand Lo/Ld coexistence is, how do lipid-lipid interactions drive phase separation (47–52)? A favorable interaction between a lipid pair would tend to enhance the frequency of contacts between those lipids, which ought to be reflected in the simulation data. Therefore, we posed the following question: compared with a random mixture, in a given pair of lipids, are the lipids more or less likely to be neighbors? We addressed this issue by 1) generating a Voronoi polyhedral around the centers of mass of each lipid in each configuration, 2) computing the length of the boundary between each lipid pair, and 3) randomizing the labels and recalculating the boundary lengths (see Fig. S2). Enhancements or reductions of boundary lengths are reported as percentages in Tables 2, 3, and 4 for all nine lipid mixtures, with the α (smooth) and β (rough) faces of Chol treated separately. A value that is not statistically different from zero indicates that the pair behaved as it would in an ideal mixture.
Table 2.
Lipid Interactions in PSM/DOPC/Chol
| PSM/DOPC/Chol | Lo/Ld | Lo | Ld |
|---|---|---|---|
| Chol(S)-SAT | +56.8(3.3) | +14.9(0.8) | +59.4(10.8) |
| Chol(S)-USAT | −65.3(2.6) | +21.1(4.7) | −4.4(1.9) |
| Chol(R)-SAT | +65.1(1.8) | +21.1(0.9) | +53.3(8.2) |
| Chol(R)-USAT | −62.9(1.2) | +49.6(5.2) | −2.9(1.8) |
| Chol(S)-Chol(S) | −0.0(7.0) | −33.3(3.9) | −51.7(12.7) |
| Chol(S)-Chol(R) | +0.5(5.4) | −36.3(3.5) | −31.9(13.0) |
| Chol(R)-Chol(R) | −40.5(3.9) | −61.4(2.5) | −24.8(20.1) |
| SAT-SAT | +17.7(0.5) | −6.6(0.1) | +26.7(5.9) |
| SAT-USAT | −49.1(0.4) | −14.7(0.3) | −7.5(1.1) |
| USAT-USAT | +88.4(0.7) | +57.5(16.2) | +1.8(0.2) |
R and S indicate the rough and smooth faces of Chol, respectively.
Table 3.
Lipid Interactions in PSM/POPC/Chol
| PSM/POPC/Chol | Lo/Ld | Lo | Ld |
|---|---|---|---|
| Chol(S)-SAT | +27.9(1.7) | +16.8(0.9) | +26.0(3.2) |
| Chol(S)-USAT | −34.0(2.2) | +10.9(3.0) | +1.7(1.1) |
| Chol(R)-SAT | +46.7(1.1) | +23.6(0.8) | +16.7(3.8) |
| Chol(R)-USAT | −37.2(1.6) | −0.8(2.9) | +5.4(1.0) |
| Chol(S)-Chol(S) | +9.4(8.6) | −36.7(2.3) | −35.1(11.3) |
| Chol(S)-Chol(R) | −11.3(4.0) | −39.5(2.6) | −54.4(3.3) |
| Chol(R)-Chol(R) | −63.7(3.1) | −50.0(2.1) | −50.3(9.2) |
| SAT-SAT | +7.2(0.2) | −6.7(0.2) | +17.7(3.0) |
| SAT-USAT | −31.4(0.6) | −5.7(0.6) | −7.1(0.9) |
| USAT-USAT | +68.4(2.0) | +13.0(3.0) | +2.3(0.3) |
Table 4.
Lipid Interactions in DPPC/DOPC/Chol
| DPPC/DOPC/Chol | Lo/Ld | Lo | Ld |
|---|---|---|---|
| Chol(S)-SAT | +25.9(1.5) | +14.9(1.5) | +27.0(1.2) |
| Chol(S)-USAT | −16.9(0.9) | +7.8(2.4) | −4.4(1.2) |
| Chol(R)-SAT | +24.8(1.4) | +7.9(1.5) | +21.7(1.5) |
| Chol(R)-USAT | +2.6(1.1) | +46.0(3.5) | +5.5(0.7) |
| Chol(S)-Chol(S) | −7.5(2.0) | −29.8(2.0) | −20.3(8.1) |
| Chol(S)-Chol(R) | −16.2(1.1) | −32.0(2.1) | −34.9(3.0) |
| Chol(R)-Chol(R) | −41.3(0.9) | −41.2(1.8) | −61.7(4.2) |
| SAT-SAT | +11.8(1.8) | +0.6(0.8) | +2.1(1.6) |
| SAT-USAT | −29.1(1.3) | −20.4(1.0) | −4.7(0.8) |
| USAT-USAT | +35.3(1.3) | +23.0(3.2) | +2.6(0.4) |
In nearly every single case, Chol-Chol interactions are disfavored quite strongly; however, in the few exceptions to this rule (in 3 out of 27 cases), these interactions are neutral. This result is consistent with the disulfide cross-linking experiments of Wang et al. (53), which directly reported the neighbor frequencies of a chemically modified Chol probe. In every single case, a strongly favorable interaction is found between the high-Tm lipid and Chol. However, PSM favors the rough face, whereas DPPC favors the smooth one.
The interactions between Chol and the low-Tm lipid (DOPC or POPC) are more complicated. In the Lo/Ld coexistence systems, interactions between Chol and the low-Tm lipid are strongly disfavored (except for one case in which the interaction is neutral), consistent with the observation that Chol is more likely to be solvated by saturated chains (42). This observation generally holds true in the Ld phase as well, where Chol is significantly more likely to neighbor saturated chains. The Lo phases, however, display a different trend. Within Lo, the two mixtures containing DOPC are similar to each other: all four possible interactions between Chol and the lipids are favorable, with the most favorable interaction occurring between DOPC and the rough face of Chol. In contrast, in the POPC Lo phase, this interaction is neutral, and overall the interactions with PSM are favored over an interaction with POPC. In other words, in the DOPC Lo phases (with either PSM or DPPC), Chol is more likely to be solvated by DOPC, whereas in the PSM/POPC Lo phase, it is more likely to be solvated by PSM.
The interactions between (non-Chol) lipids in the Lo/Ld coexistence systems are consistent with expectation, holding to the rule that like solvates like. This also holds true (but to a lesser extent) for the Ld phase. Within the Lo phases, it is also true that the low Tm lipids have a favorable interaction with one another, and the unlike lipids have unfavorable interactions. The high Tm lipids differ, however: within the PSM Lo phases, these interactions are in fact disfavored relative to a randomly mixed control, whereas in the DPPC Lo phase, these interactions are neutral. In other words, given the composition of PSM Lo phases, one would expect to see even more PSM-PSM interactions than are observed. This appears to be because Chol competes for PSM amide hydrogen bonds, resulting in a stronger PSM-Chol interaction. Amide hydrogen bonding is discussed in the following sections. The focus is on hydrogen bonding in the Lo phase of PSM/POPC/Chol, which is expected to the most relevant to the raft fraction of the outer leaflet of the mammalian plasma membrane.
Amide hydrogen bonding among SMs in the Lo phase
The structure of the ceramide backbone lends itself to hydrogen bonding between SMs. The amide of the backbone can both donate and accept a hydrogen bond, and these hydrogen bonds are located deep in the interfacial region, almost in the hydrocarbon core. As such, they tend to be long-lived hydrogen bonds (36) and may well contribute to the stability of SM Lo phases. It is therefore of interest to examine the structure of hydrogen bonding among the SM amides and how it is modified in the presence of Chol.
To form tight hydrogen bonds, SM must tilt the bulky and negatively charged phosphocholine group to the side. This results in a pattern of back-and-forth tilting of the amide plane in a chain of hydrogen-bonded SMs (for details, see Figs. 2 and S3). In the simulations of ternary mixtures reported here, hydrogen-bonded chains of SMs ranging in length from two to nine lipids were observed. In the pure fluid phase, chains of up to 12 SMs were observed in a 100 ns simulation with 324 lipids per leaflet.
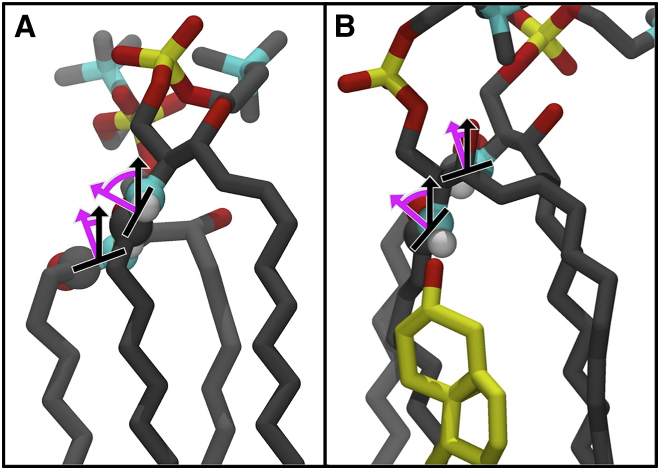
Figure 2.
Amide plane hydrogen bonding in the SM Lo phase. An illustration of how the amide plane tilts during hydrogen bonding between PSM when the amide accepts or donates a hydrogen bond is shown in (A). Purple arrows show the tilt of the plane away from the membrane normal (black arrow), approximately looking down the path of hydrogen bonding. Chol-PSM hydrogen bonding is similar; an example is shown at right in (B). Histograms of these angles are shown in Fig. 3. To see this figure in color, go online.
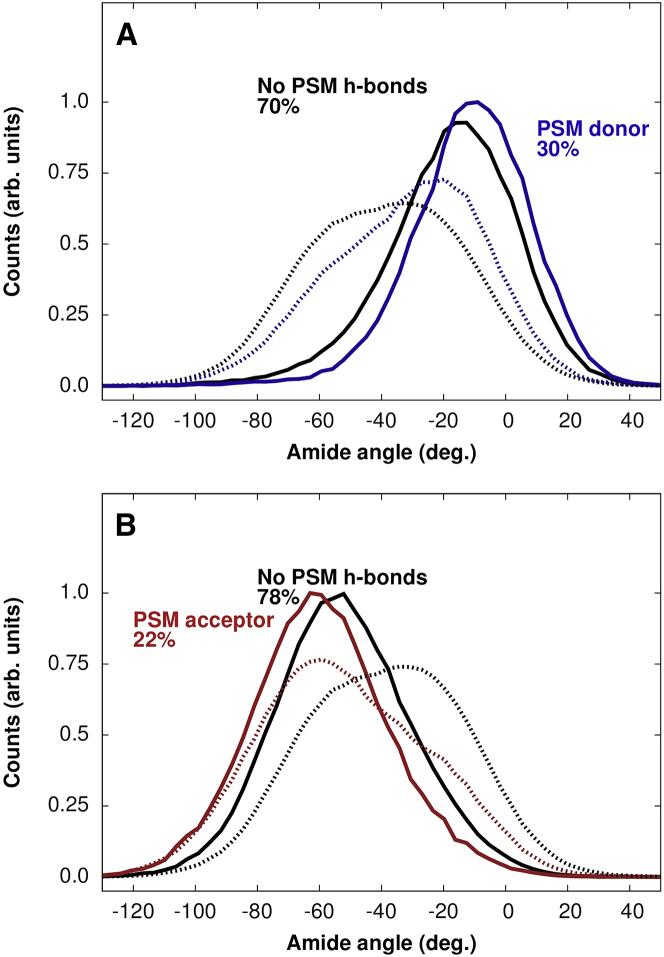
In addition to the back-and-forth pattern of amide plane tilting, there is a bias in the orientation of the amide plane depending on whether the amide is donating or accepting a hydrogen bond. This is shown by the dashed curves in Fig. 3, A and B, which show a clear shift in the distribution of the angle that the amide plane normal makes relative to the bilayer normal. When PSM donates an amide hydrogen bond, the distribution shifts to the right (Fig. 3A); when it accepts a hydrogen bond, the distribution is shifted to the left (Fig. 3B).
Figure 3.
SM amide plane angle in the Lo phase. Histograms of the angle formed between the normal to the SM plane and the bilayer normal for different amide hydrogen-bonding scenarios are shown. A comparison of the dashed curves and their solid counterparts shows how the distribution shifts when the amide plane forms a hydrogen bond with Chol (dashed line: no-Chol hydrogen bond). (A and B) The distributions shift to the right when the amide accepts a hydrogen bond from Chol (A), and to the left when it donates a hydrogen bond to Chol (B). These shifts are enhanced further when the PSM amide then donates a hydrogen bond (blue curves, A) or accepts (red curves, B) a hydrogen bond to or from another PSM amide. The percentages are the fraction contributed by that class to the total population. To see this figure in color, go online.
Chol primes the amide plane for strong hydrogen bonding among SMs
The hydroxyl group of Chol is also well positioned to hydrogen bond with the amide of SMs. In a mixture containing both Chol and SM, there are three possibilities for hydrogen bonding between the amide of a particular SM and Chol: the SM may not form a hydrogen bond with Chol, it might donate a hydrogen bond to a Chol, or it might accept a hydrogen bond from Chol. How each possibility influences the distribution of amide plane angles is shown by the solid curves in Fig. 3, A and B.
In the absence of a hydrogen bond to Chol, the amide plane distribution looks much like that observed in the pure SM case: the plane tilts one way or the other, depending on whether the SM accepts or donates a hydrogen bond to another SM. However, when SM donates a hydrogen bond to Chol, its amide plane angle shifts to the left, readying it to accept a hydrogen bond from another SM. Conversely, when SM accepts a hydrogen bond from Chol, the distribution of the amide plane angles shifts to the right, readying the SM to donate a hydrogen bond to another SM. We refer to this shifting of the amide plane tilt distribution by Chol as priming of the amide plane. An example is shown in Fig. 2B.
These shifts are quite dramatic and suggest that the SM-SM hydrogen-bonding propensities depend on these interactions with Chol. The likelihood of SM donating a hydrogen bond to another SM is unchanged when the amide plane is primed by Chol, as shown in the first column of Table 5. In marked contrast, SM is 30% more likely to accept a hydrogen bond from another SM when it is primed by donating a hydrogen bond to Chol. Thus, despite competition with Chol for a hydrogen bond that might form between SM and SM, once the bond is formed, it then dramatically increases the likelihood that other SM-SM hydrogen bonds will form.
Table 5.
PSM-PSM Amide Hydrogen-Bonding Propensities
| PSM to PSM | PSM from PSM | |
|---|---|---|
| Not Chol primed | 0.25 | 0.26 |
| Chol primed | 0.26 | 0.34 |
Chol perturbs local hexagonal order
Chol breaks the hexagonal order of the PSM chains in the Lo phase, as shown in Fig. 4. The balance between hexagonal regions and Chol-rich interstitial regions varies according to the composition, with the size of local hexagonal regions varying from a few lipids to as many as 40 (see Fig. S4 for the distribution of chains per cluster).
Figure 4.
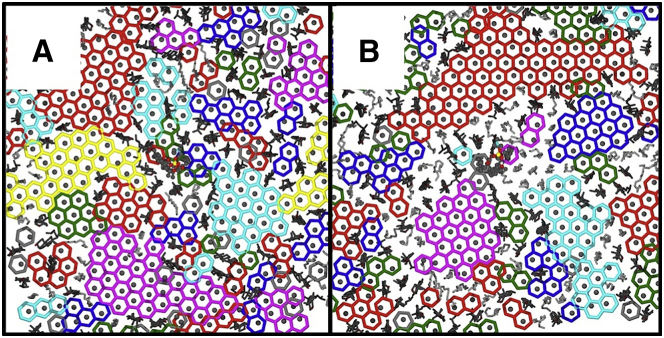
(A and B) Hexagonal substructure in PSM/POPC/Chol (A) and DPPC/DOPC/Chol (B) Lo phases. Each saturated chain is placed on a local hexagonal lattice (see Supporting Materials and Methods), with distinct lattices indicated by an arbitrary color scheme. The centers of mass of the saturated chains are indicated by filled circles. Unsaturated chains are rendered as gray sticks and Chol is shown as black sticks. To see this figure in color, go online.
In a locally hexagonal region of the PSM Lo phase, the two saturated chains of a lipid occupy neighboring lattice sites, with a peak in the chain-chain distance distribution at 4.88 Å (Fig. S5). Interaction with Chol locally disrupts this packing. When PSM forms an amide hydrogen bond with Chol, it tends to displace one of the chains, introducing a new peak in the chain-chain distance distribution at a separation of 8.18 Å (Fig. S5), with a more pronounced effect when PSM donates a hydrogen bond to Chol.
Local chain ordering is also disrupted by Chol in mixtures containing DPPC, as shown in Fig. 4B. In this case, the mechanism is different (although the weaker DPPC-Chol hydrogen-bonding mode can still be found perturbing the lattice; Fig. 4D), as DPPC lacks the amide of PSM. In the PSM Lo phase, hydrogen bonding to Chol limits hexagonal clusters to smaller sizes, as indicated by a shift in the peak in cluster size distribution (Fig. S4). Instead, significantly more DOPC is found in the DPPC/Chol Lo phase. We hypothesize that in the absence of hydrogen bonding to PSM, Chol is available to mediate interactions between DPPC and DOPC by means of its two faces. This gives a distinctly different character to the interstitial regions between hexagonal substructures, with consequences for partitioning of transmembrane segments, as discussed below.
Discussion
The outer leaflet of the plasma membrane is rich in SM, which has been proposed to collaborate with Chol in organizing the membrane into functional domains of differing composition (54). Favorable interactions between Chol and PSM are found, as expected. This is true also for the interaction between Chol and DPPC, a saturated lipid with a Tm similar to that of PSM, and is in general agreement with a previous combined Monte Carlo/FRET study (29). However, the detailed nature of the interaction between PSM and Chol is very different from that of the DPPC-Chol interaction, due to hydrogen bonding between the amide of PSM and Chol. The amide is located significantly deeper in the bilayer than other hydrogen-bonding moieties, and therefore forms more robust hydrogen bonds with Chol and other PSMs. In fact, the presence of Chol dramatically enhances the hydrogen bonding of PSM with other PSMs by priming the tilt of the amide for optimal hydrogen bonding to other PSMs. This observation contradicts the interpretation of recently published NMR data obtained for PSM/Chol mixtures by Matsumori et al. (55). These authors observed a similar average amide orientation in PSM and PSM/Chol systems, and concluded that PSM does not form hydrogen bonds with Chol. This conclusion is based on a model in which heterogeneity in the amide plane angle is assigned to wobble of the plane around a well-defined average value, and the assumption that Chol-PSM hydrogen bonding should strongly perturb the amide orientation into a new ensemble. Our data suggest instead that the amide plane adopts two distinct orientations, as shown in Fig. 3. Robust hydrogen bonding very likely enhances the stability of the PSM Lo phases relative to the same mixtures with DPPC instead of PSM, consistent with a reduced miscibility temperature of the latter mixtures (21). Investigators should also consider these results when designing fluorescent lipid analogs to be used as probes of lipid structure and dynamics, as such probes should not perturb the amide hydrogen bonding of SM.
Recent reports (56–58) are consistent with local order in the Lo phase. In these studies, quasi-Bragg scattering from oriented bilayer stacks was observed, but only when the coherence length of the neutron source was reduced to ∼30 Å by means of a low-resolution source. The results were interpreted as indicating that the ordering that produces sharp features at well-defined wavevectors is local and thus appears only when the signal has accumulated over a relatively small area. Our data are consistent with this proposal of local order, but differ in the details. The neutron data indicate a monoclinic unit cell for the lipid tails, with a lattice spacing of 5.2 Å and γ = 130.7°. Our data indicate local hexagonal ordering with a lattice spacing of 4.88 Å and γ = 120°. We stress that these are different systems: the neutron diffraction was performed on binary mixtures of DPPC and Chol in a ratio of 2:1, whereas our simulations involved three component Lo mixtures. It will be interesting to see whether accounting for lattice distortions by, for example, in silico scattering can reconcile the data, considering that a relatively small deformation of a monoclinic lattice yields a hexagonal lattice. In the simulations, the hexagonal lattice spacing is fairly insensitive to composition, as it simply represents close packing of ordered hydrocarbon chains. The hexagonal packing is quite dynamic, and therefore may be best understood as a hexagonal lattice plus disorder, with the details of the disorder depending on composition and especially the SM amide.
The hexagonal substructure previously reported for the Lo phase of DPPC/DOPC/Chol is also observed in the PSM Lo phase, but is locally modified by hydrogen bonding of the amide plane. These Chol-mediated local defects have the effect of modulating the lateral organization within the Lo phase. We hypothesize that the balance between Chol-rich interstitial regions and hexagonally packed hydrocarbon chain controls the partitioning of integral membrane proteins. Mechanisms that modulate the balance of interstitial and hexagonally packed areas within Lo regions would therefore influence partitioning. This possibility is particularly interesting in the light of recent experiments by Diaz-Rohrer et al. (59), which demonstrated partitioning into Lo of the linker for activation of T cells in giant plasma membrane vesicles, but not in ternary GUVs. Our data are consistent with the notion that a larger interstitial area is available in more physiological mixtures (PSM/POPC/Chol) than in less physiological ones (PSM/DOPC/Chol and DPPC/DOPC/Chol). Simulations of more physiological Lo-like mixtures, with a significantly greater diversity of lipids, might provide a direct test of this idea.
In summary, our extensive simulation of ternary lipid systems reveals important differences between mixtures that incorporate SM as the high-Tm lipid instead of saturated glycerolipids such as DPPC, and POPC rather than DOPC as the low-Tm lipid. Changes in lipid-lipid interactions imply that different molecular mechanisms drive phase separation, with amide hydrogen bonding in the sphingolipid mixtures disrupting the steric-driven interaction between the smooth face of Chol and ordered saturated chains. This leads to changes in the packing structure of the Lo phase, which in turn may control the partitioning of protein anchors between lipid phases.
Author Contributions
E.L., A.J.S., and R.W.P. designed the research. A.J.S. designed and performed the analysis. E.L., A.J.S., and R.W.P. wrote the manuscript.
Acknowledgments
We thank Gerald Feigenson and Tatyana Konyakhina for providing the compositions of the PSM Lo and Ld phases.
This research was supported in part by the Intramural Research Program of the National Heart, Lung and Blood Institute (NHLBI), National Institutes of Health, and utilized the NHLBI LoBoS cluster. Anton computer time was provided by the National Resource for Biomedical Supercomputing (NRBSC), the Pittsburgh Supercomputing Center (PSC), and the Biomedical Technology Research Center for Multiscale Modeling of Biological Systems through grant P41GM103712-S1 from the National Institutes of Health. The Anton machine at NRBSC/PSC was generously made available by D.E. Shaw Research.
Editor: Scott Feller.
Footnotes
Supporting Materials and Methods, Supporting Results, five figures, and three movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00771-7.
Supporting Material
References
- 1.Oldfield E., Chapman D. Dynamics of lipids in membranes: heterogeneity and the role of cholesterol. FEBS Lett. 1972;23:285–297. doi: 10.1016/0014-5793(72)80300-4. [DOI] [PubMed] [Google Scholar]
- 2.Jain M.K., White H.B., III . Long-range order in biomembranes. In: Rodolfo P., David K., editors. Advances in Lipid Research. Elsevier; Amsterdam: 1977. [DOI] [PubMed] [Google Scholar]
- 3.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 4.Eggeling C., Ringemann C., Hell S.W. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 5.Honigmann A., Mueller V., Eggeling C. Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. Nat. Commun. 2014;5:5412. doi: 10.1038/ncomms6412. [DOI] [PubMed] [Google Scholar]
- 6.Honigmann A., Sadeghi S., Vink R. A lipid bound actin meshwork organizes liquid phase separation in model membranes. eLife. 2014;3:e01671. doi: 10.7554/eLife.01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez S.A., Tricerri M.A., Gratton E. Laurdan generalized polarization fluctuations measures membrane packing micro-heterogeneity in vivo. Proc. Natl. Acad. Sci. USA. 2012;109:7314–7319. doi: 10.1073/pnas.1118288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sezgin E., Gutmann T., Schwille P. Adaptive lipid packing and bioactivity in membrane domains. PLoS One. 2015;10:e0123930. doi: 10.1371/journal.pone.0123930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feigenson G.W. Phase boundaries and biological membranes. Annu. Rev. Biophys. Biomol. Struct. 2007;36:63–77. doi: 10.1146/annurev.biophys.36.040306.132721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veatch S.L., Keller S.L. Seeing spots: complex phase behavior in simple membranes. Biochim. Biophys. Acta. 2005;1746:172–185. doi: 10.1016/j.bbamcr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Levental I., Grzybek M., Simons K. Raft domains of variable properties and compositions in plasma membrane vesicles. Proc. Natl. Acad. Sci. USA. 2011;108:11411–11416. doi: 10.1073/pnas.1105996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vist M.R., Davis J.H. Phase equilibria of cholesterol/dipalmitoylphosphatidylcholine mixtures: 2H nuclear magnetic resonance and differential scanning calorimetry. Biochemistry. 1990;29:451–464. doi: 10.1021/bi00454a021. [DOI] [PubMed] [Google Scholar]
- 13.Ipsen J.H., Mouritsen O.G., Zuckermann M.J. Theory of thermal anomalies in the specific heat of lipid bilayers containing cholesterol. Biophys. J. 1989;56:661–667. doi: 10.1016/S0006-3495(89)82713-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis J.H., Clair J.J., Juhasz J. Phase equilibria in DOPC/DPPC-d62/cholesterol mixtures. Biophys. J. 2009;96:521–539. doi: 10.1016/j.bpj.2008.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veatch S.L., Soubias O., Gawrisch K. Critical fluctuations in domain-forming lipid mixtures. Proc. Natl. Acad. Sci. USA. 2007;104:17650–17655. doi: 10.1073/pnas.0703513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartels T., Lankalapalli R.S., Brown M.F. Raftlike mixtures of sphingomyelin and cholesterol investigated by solid-state 2H NMR spectroscopy. J. Am. Chem. Soc. 2008;130:14521–14532. doi: 10.1021/ja801789t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bunge A., Müller P., Huster D. Characterization of the ternary mixture of sphingomyelin, POPC, and cholesterol: support for an inhomogeneous lipid distribution at high temperatures. Biophys. J. 2008;94:2680–2690. doi: 10.1529/biophysj.107.112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heberle F.A., Petruzielo R.S., Katsaras J. Bilayer thickness mismatch controls domain size in model membranes. J. Am. Chem. Soc. 2013;135:6853–6859. doi: 10.1021/ja3113615. [DOI] [PubMed] [Google Scholar]
- 19.Mills T.T., Tristram-Nagle S., Feigenson G.W. Liquid-liquid domains in bilayers detected by wide angle X-ray scattering. Biophys. J. 2008;95:682–690. doi: 10.1529/biophysj.107.127910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J., Wu J., Feigenson G.W. Phase studies of model biomembranes: complex behavior of DSPC/DOPC/cholesterol. Biochim. Biophys. Acta. 2007;1768:2764–2776. doi: 10.1016/j.bbamem.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veatch S.L., Keller S.L. Miscibility phase diagrams of giant vesicles containing sphingomyelin. Phys. Rev. Lett. 2005;94:148101. doi: 10.1103/PhysRevLett.94.148101. [DOI] [PubMed] [Google Scholar]
- 22.Marsh D. Liquid-ordered phases induced by cholesterol: a compendium of binary phase diagrams. Biochim. Biophys. Acta. 2010;1798:688–699. doi: 10.1016/j.bbamem.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 23.Marsh D. Cholesterol-induced fluid membrane domains: a compendium of lipid-raft ternary phase diagrams. Biochim. Biophys. Acta. 2009;1788:2114–2123. doi: 10.1016/j.bbamem.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Heberle F.A., Wu J., Feigenson G.W. Comparison of three ternary lipid bilayer mixtures: FRET and ESR reveal nanodomains. Biophys. J. 2010;99:3309–3318. doi: 10.1016/j.bpj.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petruzielo R.S., Heberle F.A., Feigenson G.W. Phase behavior and domain size in sphingomyelin-containing lipid bilayers. Biochim. Biophys. Acta. 2013;1828:1302–1313. doi: 10.1016/j.bbamem.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veatch S.L., Polozov I.V., Keller S.L. Liquid domains in vesicles investigated by NMR and fluorescence microscopy. Biophys. J. 2004;86:2910–2922. doi: 10.1016/S0006-3495(04)74342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerl M.J., Sampaio J.L., Simons K. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J. Cell Biol. 2012;196:213–221. doi: 10.1083/jcb.201108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Almeida R.F.M., Fedorov A., Prieto M. Sphingomyelin/phosphatidylcholine/cholesterol phase diagram: boundaries and composition of lipid rafts. Biophys. J. 2003;85:2406–2416. doi: 10.1016/s0006-3495(03)74664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frazier M.L., Wright J.R., Almeida P.F.F. Investigation of domain formation in sphingomyelin/cholesterol/POPC mixtures by fluorescence resonance energy transfer and Monte Carlo simulations. Biophys. J. 2007;92:2422–2433. doi: 10.1529/biophysj.106.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sodt A.J., Sandar M.L., Lyman E. The molecular structure of the liquid-ordered phase of lipid bilayers. J. Am. Chem. Soc. 2014;136:725–732. doi: 10.1021/ja4105667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baoukina S., Mendez-Villuendas E., Tieleman D.P. Computer simulations of the phase separation in model membranes. Faraday Discuss. 2013;161:63–75. doi: 10.1039/c2fd20117h. discussion 113–150. [DOI] [PubMed] [Google Scholar]
- 32.Risselada H.J., Marrink S.J. The molecular face of lipid rafts in model membranes. Proc. Natl. Acad. Sci. USA. 2008;105:17367–17372. doi: 10.1073/pnas.0807527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aittoniemi J., Niemelä P.S., Vattulainen I. Insight into the putative specific interactions between cholesterol, sphingomyelin, and palmitoyl-oleoyl phosphatidylcholine. Biophys. J. 2007;92:1125–1137. doi: 10.1529/biophysj.106.088427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandit S.A., Vasudevan S., Scott H.L. Sphingomyelin-cholesterol domains in phospholipid membranes: atomistic simulation. Biophys. J. 2004;87:1092–1100. doi: 10.1529/biophysj.104.041939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z., Bhide S.Y., Berkowitz M.L. Molecular dynamics simulations of bilayers containing mixtures of sphingomyelin with cholesterol and phosphatidylcholine with cholesterol. J. Phys. Chem. B. 2007;111:12888–12897. doi: 10.1021/jp074037i. [DOI] [PubMed] [Google Scholar]
- 36.Venable R.M., Sodt A.J., Klauda J.B. CHARMM all-atom additive force field for sphingomyelin: elucidation of hydrogen bonding and of positive curvature. Biophys. J. 2014;107:134–145. doi: 10.1016/j.bpj.2014.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yasuda T., Kinoshita M., Matsumori N. Detailed comparison of deuterium quadrupole profiles between sphingomyelin and phosphatidylcholine bilayers. Biophys. J. 2014;106:631–638. doi: 10.1016/j.bpj.2013.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw D.E., Maragakis P., Wriggers W. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 39.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 40.Wu E.L., Cheng X., Im W. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pitman M.C., Suits F., Feller S.E. Molecular-level organization of saturated and polyunsaturated fatty acids in a phosphatidylcholine bilayer containing cholesterol. Biochemistry. 2004;43:15318–15328. doi: 10.1021/bi048231w. [DOI] [PubMed] [Google Scholar]
- 43.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 44.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 45.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 46.Shan Y., Klepeis J.L., Shaw D.E. Gaussian split Ewald: a fast Ewald mesh method for molecular simulation. J. Chem. Phys. 2005;122:54101. doi: 10.1063/1.1839571. [DOI] [PubMed] [Google Scholar]
- 47.Krause M.R., Regen S.L. The structural role of cholesterol in cell membranes: from condensed bilayers to lipid rafts. Acc. Chem. Res. 2014;47:3512–3521. doi: 10.1021/ar500260t. [DOI] [PubMed] [Google Scholar]
- 48.Krisovitch S.M., Regen S.L. Nearest-neighbor recognition in phospholipid bilayers. Probing lateral organization at the molecular level. J. Am. Chem. Soc. 1991;113:8175–8177. [Google Scholar]
- 49.Almeida P.F.F. Thermodynamics of lipid interactions in complex bilayers. Biochim. Biophys. Acta. 2009;1788:72–85. doi: 10.1016/j.bbamem.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 50.Radhakrishnan A., McConnell H. Condensed complexes in vesicles containing cholesterol and phospholipids. Proc. Natl. Acad. Sci. USA. 2005;102:12662–12666. doi: 10.1073/pnas.0506043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radhakrishnan A., McConnell H.M. Condensed complexes of cholesterol and phospholipids. Biophys. J. 1999;77:1507–1517. doi: 10.1016/S0006-3495(99)76998-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugár I.P., Chong P.L.G. A statistical mechanical model of cholesterol/phospholipid mixtures: linking condensed complexes, superlattices, and the phase diagram. J. Am. Chem. Soc. 2012;134:1164–1171. doi: 10.1021/ja2092322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C., Krause M.R., Regen S.L. Push and pull forces in lipid raft formation: the push can be as important as the pull. J. Am. Chem. Soc. 2015;137:664–666. doi: 10.1021/ja5115437. [DOI] [PubMed] [Google Scholar]
- 54.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 55.Matsumori N., Yamaguchi T., Murata M. Orientation and order of the amide group of sphingomyelin in bilayers determined by solid-state NMR. Biophys. J. 2015;108:2816–2824. doi: 10.1016/j.bpj.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Armstrong C.L., Marquardt D., Rheinstädter M.C. The observation of highly ordered domains in membranes with cholesterol. PLoS One. 2013;8:e66162. doi: 10.1371/journal.pone.0066162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rheinstädter M.C., Mouritsen O.G. Small-scale structure in fluid cholesterol–lipid bilayers. Curr. Opin. Colloid Interface Sci. 2013;18:440–447. [Google Scholar]
- 58.Toppozini L., Meinhardt S., Rheinstädter M.C. Structure of cholesterol in lipid rafts. Phys. Rev. Lett. 2014;113:228101. doi: 10.1103/PhysRevLett.113.228101. [DOI] [PubMed] [Google Scholar]
- 59.Diaz-Rohrer B.B., Levental K.R., Levental I. Membrane raft association is a determinant of plasma membrane localization. Proc. Natl. Acad. Sci. USA. 2014;111:8500–8505. doi: 10.1073/pnas.1404582111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.