Abstract
Mitochondria are highly dynamic cell organelles. Continual cycles of fusion and fission play an important role in mitochondrial metabolism and cellular signaling. Previously, a novel mitochondrial morphology, the donut, was reported in cells after hypoxia-reoxygenation or osmotic pressure changes. However, the mechanism of donut formation remained elusive. Here, we obtained the distribution of donut diameters (D = 2R) and found that 95% are >0.8 μm. We also performed highly precise measurements of the mitochondrial tubule diameters using superresolution and electron microscopy. Then, we set up a model by calculating the mitochondrial bending energy and osmotic potential during donut formation. It shows that the bending energy is increased as the radius of curvature, R, gets smaller in the process of donut formation, especially for radii <0.4 μm, creating a barrier to donut formation. The calculations also show that osmotic potential energy release can balance the rising bending energy through volume expansion. Finally, we revealed the donut formation process in a Gibbs free-energy-dependent model combining calculations and measurements.
Introduction
Mitochondria are highly dynamic cell organelles whose morphology is regulated by fusion-fission cycles. Continuous fusion-fission dynamics are required for normal mitochondrial function and its role in cellular signaling (1,2). So far, several proteins have been reported to be involved in mitochondrial fusion-fission dynamics, including Mfn1 and Mfn2, controlling outer membrane fusion, Opa1, controlling inner membrane fusion, and Drp1, which is recruited by Mff to the outer membrane for fission (3). Elongation and/or fragmentation have been reported to occur under different physiological or pathological conditions (4). It has also long been known that mitochondria can undergo swelling and shrinkage (5).
In our previous work, we have reported another type of mitochondrial formation we termed donuts, which were observed in cells after hypoxia-reoxygenation and under osmotic pressure changes. Compared with swollen mitochondria, these ringlike mitochondria donuts are more able to recover to a linear shape when the stress disappears (6). Mitochondrial swelling has been observed in many tissues during reoxygenation/reperfusion after hypoxia (7–9). Mitochondrial swelling during reoxygenation at least partially results from opening of the permeability transition pore (10), but the K+ conductance of the mitochondrial inner membrane is an alternative mechanism for matrix volume changes (11). The activities of ATP-activated K+ channels and Ca2+-activated K+ channels have been reported to change during hypoxia-reoxygenation (12–15).
In this study, we measure the diameters of mitochondria at high precision using the newly developed fluorescence superresolution imaging technologies, stochastic optical reconstruction microscopy (STORM) and photoactivated localization microscopy (PALM) (16), in combination with transmission electron microscopy (TEM). Then, to investigate the physicochemical aspects of donut formation, we set up a free-energy model of donut formation by calculating mitochondrial bending energy and osmotic potential energy during donut formation. This model suggests that the increment of bending energy is the major barrier for donut formation. Meanwhile, the release of osmotic potential energy could promote the activation of donut formation through decrement of the total Gibbs free energy. The calculation also shows that formation of donuts of diameter <800 nm is inhibited by the robust increase in the bending-energy barrier at the last moments before fusion of the two ends, especially when viewed in terms of the bending energy per unit of membrane. This calculation closely aligns with our measurement of donut and mitochondrial diameters with high-precision fluorescence imaging.
Materials and Methods
Plasmids and cell transfection
The mtGFP plasmid was constructed from a commercial plasmid, pDsRed2-mito (Clontech, Mountain View, CA), by replacing DsRed2 with GFP. It codes a fusion of the GFP gene with the mitochondrial targeting sequence of COX VIII at the N-terminus. The mEOS2-Omp25 plasmid is constructed by fusion of an mEOS2 fluorescent protein with the localization sequence of the mitochondrial outer-membrane protein Omp25 (17,18). Transfection was performed by electroporation with 106 cells and 10 μg of each plasmid in 200 μL Opti-MEM. The cells were cultured on a 35 mm dish with a glass bottom (WPI, Sarasota, FL) and imaged by confocal microscopy 24 h after transfection. Cells were treated with carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) (5 μM) for 15–30 min. The donut diameter, mitochondrial diameter, and percent of mitochondria with a tail are quantified by the imaging methods as follows.
Live-cell microscopic imaging
Confocal microscopy imaging was performed with a Zeiss (Oberkochen, Germany) LSM710 inverted microscope. Experiments were performed with a 100× oil lens (Apo, NA 1.40) recording a 1024 × 1024 pixel image with a temperature and CO2 controller system. mtGFP was visualized in a time-lapse model using a 488 nm excitation laser.
Superresolution microscope imaging
Mouse embryonic fibroblasts (MEFs) were cultured in glass-bottomed chambers (Thermo, Waltham, MA) and stained with primary antibody against TOMM20 (Abcam, Cambridge, MA) and secondary antibody labeled with AlexaFluor 647 (Life Technology, Carlsbad, CA), as in a previous work (16). STORM imaging was performed with an N-STORM microscope (Nikon, Tokyo, Japan) with a 100× TRIF lens (Apo, NA 1.49) and 200 mW 647 nm lasers. The final images were then reconstructed from 100,000 frames of 256 × 256 pixel images captured with an EMCCD iXON897 camera (Andor, Belfast, Ireland) at the rate of 50 fps. Mitochondria diameters were measured by a parallel line tool marking the edge of the mitochondria with NIS-element software (Nikon). For PALM imaging, MEF cells were transfected with mEOS2-Omp25 plasmid 48 h before being fixed with 2% paraformaldehyde (Sigma, St. Louis, MO). Cell imaging was performed using N-STORM microscopy (Nikon) with a 561 nm laser (150 mW) followed by a weak 405 nm laser to transform the mEOS2 protein from green (this channel is dark without a 488 nm laser) to red. The images were captured as with STORM.
TEM imaging
MEFs were fixed with 3% glutaraldehyde, and the experiment was carried out as described previously (19).
Figure and data calculating
We used MATLAB (The MathWorks, Natick, MA) and Graphpad Prism (GraphPad Software, La Jolla, CA) for data calculations (see the Supporting Material).
Results and Discussion
Mitochondrial donut formation
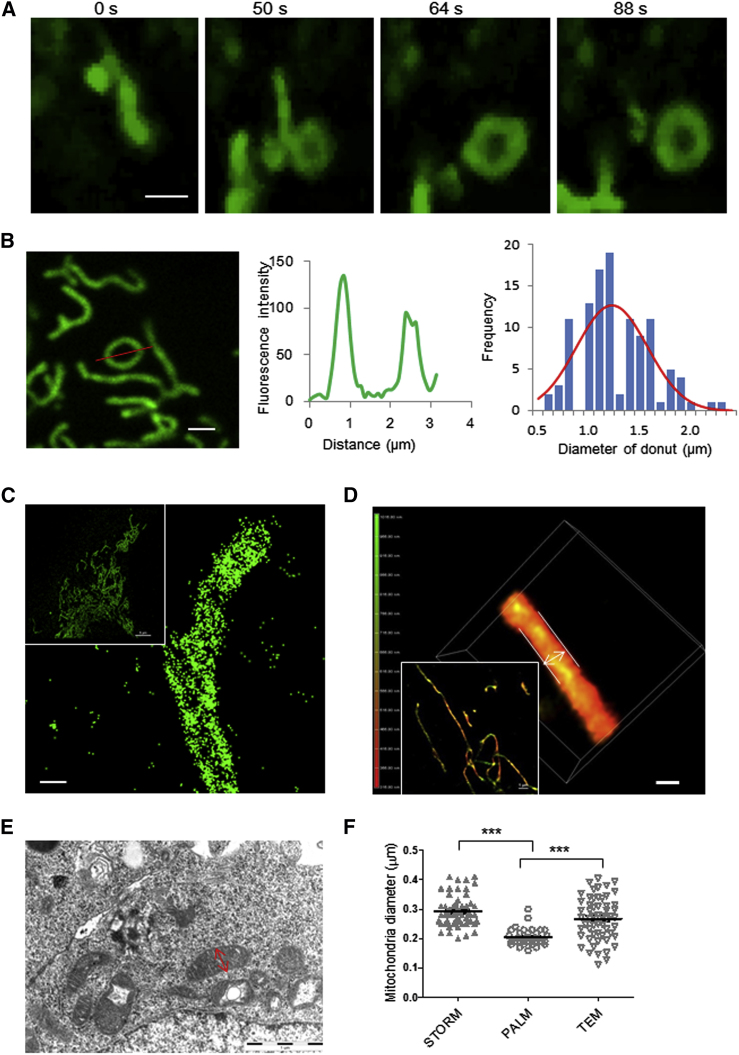
Mitochondria are tubular organelles that undergo dynamic changes in morphology, including shape changes, branching, fusion, and fission. Previously, we reported the occurrence of ring-shaped mitochondria (donuts) as determinate in the recovery of mitochondrial membrane potential after hypoxia-reoxygenation (6). Here, we describe a typical event of donut formation: one linear mitochondrion bends, self-fuses, and eventually forms a donut without a tail (Fig. 1A).
Figure 1.
Mitochondrial donut formation and diameter measurements. (A) A typical event of donut formation. The mitochondrion was tubular at 0 s, then bent to self-fusion at 50 s and finally formed a donut shape at 64 s. Scale bar, 1 μm. (B) Donut diameter was measured with images taken with a confocal microscope. The mitochondria were marked with mtGFP. The fluorescence profile along the red line (left) is shown in the middle image. The distribution of measured diameters is shown at right, and the red line shows the Gaussian fit (n = 111). Scale bar, 1 μm. (C) STORM imaging of mitochondria in MEF cells. Mitochondria were marked with TOMM20 antibody, and the image was reconstructed from 100,000 images. Scale bar, 0.2 μm. (D) 3D PALM imaging of mitochondria. The parallel lines were used to measure the diameter. The color (green-red) indicates the z-axis depth of the structure. Scale bar, 0.2 μm. (E) TEM imaging of mitochondria. The arrow shows the diameter. (F) Distributions of mitochondrial diameters measured by STORM (n = 55), PALM (n = 33), and TEM (n = 57). ∗∗∗p < 0.001 using the two-tailed t-test.
To better understand the physiological phenomenon of donut formation, we sought, first, to quantify their physical dimensions. The diameter of the resulting ring shapes (D = 2R) in MEFs treated with uncoupling FCCP were quantified using ImageJ by measuring the distances between the fluorescence peaks of a profile plot (Fig. 1B, left and middle). We found donut diameters in the range 0.5–2.4 μm with a nearly Gaussian distribution. The mean diameter was 1.17 μm, very near the 1.33 μm average reported in a different cell type (6). Ninety-five percent of donut diameters were between 0.8 and 1.8 μm, which we consider the typical range.
Mitochondrial diameter measurement
The diameter of the mitochondrial tubule (d = 2r) is also an important factor in donut formation. However, this has been difficult to reliably measure, as it is near the diffraction limit of optical microscopy resolution, in the range 200–500 nm according to previous reports (20–22). To obtain greater accuracy in the measurement of mitochondrial widths, we compared the results obtained by three methods of superresolution: fluorescence localization microscopy (STORM and PALM) and TEM.
Localization microscopy demands minimal manipulation of the sample, especially for PALM, in which one uses photoswitchable fluorescent protein tags, and the resulting resolution can be as little as 20–30 nm. For STORM imaging, mitochondria in MEFs were labeled by immunofluorescence detection of TOMM20, an outer mitochondrial membrane protein. Reconstructions were made from 100,000 frames, resulting in an effective resolution of 30 nm (Fig. 1C). For PALM, mitochondria were labeled by overexpression of a fusion construct of photoswitchable mEOS2 and the transmembrane domain of the outer membrane protein Omp25, and cells were imaged after fixation in 2% paraformaldehyde (Fig. 1D). Measurements of mitochondrial diameters from these reconstructions were compared to those from TEM (Fig. 1, E and F). The mean diameter measured by TEM is 260 ± 70 nm, whereas the STORM and PALM analyses gave values of 290 ± 50 nm and 210 ± 30 nm, respectively. Thus, from these measurements we conclude that the typical diameter of a mitochondrial tubule in MEFs is ∼200–300 nm. The difference between these measurements may be due to the methods themselves, such as the size of the antibodies in STORM and embedding and sectioning in TEM, and the fact that PALM gives the smallest radius may reflect the precision of this method due to no torsions.
Mitochondrial donut modeling
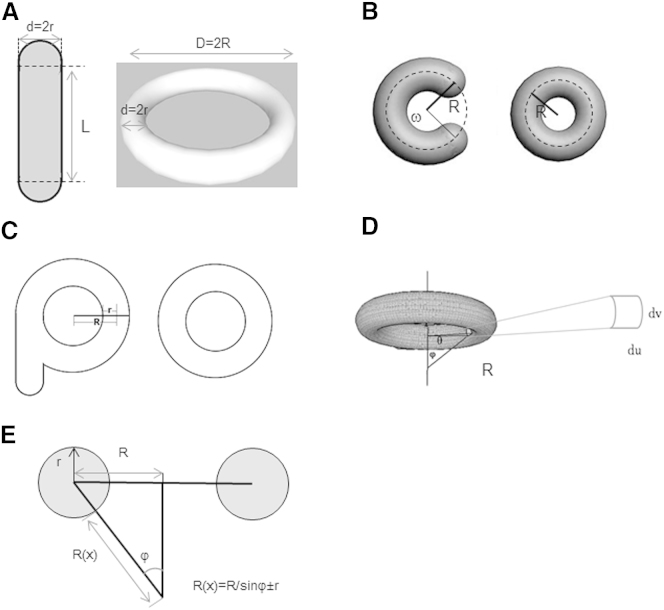
The morphology of mitochondria was reconstructed with 3D-deconvoluted images, and the caps of the cylinder mitochondria were shown to be close to hemispheric (23). We proposed a model of donut formation wherein the fundamental state of the mitochondrion is a linear rod-like cylinder with a hemispheric cap on each end (Fig. 2A). The length of the cylinder is L, the radius of both the cylinder and the hemispheres is r, and the radius of the donut is R. We defined the unclosed donut as an arc in angle ω with radius Rω = L/ω, ω∈(0, 2π), whereas the cylinder is an unclosed donut with Rω equal to ∞. Thus, the process of donut formation involves the reduction of the donut radius, Rω (Fig. 2B).There are two kinds of donuts, those with a tail and those without a tail (Fig. 2C). We defined the donuts with a tail as model A and those without a tail as model B.
Figure 2.
Geometric models of donuts. (A) A 2D model of a mitochondrion in the fundamental state (rod) and as a donut without a tail. (B) Geometric description of the process of donut formation. (C) Model of a donut with a tail. (D) A 3D model donut. (E) Donut longitudinal section.
The free energy of donut formation
Donut formation happens under particular conditions, as osmotic pressure is increased by K+ influx or some other stress conditions (6). There must be some energy driving this process, so we modeled the donut and calculated the free-energy increment of this process.
There are three kinds of structural energy that can be defined in donut formation: the surface energy (Es), the bending energy (Eb), and the osmotic energy (Ep). Thus, the total free energy of a closed lipid vesicle is
| (1) |
Surface energy (Es) can be defined as
The surface area of the mitochondrial membrane was reported to be invariant during donut formation after FCCP treatment or in swelling, as measured by 3D reconstructions of deconvolved confocal images (22,23). Donut formation and swelling are similar processes in that both happen in a relatively short time frame of several minutes. Es depends only on the surface area; therefore, we conclude that the change in surface energy is zero during donut formation (ΔEs = 0). FCCP is a weak lipid-soluble acid with the role of protein-independent uncoupling, which could specifically increase the permeability of H+ in mitochondria inner membranes (24). In our previous study, FCCP-induced donut formation is prevented by mitochondrial permeability transition pore (PTP) inhibitor, indicating the dependence of donut formation on PTP opening (6). There is also a side effect of opening the mitochondrial ATP-dependent potassium channel of the inner membrane in low-dose FCCP treatment (23,25,26) or reactive oxygen species level in long-time treatment (26). In our system, FCCP treatment takes place within 30 min, and the composition of mitochondrial outer-membrane proteins should have little chance to change. We have shown that donut formation is triggered by opening of the PTP or K+ channels, which in turn causes mitochondrial osmotic pressure change (4). FCCP treatment has the potential to influence some channel conductance in the mitochondrial inner membrane, but this occurs in the inner system of our object and thus should have no direct effect on the osmotic pressure of whole mitochondria.
Bending energy
A theoretical description of the free energy of cell membranes was first proposed by Helfrichin (15,27). The bending energy of the membrane can be described by the equation
| (2) |
Here, C1 and C2 represent the curvature of the membrane, and C0 is the curvature of the membrane with a bilayer of asymmetric chemical composition or asymmetry of the microenvironment around the membrane. The constant Kb is the bending rigidity, which is dependent on the temperature, and Kg is the elastic modulus of the Gaussian curvature. Kb is equal to 10−19 J according to a previous report (27), whereas Kg is not easily obtained, as it varies with the components of the membrane, especially the proteins located in the membrane. However, it has been reported to be <0.8 × 10−19 J (28).
Before bending to form a donut, the mitochondrial membrane is in the fundamental state. The bending energy of the whole mitochondrion increases in either donut model, A or B, after it bends to form a donut. Specifically, in model A, the tail section of the donut has no morphological change from the fundamental state, so its bending energy remains unchanged and we can ignore the tail for the purposes of our calculation. We established above our coordinate system for donut formation and defined the unclosed donut as an arc of radius Rω with the angle ω (Rω > ) (Fig. 2C).
According to our calculations in Eqs. 26–29 in the Supporting Material, the bending energy of a tubular mitochondrion is
| (3) |
The bending energy of a forming donut of radius Rω is
| (4) |
Here, Rω > .
For a perfect donut, the bending energy is
| (5) |
The change of bending energy in the donut forming process is
| (6) |
The difference between the bending energy of a tubular mitochondrion and that of a perfect donut is
| (7) |
Osmotic energy
The inflow of K+ leads to an increase of the osmotic pressure on mitochondria, which results in donut formation (6). During the process of mitochondrial swelling, the osmotic potential energy gained from K+ inflow leads to increased volume of the mitochondria. Therefore, we use the volume work (W) to represent the change of the osmotic potential energy.
| (8) |
It is equal to the external work (W) in the process of donut formation.
In our model, mitochondria resist the constant outer pressure and expand their volume by ΔV. The constant pressure consists of two parts. The first is the cytoplasm osmotic pressure, P0, which is constant (the value of cell cytoplasm osmotic pressure is equal to 800 kPa (12)), and the second is the additional pressure, ΔP, produced by the curving membrane, which is equal to 2λ/r. Here, the surface tension (λ) of mitochondria is 10−4 N/m, as reported previously (29,30). The surface area and volume change of donut formation is calculated in detail in the Supporting Material (Eqs. 14–21). According to the change of the volume, the osmotic potential can be calculated as
| (9) |
In model A, the tailed donut, the osmotic potential energy change is
| (10) |
For model B, the decrease of osmotic potential energy of the donut without a tail is
| (11) |
The change of the osmotic potential energy, ΔEp, is determined by r, but not by R:
| (12) |
where k = 1 in model A and k = 2 in model B.
The change of total free energy
As described in Eq. 1, the change of total energy is the sum of the changes of the surface, bending, and osmotic energies:
The mitochondrial diameter is a fundamental characteristic of the organelle, and is not changed during donut formation. The relationship of total free energy, ΔG, to donut diameter is as follows for mitochondria with defined r:
| (13) |
where k = 1 in model A and k = 2 in model B.
Bending energy, ΔEb, is the major barrier to donut formation
There are two steps for transforming a tubular mitochondrion into a donut. First, it bends into an unclosed donut; second, the two ends fuse together to form a complete donut. There may be a donut with a tail, but we only consider the donut part at this moment, as there is no change of energy in the tail part, as discussed above.
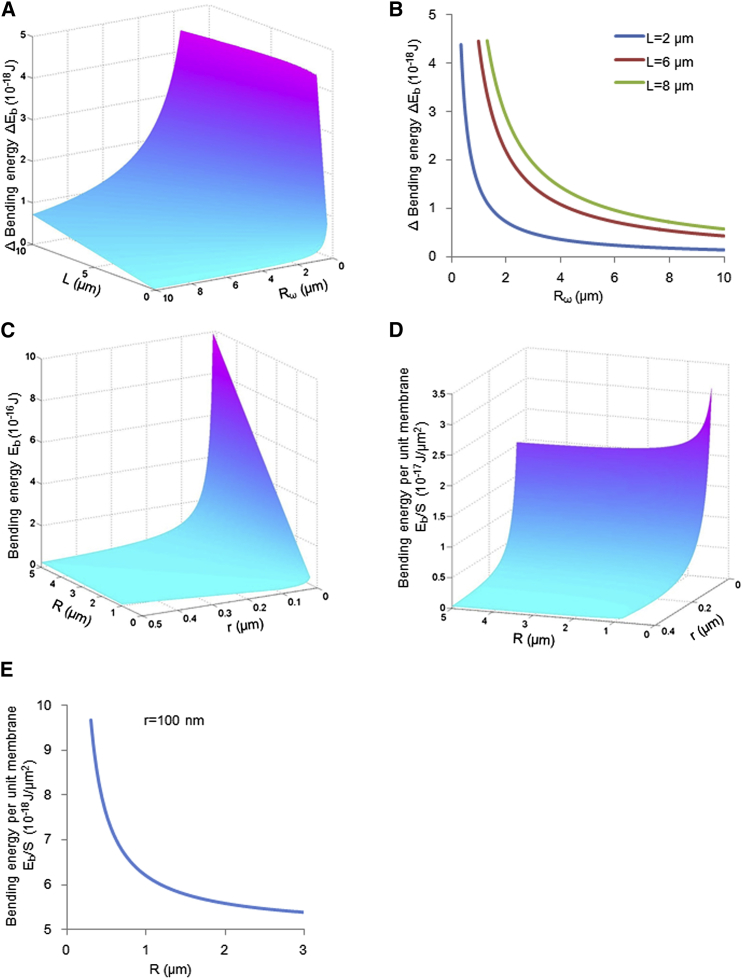
At the first step, ΔEp is equal to zero, with no change of volume. Thus, the change of free energy, ΔG, is equal to ΔEb, which was described in Eq. 4 and is plotted as a function of Rω and L in Fig. 3, A and B, with a defined mitochondrial radius of r = 100 nm. The bending energy naturally increases as the radius, R, decreases (i.e., the mitochondrion becomes more tightly bent), and it reaches the peak just before the fusion of the two ends. The bending energy increases suddenly in the last part of bending before the fusion event, which makes a barrier for donut formation, especially for short mitochondria forming with a smaller radius R (Fig. 3B).
Figure 3.
The change in bending energy during donut formation. (A) The change of bending energy, ΔEb, as a function of Rω and L in the ideal donut-formation process described in Fig. 2B under the condition r = 100 nm. (B) The change of bending energy, ΔEb, during the process of donut formation (as radius Rω gets smaller) for three typical-lengths, L. (C) The bending energy, Eb, as a function of R and r in different-sized donut mitochondria. (D) The bending energy per unit membrane, Eb/S, as a function of R and r. (E) The relationship between Eb/S and R at fixed r = 100 nm.
Meanwhile, at the fusion step, a large amount of potential energy will be released due to the release of the osmotic pressure (Eq. 12). However, it could also be visualized that the longer the mitochondrion taking part in donut formation, the more the bending energy increases, but that may be because the comparison is among different-sized mitochondria.
Donut radius, R, is a key determining factor for the bending-energy barrier in donut formation
Bending energy is the major barrier to donut formation, and the size of mitochondria may be a key factor in determining the bending energy of donut formation. There is a sudden increase of ΔEb as Rω approaches its maximum in the final stage of donut bending, especially for a donut with radius R ∼ 0.5 μm (Fig. 3B). Thus, the donut radius, R, may be a determining factor in donut formation.
To compare the donut radius, R, and the bending energy at the last moment, we plot the bending energy as a function of the donut radius, R, and the mitochondrial radius, r. It is obvious that bending energy is positively correlative with both R and r (Fig. 3C). However, when instead viewed in terms of the increment of bending energy per membrane unit (Eb/S) (Fig. 3D), it is clear that much more energy is needed to form a smaller donut on a per-membrane basis, especially for a donut with radius R < 400 nm when r is 100 nm (Fig. 3E).
The change of total free energy, ΔG, during donut formation
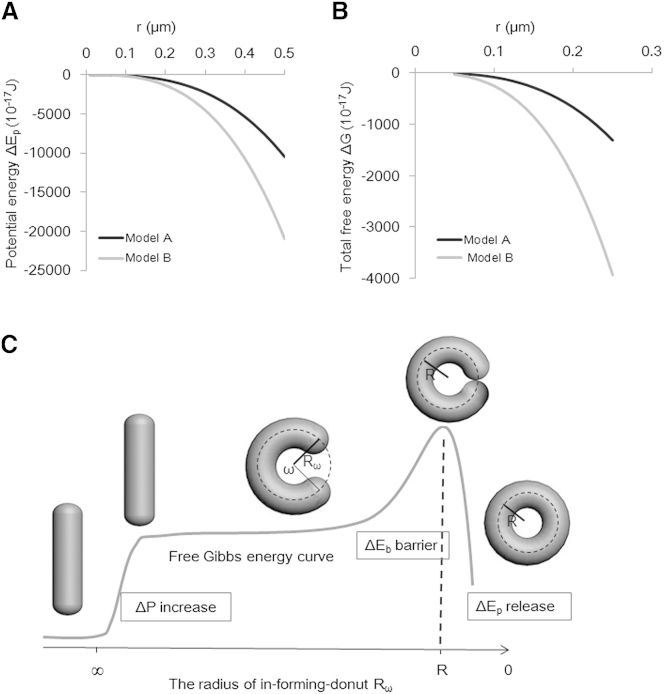
The overall process of donut formation should be energetically favorable in cases where the bending energy barrier is surmountable. We supposed that the processes of K+ influx and water flow in donut formation are carried out simultaneously, and that the expanding of the mitochondria could be regarded as volume change, ΔV, under constant pressure, P. The release of potential energy is equal to the volume work, as described in Eq. 12. The value of cell cytoplasm osmotic pressure is equal to 800 kPa (12). Therefore, we set P0 at 800 kPa and get the released potential energy (ΔEp = W) in donut formation as a function of the mitochondrial radius, r (Fig. 4A).
Figure 4.
Potential energy drives donut formation. (A) The relationship between potential energy, ΔEp (equal to external work, W), and r in the two models of donut formation. (B) The change of total free energy, ΔG, and r in donut formation at the condition R = 500 nm. (C) The Gibbs energy model of a donut-formation event.
Obviously, the released potential energy is much more than the increment of bending energy, ΔEb (Fig. 4B). Thus, donut formation ultimately reduces the Gibbs free energy of the system, which helps explain their existence (Fig. 4B). We can also conclude from Eq. 7 that the formation of donuts without tails will release twofold the potential energy of formation for tailed donuts, which means that the perfect donuts (i.e., those without tails) are much more stable in theory. Indeed, our experimental data of donuts post-FCCP treatment show that nearly half have tails at the beginning, and later most change into perfect donuts (Fig. S1). Meanwhile, it is a time-course event. The bending-energy barrier is increased robustly before the last moment of donut formation (Fig. 3B). The fusion of the two ends occurs in the final moments near the peak of bending energy. Thus, few donuts were observed in a normal condition, without swelling as a source of potential energy. However, the combining of potential energy release still has important effects on promoting donut formation.
In summary, we constructed a Gibbs energy model of a donut-formation event as shown in Fig. 4C. The increment of osmotic potential in the stress condition initiates the donut formation. Then, the mitochondrion bends to an unclosed donut, in which process the bending energy increases gradually as the radius, Rω, decreases. Near completion of the donut shape, the bending energy increases steeply ahead of fusion of the two ends, which creates a barrier for donut completion that is especially high for small donuts with R 400 nm. After the fusion of the two ends, the donut mitochondrion releases energy totaling much more than that obtained in the bending process, leaving the organelle in a lower-energy state than when it was tubular with increased potential. From this state, it can recover to the tubular structure after the stress is eliminated (6).
Author Contributions
X.L., X.W., and Q.L. designed the research. X.L., X.W., Q.L., D.Z., W.F., and Y.Z. performed the research and analyzed the data. L.Y., Y.Z., and J.Q. contributed analyses. X.L., X.W., Q.L., and D.Z. wrote the article.
Acknowledgments
This work was financially supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01020108), the Ministry of Science and Technology 973 program (2013CB967403 and 2012CB721105), the Ministry of Science and Technology 863 Program (2012AA02A708), the National Natural Science Foundation projects of China (31271527), the Guangzhou Science and Technology Program (2014Y2-00161), the Guangdong Natural Science Foundation for Distinguished Young Scientists (S20120011368), the Guangdong Natural Science Foundation for PhD Start-up (2014A030310071), the One Hundred Talents Project for Professor Xingguo Liu from the Chinese Academy of Sciences, and the Guangdong Province Science and Technology Innovation Young Talents Program (2014TQ01R559).
Editor: James Sneyd.
Footnotes
Qi Long and Danyun Zhao contributed equally to this work.
Supporting Materials and Methods, one figure, and MATLAB are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00774-2.
Contributor Information
Xin Wang, Email: wangxin80@gmail.com.
Xingguo Liu, Email: liu_xingguo@gibh.ac.cn.
Supporting Material
References
- 1.Chan D.C. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Twig G., Hyde B., Shirihai O.S. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim. Biophys. Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao J., Lendahl U., Nistér M. Regulation of mitochondrial dynamics: convergences and divergences between yeast and vertebrates. Cell. Mol. Life Sci. 2013;70:951–976. doi: 10.1007/s00018-012-1066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X., Su B., Zhu X. The role of abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J. Neurochem. 2009;109(Suppl 1):153–159. doi: 10.1111/j.1471-4159.2009.05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster K.A. Mitochondrial membrane permeabilization and cell death during myocardial infarction: roles of calcium and reactive oxygen species. Future Cardiol. 2012;8:863–884. doi: 10.2217/fca.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X., Hajnóczky G. Altered fusion dynamics underlie unique morphological changes in mitochondria during hypoxia-reoxygenation stress. Cell Death Differ. 2011;18:1561–1572. doi: 10.1038/cdd.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saris N.E., Eriksson K.O. Mitochondrial dysfunction in ischaemia-reperfusion. Acta Anaesthesiol. Scand. Suppl. 1995;107:171–176. doi: 10.1111/j.1399-6576.1995.tb04353.x. [DOI] [PubMed] [Google Scholar]
- 8.Weinberg J.M., Venkatachalam M.A., Nissim I. Mitochondrial dysfunction during hypoxia/reoxygenation and its correction by anaerobic metabolism of citric acid cycle intermediates. Proc. Natl. Acad. Sci. USA. 2000;97:2826–2831. doi: 10.1073/pnas.97.6.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoppel C.L., Tandler B., Riva A. Dynamic organization of mitochondria in human heart and in myocardial disease. Int. J. Biochem. Cell Biol. 2009;41:1949–1956. doi: 10.1016/j.biocel.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Lisa F., Canton M., Bernardi P. Mitochondria and cardioprotection. Heart Fail. Rev. 2007;12:249–260. doi: 10.1007/s10741-007-9028-z. [DOI] [PubMed] [Google Scholar]
- 11.Kaasik A., Safiulina D., Veksler V. Regulation of mitochondrial matrix volume. Am. J. Physiol. Cell Physiol. 2007;292:C157–C163. doi: 10.1152/ajpcell.00272.2006. [DOI] [PubMed] [Google Scholar]
- 12.Xu W., Liu Y., O’Rourke B. Cytoprotective role of Ca2+- activated K+ channels in the cardiac inner mitochondrial membrane. Science. 2002;298:1029–1033. doi: 10.1126/science.1074360. [DOI] [PubMed] [Google Scholar]
- 13.Murphy E., Steenbergen C. Mechanisms underlying acute protection from cardiac ischemia-reperfusion injury. Physiol. Rev. 2008;88:581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juhaszova M., Zorov D.B., Sollott S.J. Role of glycogen synthase kinase-3β in cardioprotection. Circ. Res. 2009;104:1240–1252. doi: 10.1161/CIRCRESAHA.109.197996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng Y., Gu X.Q., Siemen D. Hypoxia increases activity of the BK-channel in the inner mitochondrial membrane and reduces activity of the permeability transition pore. Cell. Physiol. Biochem. 2008;22:127–136. doi: 10.1159/000149790. [DOI] [PubMed] [Google Scholar]
- 16.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKinney S.A., Murphy C.S., Looger L.L. A bright and photostable photoconvertible fluorescent protein. Nat. Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horie C., Suzuki H., Mihara K. Characterization of signal that directs C-tail-anchored proteins to mammalian mitochondrial outer membrane. Mol. Biol. Cell. 2002;13:1615–1625. doi: 10.1091/mbc.01-12-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W., Long Q., Liu X. Mitochondrial metabolism transition cooperates with nuclear reprogramming during induced pluripotent stem cell generation. Biochem. Biophys. Res. Commun. 2013;431:767–771. doi: 10.1016/j.bbrc.2012.12.148. [DOI] [PubMed] [Google Scholar]
- 20.Jakobs S., Stoldt S., Neumann D. Light microscopic analysis of mitochondrial heterogeneity in cell populations and within single cells. Adv. Biochem. Eng. Biotechnol. 2011;124:1–19. doi: 10.1007/10_2010_81. [DOI] [PubMed] [Google Scholar]
- 21.Frey T.G., Mannella C.A. The internal structure of mitochondria. Trends Biochem. Sci. 2000;25:319–324. doi: 10.1016/s0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- 22.Ponnuswamy A., Nulton J., Baljon A.R. Modeling tubular shapes in the inner mitochondrial membrane. Phys. Biol. 2005;2:73–79. doi: 10.1088/1478-3967/2/1/009. [DOI] [PubMed] [Google Scholar]
- 23.Safiulina D., Veksler V., Kaasik A. Loss of mitochondrial membrane potential is associated with increase in mitochondrial volume: physiological role in neurones. J. Cell. Physiol. 2006;206:347–353. doi: 10.1002/jcp.20476. [DOI] [PubMed] [Google Scholar]
- 24.Benz R., McLaughlin S. The molecular mechanism of action of the proton ionophore FCCP (carbonylcyanide p-trifluoromethoxyphenylhydrazone) Biophys. J. 1983;41:381–398. doi: 10.1016/S0006-3495(83)84449-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennan J.P., Southworth R., Shattock M.J. Mitochondrial uncoupling, with low concentration FCCP, induces ROS-dependent cardioprotection independent of KATP channel activation. Cardiovasc. Res. 2006;72:313–321. doi: 10.1016/j.cardiores.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 26.To M.S., Aromataris E.C., Rychkov G.Y. Mitochondrial uncoupler FCCP activates proton conductance but does not block store-operated Ca2+ current in liver cells. Arch. Biochem. Biophys. 2010;495:152–158. doi: 10.1016/j.abb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 28.Kozlov M.M., McMahon H.T., Chernomordik L.V. Protein-driven membrane stresses in fusion and fission. Trends Biochem. Sci. 2010;35:699–706. doi: 10.1016/j.tibs.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghochani M., Nulton J.D., Baljon A.R. Tensile forces and shape entropy explain observed crista structure in mitochondria. Biophys. J. 2010;99:3244–3254. doi: 10.1016/j.bpj.2010.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S.Q., Jiang C.S., Long M. Membrane deformability and membrane tension of single isolated mitochondria. Cell. Mol. Bioeng. 2008;1:67–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.