Abstract
The sensitivity and selectivity of surface plasmon resonance imaging (SPRI) biosensing with nucleic acid microarrays can be greatly enhanced by exploiting various nucleic acid ligases, nucleases, and polymerases that manipulate the surface-bound DNA and RNA. We describe here various examples from each of these different classes of surface enzyme chemistries that have been incorporated into novel detection strategies that either drastically enhance the sensitivity of or create uniquely selective methods for the SPRI biosensing of proteins and nucleic acids. A dual-element generator–detector microarray approach that couples a bioaffinity adsorption event on one microarray element to nanoparticle-enhanced SPRI measurements of nucleic acid hybridization adsorption on a different microarray element is used to quantitatively detect DNA, RNA, and proteins at femtomolar concentrations. Additionally, this dual-element format can be combined with the transcription and translation of RNA from surface-bound double-stranded DNA (dsDNA) templates for the on-chip multiplexed biosynthesis of aptamer and protein microarrays in a microfluidic format; these microarrays can be immediately used for real-time SPRI bioaffinity sensing measurements.
I. Introduction
The simultaneous detection of multiple DNA, RNA, and proteins by bioaffinity adsorption onto biopolymer microarrays has become one of the premier tools of modern biochemical, bioanalytical, and biomedical research.1−3 Moreover, the multiplexed microarray–microfluidic format also serves as a key method for the rapid and efficient sensing of various biomarkers for disease diagnostics. Applications of multiplexed biomarker detection in healthcare include protein biomarker screening for specific cancers,4 messenger RNA gene expression profiling,5 and antibody screening.6 These applications often require the ultrasensitive multiplexed detection of nucleic acids and protein biomarkers at extremely low (femtomolar) concentrations from total sample volumes in the microliter range.
Surface plasmon resonance imaging (SPRI) is an optical detection method7,8 that has been successfully used to quantitatively measure the multiplexed bioaffinity adsorption of proteins and nucleic acids onto biopolymer microarrays.9−12 Figure 1a displays a schematic diagram of the experimental setup for SPRI measurements of microarrays.13 SPRI directly detects the bioaffinity adsorption of proteins and nucleic acids upon binding onto a biofunctionalized gold thin film via changes in the local surface refractive index. SPRI differential reflectivity images of biopolymer microarrays attached to gold thin films have been used for multiplexed, real-time measurements of bioaffinity adsorption at nanomolar concentrations. SPRI measurements are easily integrated with both simple10,14 and complex15,16 microfluidic delivery methods, enabling rapid detection of target biomolecules at nanomolar concentrations in 100–200 s.17 An example of a 16-element SPRI microfluidic chamber is shown in Figure 1b.
Figure 1.
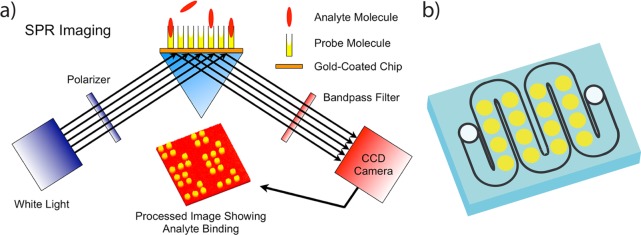
Schematic diagram of the SPRI experimental setup. (a) Bioaffinity adsorption onto a 45 nm gold thin film results in a local refractive index change. This leads to a difference in the monochromatic reflectivity image of a biopolymer microarray, which is obtained in the prism-coupled internal reflection geometry at the SPRI angle. (b) An example of a 16-element microfluidic SPRI chamber. The microfluidic channel has a cross-sectional area of 1 mm × 100 μm. The total cell volume is approximately 10 μL.
The sensitivity and selectivity of SPRI biosensing can be greatly enhanced to picomolar and even femtomolar concentrations through the action of various nucleic acid ligases, nucleases, and polymerases on nucleic acid probe microarrays.18−23 In our 2006 Langmuir Feature Article, we detailed a relevant mathematical model of these surface enzyme kinetics.24 In a number of subsequent papers, we have developed a variety of enzymatically amplified detection strategies for miRNA profiling, single nucleotide polymorphism (SNP) genotyping, and DNA, RNA, and protein biomarkers.
In this article, we discuss how ligases, nucleases, polymerases, and DNAzymes can all be used for the ultrasensitive detection of nucleic acid and protein targets in a microfluidic format with SPRI. For protein bioaffinity measurements, the additional incorporation of nucleic acid aptamers into the microarrays is also an essential component. Furthermore, DNA-functionalized gold, silica, and polymer nanoparticles are often incorporated into these detection strategies.22,23,25,26 A unique generator–detector dual microarray element approach enables the combination of both enzymatic and nanoparticle enhancement to improve the SPRI sensitivity down to femtomolar concentrations. Additionally, this dual microarray element format can be used for the on-chip templated biosynthesis of RNA and protein microarrays by surface transcription and translation in a microfluidic volume immediately prior to SPRI monitoring.
While this article focuses on the amplification of SPRI biosensing measurements with nucleic acid enzymes, there are also many other methods for the amplified detection of DNA and proteins. For example, the detection of DNA and proteins can be enhanced by nanoparticle assembly,27,28 horseradish peroxidase enzymatic amplification,29 rolling circle amplification,30 protein and DNA self-assembly,31 and coupling to plasmonic nanoparticles.28,32,33 In addition to SPRI, all of these amplification methods can be used with fluorescence, surface-enhanced Raman scattering (SERS), and electrochemical biosensors.2,28,33,34
II. Results and Discussion
A. Surface Ligation Chemistries for SPRI Microarray Biosensing
DNA and RNA ligases can be used to facilitate the biopolymer microarray fabrication process by enzymatically attaching oligonucleotides to surfaces.35−39 Two common surface ligation chemistries are shown in Figure 2. In Figure 2a, the enzyme T4 RNA ligase fastens the 3′ hydroxyl end of a single-stranded RNA (ssRNA) oligonucleotide onto a 5′ phosphate-terminated single-stranded DNA (ssDNA) oligonucleotide already bound to the surface. This surface enzymatic reaction is termed “untemplated” ligation because it involves a T4 DNA ligase or T4 RNA ligase that does not require a DNA or RNA template to form RNA microarrays.35
Figure 2.
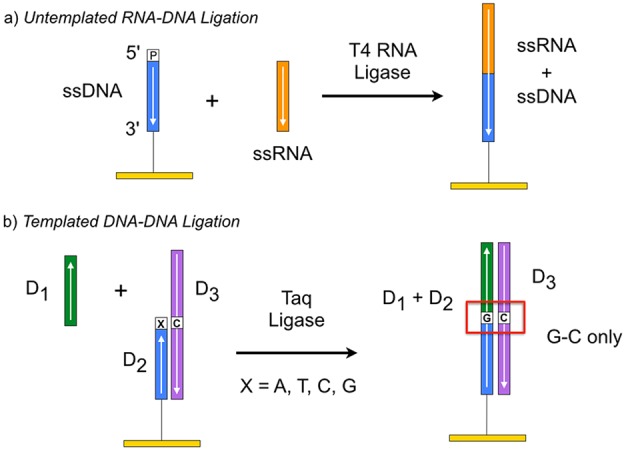
Two surface ligation chemistries. (a) Untemplated ligation utilizes T4 RNA ligase to link ssRNA with ssDNA bound on an array surface. (b) Templated ligation uses Taq ligase to conjugate surface-bound oligonucleotide D2 to oligonucleotide D1 from solution solely in the presence of complementary oligonucleotide D3 that forms a surface double-stranded DNA (dsDNA) complex. Ligation will occur only if the nucleotide on the 3′ terminus of D2 (labeled X) is complementary to the paired nucleotide in D3.
On the other hand, the enzyme Taq ligase shown in Figure 2b connects an ssDNA oligonucleotide (D1) from solution to a surface-bound ssDNA oligonucleotide (D2). This “templated” surface ligation chemistry requires the presence of a complementary ssDNA sequence (D3)—the template DNA. Such templated ligation occurs only when the final nucleotide on the 3′ end of D2 matches its complementary nucleotide in D3 (Figure 2b). This single base sensitivity has been exploited to create an SPRI-based method for SNP genotyping, specifically for the identification of a point mutation in the BRCA1 gene associated with breast cancer.37
Untemplated ligation of ssRNA to ssDNA, the process described in Figure 2a, has been a useful tool for RNA microarray formation. These recyclable RNA microarrays are built upon a monolayer of 3′-thiolated ssDNA previously spotted onto gold array elements. T4 DNA or RNA ligase plays a crucial role in the formation of phosphodiester bonds, linking the 3′-hydroxyl end on the ssRNA strands in solution to the 5′-phosphate-terminated, surface-bound ssDNA. This surface ligation reaction has a 90 ± 5% efficiency, as estimated from the SPRI signal of ssDNA hybridizing onto the RNA microarray.40 Such a ligation-aided attachment technique holds several advantages over other fabrication methods that involve amino-functionalized, alkyne-terminated, or biotin-modified ssRNA. It requires only unmodified ssRNA strands that directly attach to the ssDNA-functionalized microarray element. This avoids the costly chemical alteration of ssRNA strands, which may result in the loss of their integrity and functionality. The highly efficient ligation not only overcomes the need to introduce any additional chemical moieties or proteins (such as streptavidin) into the microarray element surface but also provides a DNA–RNA chimera that can be utilized in subsequent enzymatic reactions.
RNA aptamer microarrays, a subset of ssRNA microarrays created by untemplated ligation, are valuable tools for the study of nucleic acid–protein binding and for the fabrication of SPRI protein biosensors. An RNA aptamer is an ssRNA sequence with a unique binding affinity toward a specific ligand biomolecule; RNA aptamers are considered to be potential alternatives to antibodies for protein capture.41−44 The aptamers are typically identified by a combinatorial systematic evolution of ligands by an exponential enrichment (SELEX) screening process;42,43 quantitative SPRI measurements of the aptamer–protein binding strength can be used subsequently to select the best aptamer sequences for biosensor applications. For example, a surface-ligated RNA microarray was used to obtain quantitative SPRI measurements of the aptamer–protein binding strength (Langmuir adsorption coefficient) for a set of potential RNA aptamer sequences for blood disease biomarker protein factor IXa.36 Surface-ligated aptamer microarrays have also been used for the enzymatically amplified SPRI detection of two other biomarkers, human thrombin (hTh) and vascular endothelial growth factor (VEGF).38
Aptamer microarrays are an alternative to antibody microarrays. With high target specificity and binding strength, antibodies are commercially available and widely used as capture molecules for microarrays. However, antibody microarrays can sometimes be prone to antibody–antibody adsorption crosstalk and can denature if accidentally dried. In contrast, aptamer microarrays have less aptamer–aptamer crosstalk and maintain their functionality if dried and reconstituted.45,46 Additionally, the protein-bound aptamer microarrays can be selectively released by hybridization with complementary ssDNA, and the aptamer oligonucleotides can also include additional sequences for use in further enzymatic amplification.
Another application of the surface ssRNA–ssDNA ligation is for the detection of multiple miRNA sequences at picomolar concentrations. These short RNA strands are potential targets of interest due to their regulatory role in gene expression as they interact with messenger RNAs (mRNA).47 Profiling miRNA expression at picomolar to femtomolar concentrations can pinpoint various disease conditions, including cancers, cardiovascular diseases, and liver damage.48 One method of miRNA detection with SPRI biosensing is shown in Figure 3.23
Figure 3.
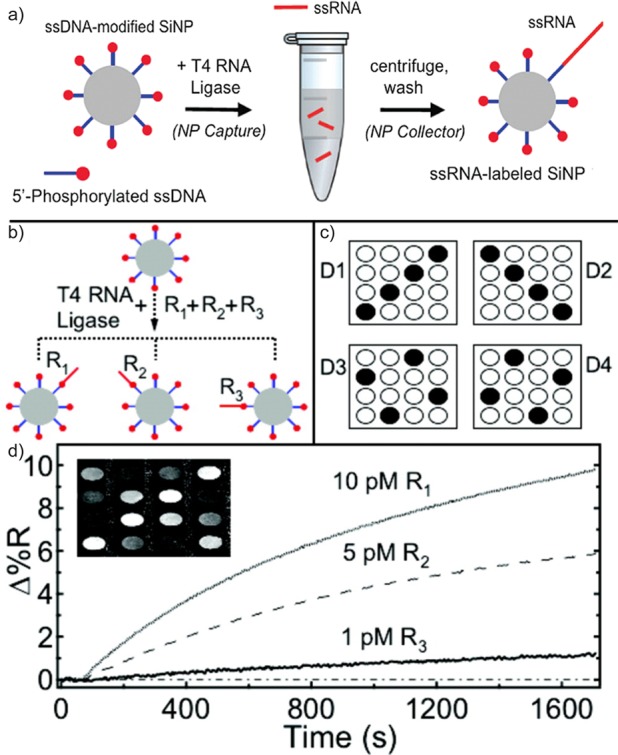
Detection of miRNA with surface ligation attachment chemistry on SiNPs. (a) Schematic of enzymatic ligation performed on the nanoparticles to prepare miRNA-modified SiNPs. (b) Diagram of multiplexed miRNA detection by these miRNA-functionalized SiNPs. (R1, R2, and R3 represent different miRNA sequences.) (c) Layout of the ssDNA microarray (D1, D2, and D3 as detector sequences and D4 as the control). (d) SPRI adsorption curves resulting from these SiNPs coated with R1, R2, and R3 miRNAs. The inset displays an SPR difference image.
In this SPRI biosensing measurement, the target single-stranded miRNA is captured by untemplated enzymatic ligation on ssDNA-functionalized silica nanoparticles (SiNPs) instead of a planar gold thin film microarray surface.23 The key miRNA ligation attachment step occurs in 10 to 100 μL of solution, eventually yielding miRNA targets labeled with SiNPs. This novel method of miRNA detection has several advantages. Rather than capturing the target miRNA on a planar microarray surface, utilizing SiNPs accelerates the attachment ligation chemistry due to the greater total surface area of the nanoparticles. Also, the radial diffusion of miRNA strands to the SiNPs requires less time than planar diffusion to a microarray surface. Once the target miRNA is securely ligated onto the SiNP surface, it is possible to centrifuge these miRNA–SiNPs in order to isolate, collect, and concentrate them for their subsequent SPRI measurements. Finally, the SPRI signal (reflectivity change) that is observed due to the hybridization adsorption of miRNA–SiNPs onto a microarray surface is greatly enhanced due to the large change in local refractive index caused by the presence of the SiNPs. This method can be used to detect multiple miRNA sequences at concentrations as low as 1 pM.
B. Surface Nuclease Reactions for SPRI Microarray Biosensing
Nucleases can remove oligonucleotides attached to microarray surfaces by the hydrolysis of phosphodiester bonds. This detachment of bound biomolecules results in a decrease in the SPRI signal (negative reflectivity change). Each nuclease exhibits a cleaving selectivity with specific directional and nucleic acid conformational preferences. For example, enzyme Exonuclease I (Exo I) digests ssDNA in the 3′ to 5′ direction but does not degrade dsDNA. Exo I has been employed in DNA computing on surfaces to eliminate ssDNA in the presence of dsDNA.49 In contrast, enzyme Exonuclease III (Exo III) hydrolyzes one strand of a dsDNA duplex in the 3′ to 5′ direction, starting at either a blunt end or a 5′ overhang (but not a 3′ overhang). As shown in Figure 4, this exonuclease activity can be used to manipulate DNA on surfaces.19
Figure 4.
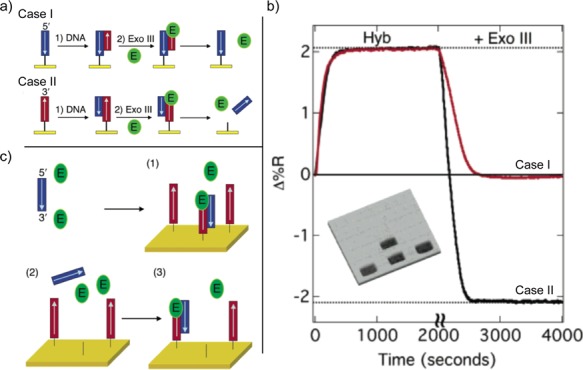
Exo III activity on surfaces. (a) Exo III activity on dsDNA attached via the 3′ and 5′ ends (labeled case I and case II, respectively). (b) SPRI image and reflectivity changes during Exo III activity for case I and case II. (c) Scheme for the amplified SPRI detection of ssDNA by Exo III activity on a DNA microarray element.
Figure 4a presents two possible scenarios (labeled as case I and case II) of Exo III activity on surface-bound dsDNA. In case I, after connecting the 3′ thiol-modified ssDNA strands to the gold surface, hybridization adsorption of complementary ssDNA sequences forms dsDNA on the surface. This leads to an increase in the SPRI signal, as shown in Figure 4b. Subsequent exposure of this microarray element to Exo III digests the hybridized complementary oligonucleotide, returning the SPRI signal to its original level. Meanwhile, the 3′ thiolated ssDNA remains intact. As for case II, with 5′ thiol-terminated oligonucleotides already linked to the array surface, dsDNA also forming via hybridization with the complementary sequences occurs. However, exposing the array element to Exo III has two consequences: as Exo III digests the surface-bound 5′ thiol-modified ssDNA, the intact complementary sequence departs from the element surface as well. The SPRI data reported in Figure 4b confirms this desorption, as the signal in case II displays twice the loss of that in case I.
Such asymmetry in the digestion of dsDNA sequences on the surface can be exploited to facilitate an amplified SPRI method for detecting DNA, as depicted in Figure 4c. This enables a minute amount of target ssDNA to trigger the destruction of multiple complementary 5′ thiolated ssDNA copies previously attached to the gold surface. After the target’s hybridization adsorption, Exo III selectively consumes the surface-bound ssDNA probe, which has its 3′ end exposed. The released target then readsorbs onto a neighboring probe, which is also digested by Exo III. This repeated cycle of target hybridization adsorption, ExoIII probe hydrolysis, and subsequent target release results in a large net amplified SPRI signal loss from a miniscule target concentration, which has been used to detect ssDNA at concentrations as low as 10 pM.19 Note that although this detection strategy is described as enzymatic amplifiied SPRI, an enlarged loss of SPRI signal is actually being measured.
Analogously, Figure 5 describes an enzymatically amplified DNA sensing method that employs the ribonuclease RNase H to react with an ssRNA microarray platform for detecting ssDNA targets at subpicomolar concentrations.40
Figure 5.
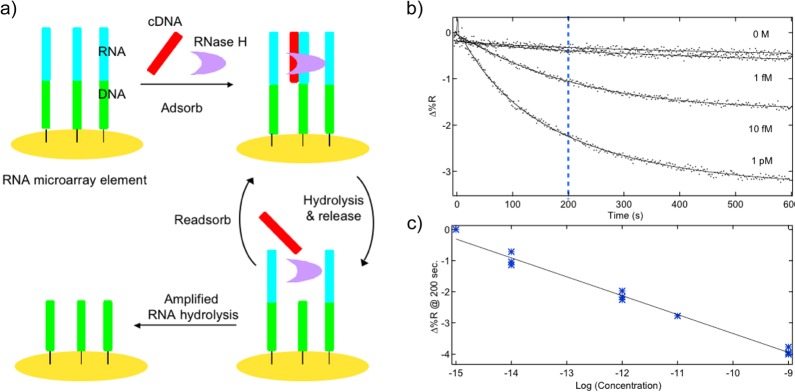
RNase H for ultrasensitive DNA microarray detection with SPRI. (a) RNase H amplification schematic. b) RNase H amplified SPRI detection of ssDNA solution from 0 M to 1 pM. (c) SPRI signal (Δ%R) at 200 s after target exposure vs log of the target ssDNA concentration.
RNase H specifically digests RNA oligonucleotides in an RNA–DNA heteroduplex, without the digestion of ssRNA, ssDNA, dsRNA, or dsRNA. As seen in the scheme in Figure 5a, after a target ssDNA oligonucleotide hybridizes with a surface-bound RNA oligonucleotide, the RNase H will digest the RNA in the RNA–DNA duplex and release the target ssDNA back into solution. The same strand of target ssDNA can then hybridize with another ssRNA on the microarray surface and induce another hydrolysis event. This repeatable cycle leads to enzymatic amplification, and only a minute amount—1 to 10 fM—of target ssDNA is needed to produce a detectable SPRI signal loss. The speed and sensitivity of this enzymatic enhancement technique have been attributed to both the enzyme’s great selectivity and RNase H being an endonuclease, which cuts the ssRNA all along the heteroduplex. This method is also capable of detecting longer, unpurified PCR products from genomic ssDNA samples.50 In this case, the ssRNA probes attached to the microarray surface are complementary to only a certain section of the target ssDNA; however, RNase H amplification still delivers a rapid and full SPRI signal loss. Meanwhile, this experimental design can also detect specific DNA sequences in human genomic samples that have not been amplified by PCR.18
C. Surface Polymerase Reactions for SPRI Microarray Biosensing
DNA and RNA polymerases are a powerful class of enzymes that can be used in multiple ways to create enzymatically amplified SPRI measurements. The word “amplification” indicates how these enzymes not only augment the SPRI signal but also synthesize copies of nucleic acid biopolymers from nucleotides, which is how the word is used in the more traditional biochemical sense. DNA and RNA polymerases have been used in a variety of amplification methods for ultrasensitive SPRI microarray biosensing measurements of nucleic acids.
The polynucleotide adenyltransferase poly(A) polymerase has been used to greatly amplify the nanoparticle-enhanced SPRI microarray detection of miRNA.21 Poly(A) polymerase adds adenosine nucleotides to the 3′ end of ssRNA in a continual fashion, engendering a poly(A) tail. Polyadenylation consumes adenosine triphosphate (ATP), releasing a diphosphate in exchange for each adenosine monophosphate added to the ssRNA. The addition of a poly(A) tail to mRNA in eukaryotes has multiple functions during translation.51 Figure 6 demonstrates how this poly(A) tail addition becomes a highly sensitive miRNA detection strategy when coupled to nanoparticle-enhanced SPRI.21 This joint process involves two steps to detect the hybridization adsorption of miRNA onto an SPRI microarray element. Poly(A) polymerase first generates adenosine units on the 3′ end of the surface-bound miRNA, which then provides a template for the subsequent hybridization adsorption of multiple 13 nm gold nanoparticles functionalized with T30 ssDNA (T30 AuNPs). This amplification method is an example of nanoparticle-enhanced SPRI, where the specific adsorption of one or more biofunctionalized nanoparticles is used to greatly increase the SPRI reflectivity response. Nanoparticle-enhanced SPRI was first used for SPRI by He et al. to detect DNA at picomolar concentrations.26 Subsequently, gold, silica, and polystyrene nanoparticles have all been used for the enhanced detection of nucleic acid and protein biomarkers with SPRI and other SPR methods.22,23,25,26
Figure 6.
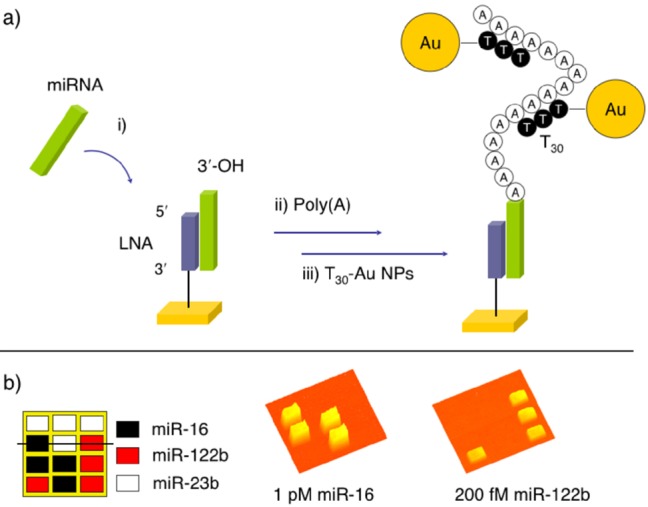
Surface poly(A)polymerase reactions for ultrasensitive miRNA detection. (a) Schematic of miRNA detection by polyadenylation and then coupling with nanoparticle-enhanced SPRI by the adsorption of T30 AuNPs to the poly(A) tail. (b) SPR difference images taken by drawing a line profile across the microarray image (shown as a solid line).
As seen in Figure 6, the enzymatic polyadenylation reaction requires that the 3′ terminus of the hybridized target miRNA be exposed to solution. For this reason, miRNA is captured with a shorter complementary locked nucleic acid (LNA) that is linked to the gold surface by the 3′ terminus. LNAs provide for stronger hybridization adsorption binding with the miRNA,52 and using a shorter LNA creates a 3′ overhang of six or more bases to further enhance the poly(A) polymerase’s initial binding efficiency. Figure 6b illustrates the microarray detection of miRNA at femtomolar concentrations by this combined approach of polyadenylation coupled to nanoparticle-enhanced SPRI. Through additional experiments, it was estimated that the poly(A) polymerase attaches approximately 250 adenosine units to the hybridized miRNA, creating multiple binding sites for the T30 AuNPs on each adsorbed RNA.21 The general concept of combining enzymatic amplification with nanoparticle enhancement has become one of the primary methods for ultrasensitive SPRI biosensing.
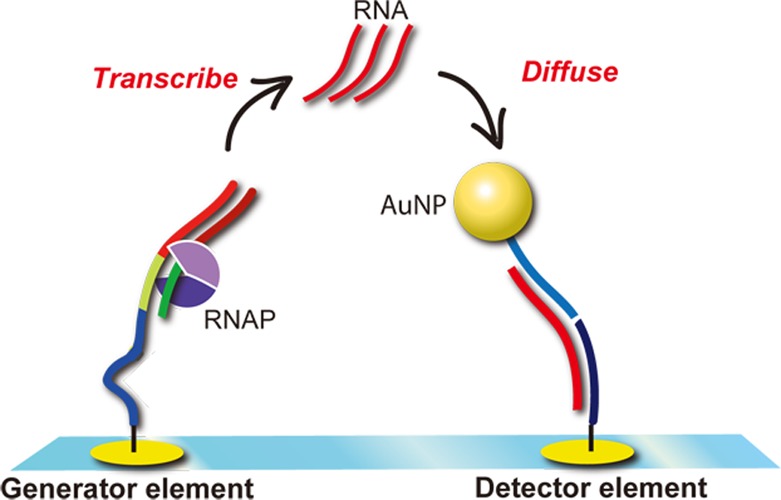
A second example of this combined signal amplification is the use of the enzyme T7 RNA polymerase with DNA-functionalized AuNPs to detect ssDNA. T7 RNA polymerase creates multiple ssRNA copies from a dsDNA template by first binding to a specific T7 promoter sequence and then synthesizing ssRNA copies in the 5′ to 3′ direction.53 This RNA transcription reaction is the initial step in gene expression. Figure 7 depicts how a surface RNA transcription reaction can be coupled to nanoparticle-enhanced SPRI to detect ssDNA.22
Figure 7.
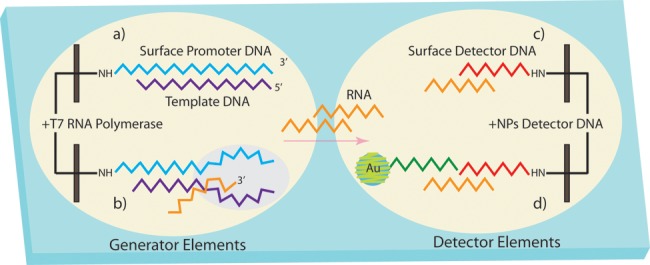
Diagram of dual microarray element strategy. (a) Template DNA hybridizes with the bound surface promoter DNA on the generator element. (b) Surface transcription occurs on the generator element. (c) After diffusing to the detector element, surface transcribed RNAs get captured by hybridizing with the surface detector DNA. (d) SPRI sensing determines the quantity of DNA from the measured sum of transcripts hybridized with DNA-decorated AuNPs.
The key design feature of Figure 7 is the use of two adjacent microarray elements on the same chip for RNA transcription and the nanoparticle-enhanced detection of the transcribed RNA; these elements are denoted as the generator and detector elements, respectively. The interelement distance is roughly 1 mm. Attached to the generator element are 5′ amino-modified ssDNA probes that include a T7 promoter sequence (denoted in Figure 7 as Surface Promoter DNA). This probe DNA hybridizes with a complementary target ssDNA to create a dsDNA template for the synthesis of numerous ssRNA copies via in vitro surface transcription (Figure 7b). The T7 RNA polymerase reaction occurs on the chip in a 25 μL volume of a reaction mixture that includes T7 RNA polymerase and ribonucleoside triphosphate building blocks. The reaction proceeded for 120 min, during which time the newly synthesized RNA transcripts diffuse to the detector elements (step (c) in Figure 7), where they are captured by a second ssDNA that is complementary to a sequence on the 3′ end of the ssRNA (shown as the Surface Detector DNA). Hybridization of the ssRNA transcript to the surface detector DNA still leaves half of the ssRNA sequence available for hybridization (Figure 7c). Exposing the microarray to a 4 nM solution of ssDNA-functionalized AuNP solution that is complementary to the other half of the transcribed RNA completes the last step in this scheme. The biofunctionalized AuNPs adsorb onto the detector elements, creating a real-time SPRI signal increase that can be used to quantify the amount of original target template ssDNA.
This dual amplification–detection method was used to detect ssDNA at concentrations as low as 1 fM. Additional experiments revealed that the T7 RNA polymerase reaction synthesized roughly 2000 ssRNA transcripts from each adsorbed target DNA and was limited by the quantity of reagents in the incubation chamber.22 A caveat of this method is that the target ssDNA sequence must be 3′-labeled with the T7 promoter sequence for the transcription reaction to occur.
As a final example of the use of surface polymerase reactions for enzymatically amplified SPRI, surface extension reactions using Klenow fragment DNA polymerase have been employed in conjunction with nanoparticle-enhanced SPRI detection for the detection target ssDNA at concentrations as low as 400 fM.54 This method provides the unique advantage of being able to detect relatively long, unknown ssDNA sequences that are labeled on each end with a known 20 base sequence, one to hybridize to the surface-bound probe ssDNA and the other to capture the ssDNA-functionalized AuNPs.
D. DNAzymes for SPRI Microarray Biosensing
A final set of enzymes that can be incorporated into enhanced SPRI biosensing measurements are ribozymes and DNAzymes (short for DNA-cleaving deoxyribozymes), which are single-stranded RNA or DNA oligonucleotides that can catalyze nucleic acid hydrolysis, ligation, or other biochemical reactions.55,56 We recently described the concept of DNAzyme footprinting for the detection of protein-aptamer bioaffinity adsorption and the amplified detection of protein biomarkers.57 This methodology is shown in Figures 8 and 9.
Figure 8.
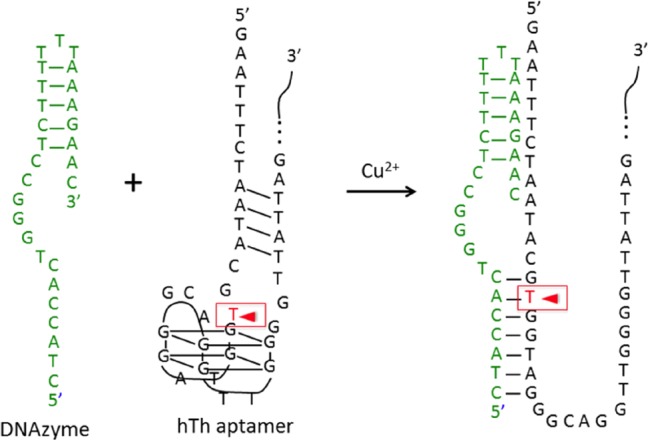
DNAzyme sequence that can cleave the thrombin aptamer sequence in the absence of thrombin–aptamer binding.
Figure 9.
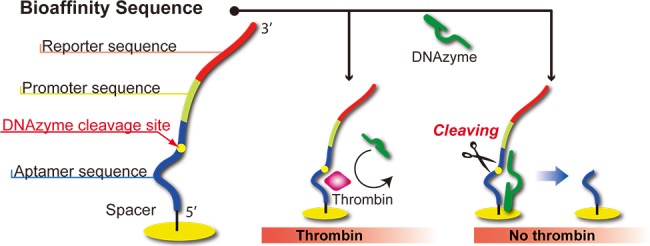
Schematic diagram of DNAzyme footprinting. The presence of thrombin that is adsorbed to an ssDNA monolayer can block the surface enzymatic endonuclease activity of the DNAzyme. After exposure to the DNAzyme, the number of reporter sequences remaining on the microarray element is equal to the number of adsorbed thrombin molecules.
Figure 8 depicts the two ssDNA sequences employed in our initial DNAzyme footprinting measurements.57 The ssDNA DNAzyme sequence in Figure 8 has been designed to hybridize to the ssDNA human thrombin (hTh) aptamer sequence, which normally folds into the G quartet structure shown in the figure. Once hybridized, the DNAzyme sequence will cleave the hTh aptamer at the thymine base location denoted in Figure 8 with the red arrow in the presence of Cu2+ ions. This DNAzyme sequence has been designed to have a weaker hybridizing strength (ΔG° = −43.1 kJ/mol) toward the hTh aptamer sequence than the thrombin’s binding strength (ΔG° = −51.8 kJ/mol) to the same aptamer. Thus, in the presence of thrombin, this DNAzyme will not bind to the aptamer, and the cleavage reaction will be blocked. This is the essence of DNAzyme footprinting, which conceptually is similar to the process of DNase footprinting for the detection of protein–dsDNA complexes.
Figure 9 shows how DNAzyme footprinting can be used in an aptamer microarray format to detect proteins.57 When a microarray element modified with an ssDNA monolayer of the thrombin aptamer sequence is first exposed to a thrombin solution and then to a DNAzyme solution, the DNAzyme will cleave all of the aptamer sequences that are not interacting with thrombin proteins. If the aptamer oligonucleotides attached to the microarray element also include a reporter sequence as shown in Figure 9, then the number of reporter sequences that remain on the surface after DNAzyme cleavage will be proportional to the number of adsorbed thrombin molecules. These reporter sequences begin with a T7 promoter sequence and therefore can be detected by the hybridization of a complementary ssDNA, followed by enzymatically amplified detection of the dsDNA surface duplex by the dual-element RNA polymerase and nanoparticle-enhanced SPRI detection strategy described in section C. This DNAzyme footprinting technique, when coupled with transcription–nanoparticle enhanced SPRI, can be used to detect thrombin at concentrations as low as 100 fM and serves as an example of how to pair surface enzymatic amplification with protein–aptamer interactions to develop novel SPRI protein biomarker sensing methods.
E. On-Chip Templated Biosynthesis of RNA and Protein Microarrays
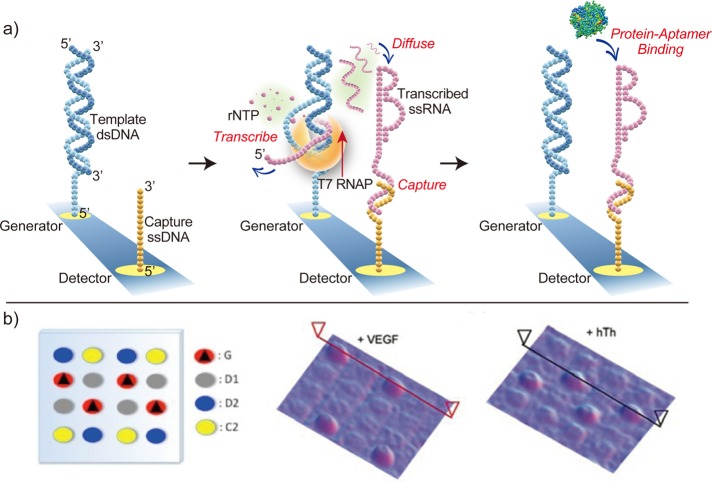
A final important application of surface RNA polymerase chemistry is the on-chip templated biosynthesis of RNA and protein microarrays. On-chip biosynthesis offers a clean, fast, and efficient microarray construction method that eliminates the need for postsynthesis purification and the use of a complicated spotting apparatus for array fabrication. For the on-chip biosynthesis of RNA aptamer microarrays, a particularly important advantage is that the on-chip biosynthesis minimizes any possible contamination or degradation of the transcribed RNA by confining the transcription reaction to just the microfluidic volume in contact with the microarray chip.58
Figure 10a displays the steps involved in constructing an RNA aptamer microarray in an on-chip biosynthesis format.58 Employing the dual microarray element generator–detector strategy detailed in section C, the dsDNA template now encodes an ssRNA aptamer sequence along with a unique capture sequence. Introducing the transcription mixture with T7 RNA polymerase and ribonucleoside triphosphate units triggers the production of aptamer transcripts. Diffusion delivers the aptamers from the generator site to the adjacent detector element, which holds a complementary ssDNA capture sequence that secures the aptamer by hybridization adsorption. Because each transcribed aptamer is paired with a unique capture tag, each aptamer sequence in the multiplexed on-chip transcription will bind only to a specific, designated detector element modified with the complementary ssDNA capture sequence. The resultant self-assembled RNA aptamer microarray can be immediately used in SPRI biosensing measurements of protein biomarkers. As a demonstration, this on-chip biosynthesis method was used to fabricate a two-component ssRNA aptamer microarray for the SPRI detection of vascular endothelial growth factor (VEGF) and human thrombin. Figure 10b presents SPRI reflectivity difference images of this microarray after its exposure to nanomolar solutions of the proteins.58
Figure 10.
On-chip synthesis of RNA microarrays. (a) Schematic of on-chip RNA microarray synthesis. RNA aptamers are transcribed from a dsDNA template on a generator element and then captured on adjacent detector elements. (b) SPRI difference images of a two-component aptamer microarray after 10 min of exposure to solutions containing 40 nM VEGF and 25 nM thrombin. The diagram of the microarray layout shows the four different components of the microarray: generator elements, detector elements for VEGF and thrombin, and control elements that are labeled G, D1, D2, and C2, respectively.
A second, more ambitious application of RNA polymerase surface transcription is the on-chip templated biosynthesis of protein microarrays for SPRI biosensing utilizing the process of in vitro transcription and translation (IVTT). Among the established on-chip protein microarray fabrication techniques,59,60 Figure 11 illustrates the first attempt at the on-chip templated biosynthesis of a protein microarray for the SPRI detection of antibodies.61
Figure 11.
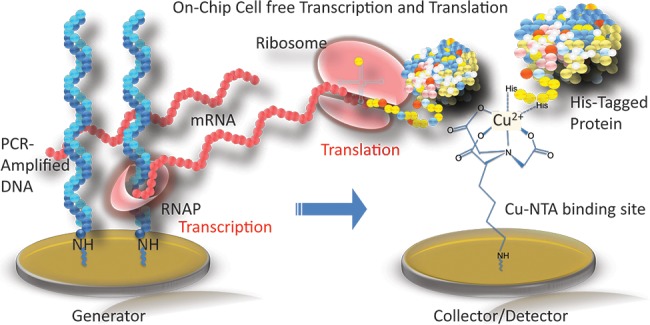
Schematic showing the synthesis of a protein microarray from a DNA microarray via surface transcription and translation. The dsDNA templates on the generator elements encode His6-tagged proteins that are captured by Cu(II)-nitrilotriacetic acid (NTA) attached to detector elements.
For on-chip protein biosynthesis, template dsDNA sequences were synthesized by PCR amplification from expression plasmids that included the T7 promoting and terminating regions, a ribosomal binding sector, a hexhistidine (His6) tag, and a code for the desired proteins. The forward primer for the PCR was 5′ amino modified in order to chemically attach the template dsDNA to the generator microarray elements.
After the dsDNA templates were attached to the generator elements, the microarray was exposed to 25 μL of the IVTT solution in a microfluidic format. The T7 RNA polymerase within the IVTT mix transcribed multiple mRNA strands that were subsequently translated into proteins by the ribosomes in the IVTT solution. To capture the His6-labeled proteins, the detector elements were modified with a Cu(II)-nitrilotriacetic acid monolayer. All synthesis steps occur isothermally within the microfluidic volume directly above the chip, creating a protein microarray from a dsDNA microarray. Unfortunately, this microarray fabrication technology is not self-assembling as there is only one surface attachment mechanism (NTA capture of the His6 tag), thus separate microfluidic channels need to be employed for each protein. In these initial experiments, this on-chip templated IVTT protein biosynthesis was used to create two-component protein microarray chips for real-time SPRI measurements of the specific adsorption of antibodies onto green fluorescent protein (GFP) and luciferase monolayers.61
III. Future Directions
We expect the use of isothermal surface enzyme reactions in SPRI biosensing for the enhanced selection, capture, and amplification of DNA, RNA, and protein biomarkers to continue to expand. The integration of metal, polymer, and magnetic nanoparticles into these ultrasensitive surface enzyme chemistries will also increase as the methods for nanoparticle biofunctionalization improve. Note that many of these enzymatic amplification methods can be easily adapted for many other traditional and newer biosensing techniques: fluorescence, SERS, electrochemical detection, quartz crystal microbalance with dissipation monitoring and surface acoustic wave devices, localized surface plasmon resonances including nanohole and nanoring arrays, high quality factor (high-Q) optical sensors, and surface plasmon resonance phase imaging.2,28,33,34,62−69
We also expect that the incorporation of aptamer bioaffinity processes into enzymatically amplified SPRI biosensing will become more prevalent. The DNAzyme footprinting methodology described in this article is just the first example of how aptamer–enzyme coupled methods can be used for ultrasensitive protein biomarker detection. There are a significant number of catalytic aptamers, such as ribozymes, riboswitches, and riboregulators, that can potentially be coupled to nanoparticle-enhanced SPRI for the ultrasensitive detection of both proteins and metabolites.70
Finally, we expect that multiplexed surface RNA polymerase transcription reactions and on-chip templated biosynthesis will eventually become the preferred methods of RNA and protein microarray fabrication. One powerful feature of these array fabrication methods is that they require only a miniscule quantity of transcribed RNA probes or synthesized proteins—approximately 10 to 50 femtomoles per element. Both the in situ transcription and self-assembly of RNA microarrays from DNA microarrays and the on-chip multiplexed IVTT to build protein microarrays from DNA template microarrays offer simple, highly efficient surface enzymatic approaches to the rapid implementation of multiplexed bioaffinity adsorption measurements in a microfluidic format, with SPRI or other detection methods.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM059622). R.M.C. thanks all of the students and postdocs who have contributed to these papers over the last 10 years.
Biographies

Jennifer B. Fasoli received her B.S. degree in chemical biology from the University of California, Berkeley in 2010. She is currently a Ph.D. candidate in chemistry at the University of California, Irvine. Her research interest is the fabrication of nucleic acid microarrays for SPRI biosensing.

Robert M. Corn is a professor of chemistry at the University of California, Irvine. Prof. Corn received a B.A. in chemistry from the University of California, San Diego in 1978 and a Ph.D. from the University of California, Berkeley in 1983 and was a postdoctoral scientist at IBM San Jose Research Laboratory from 1983 to 1984. In 1985, he joined the faculty in the Department of Chemistry at the University of Wisconsin—Madison, where he was a professor from 1985 to 2004. In 2004, Prof. Corn moved to the Department of Chemistry at the University of California, Irvine, where he also holds a joint appointment in the Department of Biomedical Engineering. With over 150 publications and patents and various awards, his research centers on the creation of new interfacial chemistries, spectroscopies, and biochemistries. Prof. Corn has dedicated his career to the development and application of surface-sensitive spectroscopies such as surface plasmon resonance imaging (SPRI), optical second-harmonic generation (SHG), and polarization-modulation FTIR spectroscopy for the characterization of solid–liquid and liquid–liquid interfaces. In addition to the development of multiplexed biosensing with microarrays, his research interests include the fabrication of nanostructured interfaces with unique optical and physical properties, the on-chip templated biosynthesis of nucleic acids and proteins, the synthesis of magnetic nanomaterials for high-frequency inductor applications, and the use of nucleic acids for DNA computing and other nonbiological applications.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Haab B. B.; Dunham M. J.; Brown P. O. Protein Microarrays for Highly Parallel Detection and Quantitation of Specific Proteins and Antibodies in Complex Solutions. Genome Biol. 2001, 2, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson K. L.; Steemers F. J.; Lee G.; Mendoza L. G.; Chee M. S. A Genome-Wide Scalable SNP Genotyping Assay Using Microarray Technology. Nat. Genet. 2005, 37, 549–554. [DOI] [PubMed] [Google Scholar]
- Stoughton R. B. Applications of DNA Microarrays in Biology. Annu. Rev. Biochem. 2005, 74, 53–82. [DOI] [PubMed] [Google Scholar]
- Wulfkuhle J. D.; Liotta L. A.; Petricoin E. F. Proteomic Applications for the Early Detection of Cancer. Nat. Rev. Cancer 2003, 3, 267–275. [DOI] [PubMed] [Google Scholar]
- Duggan D. J.; Bittner M.; Chen Y. D.; Meltzer P.; Trent J. M. Expression Profiling Using cDNA Microarrays. Nat. Genet. 1999, 21, 10–14. [DOI] [PubMed] [Google Scholar]
- Kingsmore S. F. Multiplexed Protein Measurement: Technologies and Applications of Protein and Antibody Arrays. Nat. Rev. Drug Discov 2006, 5, 310–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatman E.; Ash E. A. Surface-Plasmon Microscopy. Electron. Lett. 1987, 23, 1091–1092. [Google Scholar]
- Rothenhausler B.; Knoll W. Surface-Plasmon Microscopy. Nature 1988, 332, 615–617. [Google Scholar]
- Brockman J. M.; Nelson B. P.; Corn R. M. Surface Plasmon Resonance Imaging Measurements of Ultrathin Organic Films. Annu. Rev. Phys. Chem. 2000, 51, 41–63. [DOI] [PubMed] [Google Scholar]
- Kanda V.; Kariuki J. K.; Harrison D. J.; McDermott M. T. Label-Free Reading of Microarray-Based Immunoassays with Surface Plasmon Resonance Imaging. Anal. Chem. 2004, 76, 7257–7262. [DOI] [PubMed] [Google Scholar]
- Phillips K. S.; Cheng Q. Recent Advances in Surface Plasmon Resonance Based Techniques for Bioanalysis. Anal. Bioanal. Chem. 2007, 387, 1831–1840. [DOI] [PubMed] [Google Scholar]
- Spoto G.; Minunni M. Surface Plasmon Resonance Imaging: What Next?. J. Phys. Chem. Lett. 2012, 3, 2682–2691. [DOI] [PubMed] [Google Scholar]
- Blow N. Proteins and Proteomics: Life on the Surface. Nat. Methods 2009, 6, 389–392. [Google Scholar]
- Lee H. J.; Goodrich T. T.; Corn R. M. SPR Imaging Measurements of 1-D and 2-D DNA Microarrays Created from Microfluidic Channels on Gold Thin Films. Anal. Chem. 2001, 73, 5525–5531. [DOI] [PubMed] [Google Scholar]
- Ouellet E.; Lausted C.; Lin T.; Yang C. W. T.; Hood L.; Lagally E. T. Parallel Microfluidic Surface Plasmon Resonance Imaging Arrays. Lab Chip 2010, 10, 581–588. [DOI] [PubMed] [Google Scholar]
- Malic L.; Veres T.; Tabrizian M. Nanostructured Digital Microfluidics for Enhanced Surface Plasmon Resonance Imaging. Biosens. Bioelectron. 2011, 26, 2053–2059. [DOI] [PubMed] [Google Scholar]
- Wegner G. J.; Wark A. W.; Lee H. J.; Codner E.; Saeki T.; Fang S. P.; Corn R. M. Real-Time Surface Plasmon Resonance Imaging Measurements for the Multiplexed Determination of Protein Adsorption/Desorption Kinetics and Surface Enzymatic Reactions on Peptide Microarrays. Anal. Chem. 2004, 76, 5677–5684. [DOI] [PubMed] [Google Scholar]
- Goodrich T. T.; Lee H. J.; Corn R. M. Direct Detection of Genomic DNA by Enzymatically Amplified SPR Imaging Measurements of RNA Microarrays. J. Am. Chem. Soc. 2004, 126, 4086–4087. [DOI] [PubMed] [Google Scholar]
- Lee H. J.; Li Y.; Wark A. W.; Corn R. M. Enzymatically Amplified Surface Plasmon Resonance Imaging Detection of DNA by Exonuclease III Digestion of DNA Microarrays. Anal. Chem. 2005, 77, 5096–5100. [DOI] [PubMed] [Google Scholar]
- Lee H. J.; Wark A. W.; Goodrich T. T.; Fang S. P.; Corn R. M. Surface Enzyme Kinetics for Biopolymer Microarrays: A Combination of Langmuir and Michaelis-Menten Concepts. Langmuir 2005, 21, 4050–4057. [DOI] [PubMed] [Google Scholar]
- Fang S. P.; Lee H. J.; Wark A. W.; Corn R. M. Attomole Microarray Detection of microRNAs by Nanoparticle-Amplified SPR Imaging Measurements of Surface Polyadenylation Reactions. J. Am. Chem. Soc. 2006, 128, 14044–14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendroiu I. E.; Gifford L. K.; Luptak A.; Corn R. M. Ultrasensitive DNA Microarray Biosensing via in Situ RNA Transcription-Based Amplification and Nanoparticle-Enhanced SPR Imaging. J. Am. Chem. Soc. 2011, 133, 4271–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. J.; Chen Y. L.; Corn R. M. Ultrasensitive Microarray Detection of Short RNA Sequences with Enzymatically Modified Nanoparticles and Surface Plasmon Resonance Imaging Measurements. Anal. Chem. 2011, 83, 3897–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J.; Wark A. W.; Corn R. M. Creating Advanced Multifunctional Biosensors with Surface Enzymatic Transformations. Langmuir 2006, 22, 5241–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon L. A.; Musick M. D.; Natan M. J. Colloidal Au-Enhanced Surface Plasmon Resonance Immunosensing. Anal. Chem. 1998, 70, 5177–5183. [DOI] [PubMed] [Google Scholar]
- He L.; Musick M. D.; Nicewarner S. R.; Salinas F. G.; Benkovic S. J.; Natan M. J.; Keating C. D. Colloidal Au-Enhanced Surface Plasmon Resonance for Ultrasensitive Detection of DNA Hybridization. J. Am. Chem. Soc. 2000, 122, 9071–9077. [Google Scholar]
- Taton T. A.; Mirkin C. A.; Letsinger R. L. Scanometric DNA Array Detection with Nanoparticle Probes. Science 2000, 289, 1757–1760. [DOI] [PubMed] [Google Scholar]
- Peng H. I.; Miller B. L. Recent Advancements in Optical DNA Biosensors: Exploiting the Plasmonic Effects of Metal Nanoparticles. Analyst 2011, 136, 436–447. [DOI] [PubMed] [Google Scholar]
- Kim M. G.; Shin Y. B.; Jung J. M.; Ro H. S.; Chung B. H. Enhanced Sensitivity of Surface Plasmon Resonance (SPR) Immunoassays Using a Peroxidase-Catalyzed Precipitation Reaction and Its Application to a Protein Microarray. J. Immunol. Methods 2005, 297, 125–132. [DOI] [PubMed] [Google Scholar]
- Zhao W. A.; Ali M. M.; Brook M. A.; Li Y. F. Rolling Circle Amplification: Applications in Nanotechnology and Biodetection with Functional Nucleic Acids. Angew. Chem., Int. Ed. 2008, 47, 6330–6337. [DOI] [PubMed] [Google Scholar]
- Li D.; Shlyahovsky B.; Elbaz J.; Willner I. Amplified Analysis of Low-Molecular-Weight Substrates or Proteins by the Self-Assembly of DNAzyme-Aptamer Conjugates. J. Am. Chem. Soc. 2007, 129, 5804–5805. [DOI] [PubMed] [Google Scholar]
- Haes A. J.; Van Duyne R. P. A Nanoscale Optical Biosensor: Sensitivity and Selectivity of an Approach Based on the Localized Surface Plasmon Resonance Spectroscopy of Triangular Silver Nanoparticles. J. Am. Chem. Soc. 2002, 124, 10596–10604. [DOI] [PubMed] [Google Scholar]
- Bantz K. C.; Meyer A. F.; Wittenberg N. J.; Im H.; Kurtulus O.; Lee S. H.; Lindquist N. C.; Oh S. H.; Haynes C. L. Recent Progress in SERS Biosensing. Phys. Chem. Chem. Phys. 2011, 13, 11551–11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y. R.; Zhao D.; Yuan T. X.; Hu J.; Zhang D. C.; Cheng W.; Zhang W.; Ding S. J. A Simple and Highly Sensitive Electrochemical Biosensor for microRNA Detection Using Target-Assisted Isothermal Exponential Amplification Reaction. Electroanalysis 2013, 25, 2354–2359. [Google Scholar]
- Lee H. J.; Wark A. W.; Li Y.; Corn R. M. Fabricating RNA Microarrays with RNA-DNA Surface Ligation Chemistry. Anal. Chem. 2005, 77, 7832–7837. [DOI] [PubMed] [Google Scholar]
- Li Y.; Lee H. J.; Corn R. M. Fabrication and Characterization of RNA Aptamer Microarrays for the Study of Protein-Aptamer Interactions with SPR Imaging. Nucleic Acids Res. 2006, 34, 6416–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. A.; Wark A. W.; Lee H. J.; Corn R. M. Single-Nucleotide Polymorphism Genotyping by Nanoparticle-Enhanced Surface Plasmon Resonance Imaging Measurements of Surface Ligation Reactions. Anal. Chem. 2006, 78, 3158–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Lee H. J.; Corn R. M. Detection of Protein Biomarkers Using RNA Aptamer Microarrays and Enzymatically Amplified Surface Plasmon Resonance Imaging. Anal. Chem. 2007, 79, 1082–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H.; Crooks R. M. Parallel Fabrication of RNA Microarrays by Mechanical Transfer from a DNA Master. Anal. Chem. 2007, 79, 8994–8999. [DOI] [PubMed] [Google Scholar]
- Seefeld T. H.; Zhou W. J.; Corn R. M. Rapid Microarray Detection of DNA and Proteins in Microliter Volumes with Surface Plasmon Resonance Imaging Measurements. Langmuir 2011, 27, 6534–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A. D.; Szostak J. W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- Tuerk C.; Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- Wilson D. S.; Szostak J. W. In Vitro Selection of Functional Nucleic Acids. Annu. Rev. Biochem. 1999, 68, 611–647. [DOI] [PubMed] [Google Scholar]
- Xing H.; Hwang K.; Li J.; Torabi S. F.; Lu Y. DNA Aptamer Technology for Personalized Medicine. Curr. Opin Chem. Eng. 2014, 4, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena S. D. Aptamers: An Emerging Class of Molecules That Rival Antibodies in Diagnostics. Clin Chem. 1999, 45, 1628–1650. [PubMed] [Google Scholar]
- Brody E. N.; Gold L. Aptamers as Therapeutic and Diagnostic Agents. J. Biotechnol. 2000, 74, 5–13. [DOI] [PubMed] [Google Scholar]
- He L.; Hannon G. J. MicroRNAs: Small RNAs with a Big Role in Gene Regulation. Nat. Rev. Genet 2004, 5, 522–531. [DOI] [PubMed] [Google Scholar]
- Calin G. A.; Croce C. M. MicroRNA Signatures in Human Cancers. Nat. Rev. Cancer 2006, 6, 857–866. [DOI] [PubMed] [Google Scholar]
- Liu Q. H.; Wang L. M.; Frutos A. G.; Condon A. E.; Corn R. M.; Smith L. M. DNA Computing on Surfaces. Nature 2000, 403, 175–179. [DOI] [PubMed] [Google Scholar]
- Goodrich T. T.; Lee H. J.; Corn R. M. Enzymatically Amplified Surface Plasmon Resonance Imaging Method Using RNase H and RNA Microarrays for the Ultrasensitive Detection of Nucleic Acids. Anal. Chem. 2004, 76, 6173–6178. [DOI] [PubMed] [Google Scholar]
- Colgan D. F.; Manley J. L. Mechanism and Regulation of mRNA Polyadenylation. Gene Dev 1997, 11, 2755–2766. [DOI] [PubMed] [Google Scholar]
- Castoldi M.; Schmidt S.; Benes V.; Noerholm M.; Kulozik A. E.; Hentze M. W.; Muckenthaler M. U. A Sensitive Array for microRNA Expression Profiling (miChip) Based on Locked Nucleic Acids (LNA). RNA 2006, 12, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F.; Groebe D. R.; Witherell G. W.; Uhlenbeck O. C. Oligoribonucleotide Synthesis Using T7 RNA-Polymerase and Synthetic DNA Templates. Nucleic Acids Res. 1987, 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford L. K.; Sendroiu I. E.; Corn R. M.; Luptak A. Attomole Detection of Mesophilic DNA Polymerase Products by Nanoparticle-Enhanced Surface Plasmon Resonance Imaging on Glassified Gold Surfaces. J. Am. Chem. Soc. 2010, 132, 9265–9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker R. R.; Joyce G. F. A DNA Enzyme That Cleaves RNA. Chem. Biol. 1994, 1, 223–229. [DOI] [PubMed] [Google Scholar]
- Silverman S. K. In Vitro Selection, Characterization, and Application of Deoxyribozymes That Cleave RNA. Nucleic Acids Res. 2005, 33, 6151–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Corn R. M. DNAzyme Footprinting: Detecting Protein-Aptamer Complexation on Surfaces by Blocking DNAzyme Cleavage Activity. J. Am. Chem. Soc. 2013, 135, 2072–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Nakamoto K.; Niwa O.; Corn R. M. On-Chip Synthesis of RNA Aptamer Microarrays for Multiplexed Protein Biosensing with SPR Imaging Measurements. Langmuir 2012, 28, 8281–8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran N.; Hainsworth E.; Bhullar B.; Eisenstein S.; Rosen B.; Lau A. Y.; Walter J. C.; LaBaer J. Self-Assembling Protein Microarrays. Science 2004, 305, 86–90. [DOI] [PubMed] [Google Scholar]
- He M.; Stoevesandt O.; Palmer E. A.; Khan F.; Ericsson O.; Taussig M. J. Printing Protein Arrays from DNA Arrays. Nat. Methods 2008, 5, 175–177. [DOI] [PubMed] [Google Scholar]
- Seefeld T. H.; Halpern A. R.; Corn R. M. On-Chip Synthesis of Protein Microarrays from DNA Microarrays via Coupled In Vitro Transcription and Translation for Surface Plasmon Resonance Imaging Biosensor Applications. J. Am. Chem. Soc. 2012, 134, 12358–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.; Tang W.; Wang D. Z.; Wu X. J.; Li N.; Liu F. Amplified QCM-D Biosensor for Protein Based on Aptamer-Functionalized Gold Nanoparticles. Biosens. Bioelectron. 2010, 26, 575–579. [DOI] [PubMed] [Google Scholar]
- Lange K.; Rapp B. E.; Rapp M. Surface Acoustic Wave Biosensors: A Review. Anal. Bioanal. Chem. 2008, 391, 1509–1519. [DOI] [PubMed] [Google Scholar]
- Gao H. W.; Yang J. C.; Lin J. Y.; Stuparu A. D.; Lee M. H.; Mrksich M.; Odom T. W. Using the Angle-Dependent Resonances of Molded Plasmonic Crystals to Improve the Sensitivities of Biosensors. Nano Lett. 2010, 10, 2549–2554. [DOI] [PubMed] [Google Scholar]
- Nakamoto K.; Kurita R.; Niwa O.; Fujii T.; Nishida M. Development of a Mass-Producible on-Chip Plasmonic Nanohole Array Biosensor. Nanoscale 2011, 3, 5067–5075. [DOI] [PubMed] [Google Scholar]
- Brolo A. G. Plasmonics for Future Biosensors. Nat. Photonics 2012, 6, 709–713. [Google Scholar]
- Huang C. J.; Ye J.; Wang S.; Stakenborg T.; Lagae L. Gold Nanoring as a Sensitive Plasmonic Biosensor for on-Chip DNA Detection. Appl. Phys. Lett. 2012, 100, 173114. [Google Scholar]
- Luchansky M. S.; Bailey R. C. High-Q Optical Sensors for Chemical and Biological Analysis. Anal. Chem. 2012, 84, 793–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. J.; Halpern A. R.; Seefeld T. H.; Corn R. M. Near Infrared Surface Plasmon Resonance Phase Imaging and Nanoparticle-Enhanced Surface Plasmon Resonance Phase Imaging for Ultrasensitive Protein and DNA Biosensing with Oligonucleotide and Aptamer Microarrays. Anal. Chem. 2012, 84, 440–445. [DOI] [PubMed] [Google Scholar]
- Famulok M.; Mayer G. Aptamer Modules as Sensors and Detectors. Acc. Chem. Res. 2011, 44, 1349–1358. [DOI] [PubMed] [Google Scholar]