Abstract
Vacuolar-type ATPase (V-ATPase) is a primary proton pump with versatile functions in various tissues. In nerve cells, V-ATPase is required for accumulation of neurotransmitters into secretory vesicles and subsequent release at the synapse. Neurons express a specific isoform (G2) of the G subunit of V-ATPase constituting the catalytic sector of the enzyme complex. Using gene targeting, we generated a mouse lacking functional G2 (G2 null), which showed no apparent disorders in architecture and behavior. In the G2-null mouse brain, a G1 subunit isoform, which is ubiquitously expressed in neuronal and non-neuronal tissues, accumulated more abundantly than in wild-type animals. This G1 upregulation was not accompanied by an increase in mRNA. These results indicate that loss of function of neuron-specific G2 isoform was compensated by an increase in levels of the G1 isoform without apparent upregulation of the G1 mRNA.
Vacuolar-type ATPase (V-ATPase) is a proton translocating pump driven by chemical energy released by ATP hydrolysis. It is present in the internal membranes of all eukaryotic cells, where it translocates protons from the cytosol into organelle lumen, resulting in an acidic environment within1. Various cellular functions of vital importance, including transport of solutes, membrane trafficking, and hydrolysis of macromolecules in digestive compartments are highly dependent on the acidic conditions and chemiosmotic potential energy created by V-ATPase2. Accordingly, loss of its activity results in embryonic lethality at the peri-implantation stage of early embryogenesis3. V-ATPase is also implicated in highly differentiated functions in various cells and tissues4,5,6. For example, V-ATPase-dependent acidification in the endocytic pathway is essential for defense against pathogens by macrophages as well as T-cell mediated antigen processing7,8. V-ATPase also contributes to establish the acidic environment outside the differentiated cells such as bone resorbing osteoclasts, epithelial cells in the kidney, epididymis, and other tissues5,9,10,11,12.
V-ATPase is a multi-subunit complex that comprises 2 major functional sectors, V1 and V013. V1 is involved in ATP hydrolysis and consists of 8 subunits (A, B, C, D, E, F, G, and H). V0 serves as a proton pathway across the membranes and consists of 5 subunits (a, c, c’, d, and e). The catalytic sites, composed of the A and B subunits are connected to the V0 proton-transporting sector by a central stalk composed of the D subunit and 3 elongated structures, made up of the subunits E and G14. These stalks are involved in the energy coupling of V1 and V0, which undergo association/dissociation cycles by which V-ATPase activity changes in response to physiological conditions15. The stalk structures play an important role in the reversible assembly/disassembly of the enzyme complex16.
The G subunit in the mouse is a small 13-kDa protein, composed of 118 amino acids. G subunit interacts with the E subunit to form a rod-like structure connecting the V1 catalytic and V0 transporting domains. Therefore, G is thought to be important for regulation of V-ATPase activities14,17. The mammalian genome contains 3 distinctive genes for this subunit, namely ATP6V1G1, ATP6V1G2, and ATP6V1G318. G1 isoform, encoded by ATP6V1G1 is expressed in various tissues and cells, while, G2 and G3 isoforms show restricted tissue-specific distribution in the brain and kidney, respectively18,19. The neurons express the unique G2 isoform in addition to the ubiquitously distributed G1 isoform, whereas other cells including astrocytes and glia only express the ubiquitous G1 isoform19.
V-ATPase is involved in multiple neuronal functions. Abundant in synaptic vesicles, it provides chemiosmotic energy for loading neurotransmitters20,21,22. Additionally, V-ATPase participates in membrane fusion between the synaptic vesicle and presynaptic plasma membrane23. Tissue-specific expression of the G2 isoform raise an intriguing argument that it may have a dedicated function in the neural system. However, the physiological significance of this unique expression pattern of the G2 isoform has not been elucidated to date. No genetic impairment of the ATP6V1G2 locus in human or any animal model has been identified to date; therefore, phenotypic consequences of the loss of neuron-specific G2 remains to be experimentally determined. In this study, we introduced a null mutation into the mouse Atp6v1g2 locus. The loss of the G2 function did not result in any developmental defect or obvious behavioral abnormality until adulthood. We found that deletion of G2 function induced the up-regulation of G1 subunit levels in the brain, suggesting that the amounts of V-type ATPase subunits are strictly regulated. This up-regulation was not accompanied by an increase in levels of the Atg6v1g1 transcripts, implying the existence of a post-transcriptional regulatory mechanism.
Results
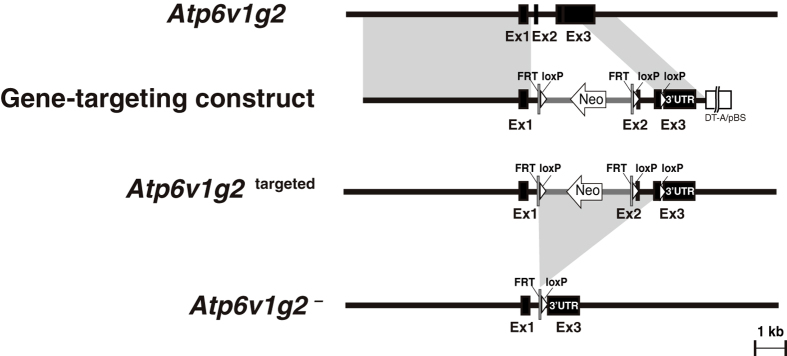
The mammalian genome contains 3 isoforms of the V-ATPase G subunit, the catalytic sector of the proton pump complex. Transcription of the Atp6v1g2 locus is highly restricted to the neurons. In order to elucidate the physiological relevance of this G subunit isoform, we created a genetically modified mouse. We generate a modified version of Atp6v1g2, in which lox P-Frt-npt-lox P-Frt was placed in intron 1–2, and the third lox P in the 3′-untranslated region (Fig. 1). Cre-mediated recombination between the remotest lox P sequences produced an allele that was missing the exons 2 and 3, resulting in a truncated coding sequence comprising only residues 1 through 27; therefore, it was highly likely to be a null allele (Fig. 1), which we designated as Atp6v1g2−.
Figure 1. Modification of the mouse Atp6v1g2 locus encoding V-ATPase G2 subunit.
The embryonic stem cell line R1 was transfected with a gene-targeting construct carrying a lox P-Frt-npt-lox P-Frt cassette and the third lox P, in intron 1–2 and the 3′-UTR of Atp6v1g2, respectively. Chimeric animals were generated by injecting the homologous recombinants into the blastocoels of C57Bl/6 embryos. Mice carrying the Atp6v1g2targeted allele were crossed with the FvB/N-Tg(EIIa-cre)C5379Lmgd/J strain, expressing Cre recombinase during early development to remove exon 2 (Ex2) and a part of exon 3 (Ex3), to generate Atp6v1g2+/− mice. Atp6v1g2+/− mice were further crossed with wild-type C57Bl/6 mice to remove the Cre transgene and a cohort of Atp6v1g2+/− mice was established. 3′-UTR; 3′-untranslated region, Neo; neomycin resistance cassette.
Mice heterozygous for the null allele (Atp6v1g2+/−) were normal and fertile. They were intercrossed to generate Atp6v1g2−/− progeny, which appeared at the expected ratio upon the genotyping at 3 weeks after birth, indicating that the loss of the G2 isoform did not result in embryonic or post-natal lethality (Table 1). They did not show any discernible behavioral differences among littermates with mixed genetic background of 129Sv (from ES cells), C57Bl6 (breeding spouse), and FvB (Cre-expressing strain). We were unable to distinguish G2-null mice from heterozygous or wild-type littermates in a cohort maintained for more than a year, unless genotypes were determined by PCR.
Table 1. Genotype of progeny from intercross of Atp6v1g2+/− mice.
|
Genotype |
|||
|---|---|---|---|
| +/+ | +/− | −/− | |
| 3–4 wks after birth | 14 | 36 | 17 |
| (21%) | (54%) | (25%) | |
Genotype of progenies were determined by PCR of genomic DNA obtained from tail-tips.
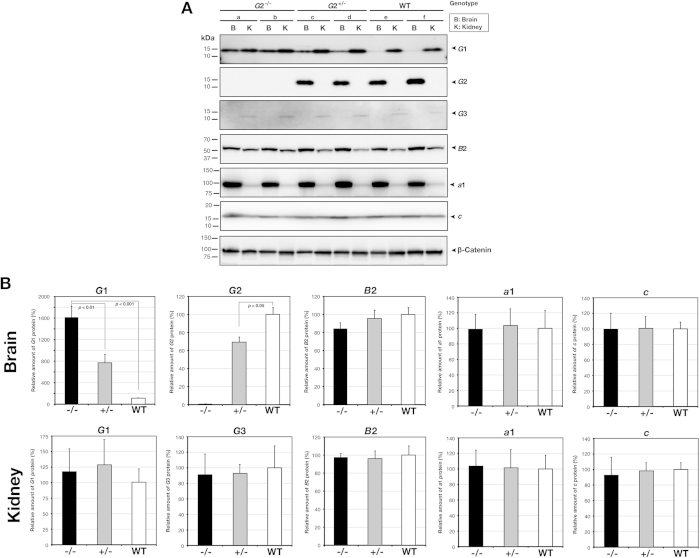
We previously reported a set of polyclonal antibodies, each specifically recognizing G1, G2, and G3 isoforms19,24. Using isoform-specific antibodies, we examined the expression of G1, G2, and G3 in the brain and kidney of wild-type and G2-null mice by immunoblot (Fig. 2A). The G1 isoform was present in both, the brain and kidneys of the wild-type mouse, but G2 was detected only in the brain and not the kidneys (Fig. 2A). As previously reported18, G3 isoform was specifically expressed in the kidney (Fig. 2A). In the mice homologous for the null allele (Atp6v1g2−/− mice), no G2 was detected in the brain lysate (Fig. 2A,B), thus underscoring that the Atp6v1g2− allele was indeed null. The quantity of V-ATPase B2 subunit, the component of the V1 sectors was similar in the wild-type and mutant brains. The amounts of subunits a1 and c, the components of the V0 membrane intrinsic sector, were accumulated similarly in the brains of all the genotypes (Fig. 2). These results implied that the total quantity of V-ATPase accumulated remained unchanged. The G1 isoform in the mutant brain appeared more abundant as compared to that in the wild-type brain (Fig. 2). In contrast, the quantity of G1 isoform in the kidney, where no G2 isoform is normally produced, was similar in the wild-type and mutants (Fig. 2). This observation indicated that an increase of G1 subunit occurred specifically in the brain, where the G2 subunit was highly expressed. The compensatory increase of G1 associated with the genetic loss of G2 isoform suggested that the absence of G2 subunit leads to an increase in G1 subunit in the brain. In contrast to the compensation between G1 and G2 isoforms, G3 remained absent in the mutant brain, showing that loss of G2 did not alter the expression specificity of the kidney-specific isoform.
Figure 2. Immunoblot analysis of G subunit isoforms in the brain and kidney.
(A) lysates (10 μg/lane) prepared from the brain and kidneys of Atp6v1g2−/− (lanes a,b) mice, Atp6v1g2+/− (lanes c,d) and wild-type (lanes e,f) mice, and then they were subjected to gel electrophoresis in the presence of sodium dodecyl sulfate. G1, G2, G3 isoforms, a1, c and B2 subunit were probed by the specific antibodies. V-ATPase a1, c, and B2 subunits represent the quantity of total V-ATPase in tissues. β-catenin was also detected for standardization of loadings. (B) the band density was quantified using a luminoimager and relative quantity of subunit isoforms was determined. The levels in the wild-type tissues were defined as 100%. The bars represent standard deviations of 3 independent experiments using 4 samples of each genotype.
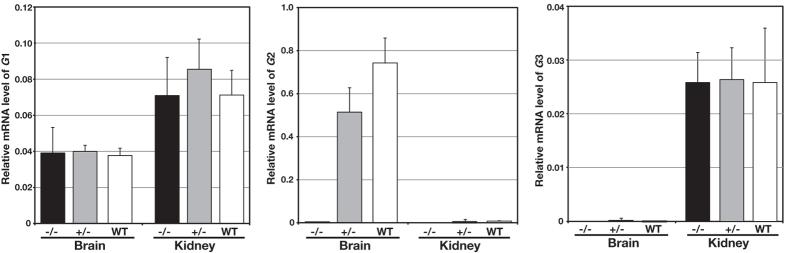
We investigated whether the observed increase in the quantity of G1 isoform reflected an up-regulation at the transcription level by examining the levels of Atp6v1g1, Atp6v1g2, and Atp6v1g3 transcripts. Total RNA was isolated from the brain and kidney of the wild-type, heterozygous (Atp6v1g2+/−), and homozygous (Atp6v1g2−/−) mutant animals, cDNAs were reverse-transcribed, and quantitative real-time PCR analysis was performed. mRNA quantity was estimated by the efficiency calibrated model25 using Gapdh transcripts as standard (Fig. 3). In the wild-type animals, G1 and G2 transcripts were detected at high levels in the brain, whereas G2 transcripts were found in the brain but only a minor fraction in the kidney. G3 transcripts were strictly restricted to the kidney, while this expression pattern and levels in the kidney were unchanged across all genotypes. In the heterozygous and homozygous mutant brains, the levels of G1 transcripts remained unchanged. The fold changes of G1 expression in heterozygous (Atp6v1g2+/−), and homozygous (Atp6v1g2−/−) mutant brains when compared with wild-type were 1.27 and 1.02, respectively, indicating that the transcription and stability of mRNA play a minor role, if any, for upregulation of G1 subunit isoform. The quantity of G2 transcripts showed a moderate decrease in the heterozygous mice (0.65-folds of that in the wild-type), indicating that transcripts levels were proportional to the gene dosage.
Figure 3. Expression of G subunit isoforms in the brain and kidneys.
The expression levels of G-subunit isoform transcripts were determined by quantitative PCR. Total RNA was obtained from the brain and kidney, reverse-transcribed, and then amplified in the presence of primer pairs, specific for each isoform. Amplification was monitored by SYBR green fluorescence using a Prism 7000 detector. Relative expression levels were determined after normalization to Gapdh transcripts. The expression levels were determined in 4, 3, and 4 mice of wild-type, Atp6v1g2+/−, and Atp6v1g2−/−, respectively. The bars represent standard deviations.
Discussion
The variety of functions performed by V-ATPase in different cellular contexts are attributed to the structural heterogeneity of the enzyme complex, comprising of multiple subunit isoforms. The higher eukaryotes, both, vertebrates and invertebrates develop such diversity by having evolved different subunit isoforms in both, V1 and V0 sectors26. This suggests that diverse structural variation is required for more elaborate physiological demands in multicellular organisms.
In mammals, there are 3 distinctive isoforms of the G subunit. In a previous study, we showed that V-ATPase complexes immunoisolated by anti-G1 and anti-G2 antibodies exhibit similar kinetic profiles for ATP hydrolysis19. However, the ubiquitously expressed G1 isoform can complement the loss of Vma10, a yeast orthologue of the G subunit, whereas the brain-specific isoform G2 does not rescue Vma10 deficiency in yeast. This difference shown in a heterologous expression system suggests that G1 and G2 interact differently with other subunits of yeast V-ATPase19.
The G subunit forms a rod-shaped structure with the E subunit. This rod constitutes a stator, connecting the membrane-peripheral V1 and membrane-intrinsic V0 sectors14. V-ATPase undergoes reversible dissociation/association of the V1 and V0 sectors, through which, ATP hydrolysis and proton translocation can be regulated in response to physiological conditions27. The stator structures play a central role in the reversible disassembly of the V0V1-ATPase complex. Physiological relevance of this dissociation/association is established in yeast and invertebrate organisms, however, its role in mammals remains to be explored. Structural signatures of G, E, and C subunits which have 3, 2, and 3 isoforms, respectively, might confer differential characteristics of assemble/disassemble cycles in mammalian enzymes. This, genetically modified animals with specific loss of the G2 subunit could provide a model to explore this possibility.
We found that the quantity of V-ATPase seemed to be maintained in spite of loss of G2 subunit isoforms, suggesting the presence of a mechanism regulating the total activity and/or quantity of V-ATPase in the cell. This argument has been repeatedly raised by previous observations with regards to specific impairment of a- and B-subunit isoforms. B1-deficient renal epithelium can be rescued by increasing activity of V-ATPase B2 isoform28. This compensation is achieved by recruiting B2-containing V-ATPase from intracellular membranes to the cell surface, not necessarily involving transcriptional and/or translational up regulation29. Acidification of phagosomes requires the a3 isoform. The lumen of phagosomes in a3-deficient mice still exhibits bafilomycin-sensitive, V-ATPase-dependent acidification, though far less efficiently than that in wild-type macrophages8. This indicates that the a1- or a2 subunit isoform can partially rescue the loss of the a3 isoform. These observations suggest that various mechanisms at multiple levels are involved in regulation of the levels of the proton pump complex and its precise membrane distribution as well, for the maintenance of acid-ion homeostasis in various tissues.
Several studies have shown the underlying mechanism how cells sense the activities of proton pumps and acidification in the various subcellular compartments. Heteronuclear ribonucleoprotein-mediated stabilization of mRNA is involved in maintaining transcription level of E1 subunit30,31. Other studies have shown that sensing of organelle acidification regulates interaction of V-ATPase and cytoskeleton thus regulates subcellular distribution of V-ATPase32,33,34,35. Further, G subunit is a substrate for the proteasome-dependent proteolysis coupled with rab7/RILP endosome regulation36. These observations suggest that multiple regulatory mechanisms are involved in the determination of stable and transient levels of V-ATPase complex in response to the extent of acidification within various subcellular compartments as well as in the external milieu. Here, the current results suggested that the quantity of G subunits is regulated at the post-transcriptional level. Further studies into the mechanism responsible for quantity regulation of G as well as other subunits are needed to improve our understanding of the versatile physiological function of V-ATPase.
Methods
Reagents
Rabbit antibodies to G1, G2, G3, and anti-V-ATPase a1, B2 and c subunits were described previously19,24,37. Mouse antibody against β-catenin was obtained from Beckton Dickinson. Polymerase chain reactions (PCR) for bacterial artificial chromosome (BAC) modification and vector construction were performed with Phusion DNA polymerase (Finnzyme). Diagnostic PCR was performed using Ex Taq DNA polymerase (TaKaRa).
Targeting construct
A targeting vector was created using recombinogenic strategy. A BAC clone (MGS1-205a4) containing the Atp6v1g2 locus of mouse 129Sv strain was obtained from Genome Systems and introduced into Escherichia coli strain DY380, carrying an inducible homologous recombination system to obtain a strain 205a4/DY38038. A lox P-Zeo-lox P fragment with arms homologous to Atp6v1g2 was amplified by PCR using the primer pair G2LZL-S1: 5′-ccaccccaactatcgggttactgtctagaaccatcgctcagggacacatcTCAACCATCATCGATCATAA-3′ and G2LZL-A1: 5′- tttgagttgggatatttctctgtggacggagctgacagaaggagtcactcAACCCTGAAGCTATCATAA-3′ (lowercase letters correspond to sequences in the 3′-untranslated region of the Atp6v1g2, and uppercase letters correspond to a template plasmid pMODloxZeo-ΔAmp3). The lox P-Zeo-lox P cassette vector39 was introduced into E. coli strain 205a4/DY380. Correct homologous recombination yielding a modified BAC containing Atp6v1g2 locus with lox P-Zeo-lox P was screened via Zeocin resistance and confirmed by PCR analysis. Next, a linear pBSDT-AII fragment39 flanked by short Atp6v1g2 segments was amplified using the primer pair G2DTAL: 5′-GCTCCTGGGTGGTAGCAGTGTGGTGCCTAGGGTTGCCGCTTACGgatcctagagcggccgccaccgcg-3′ and G2DTAR: 5′-CAGCACCTTGCCTCCTTGTGTAACAGCGGAGTGTCAATAAAATACTGGTGgatccggccggcccgggcga-3′ (lowercase and uppercase letters correspond to pBSDT-AII and Atp6v1g2 sequences, respectively). The fragment was introduced into DY380 carrying the modified BAC to retrieve an approximately 7-kb fragment in the multicopy plasmid pBSDT-AII. The resultant plasmid was propagated in E. coli strain BH25.8 expressing Cre recombinase to remove the Zeo cassette and one of the lox P. Then, the vector was further modified with a lox P-Frt-npt-lox P-Frt cassette39 using a mini-transposon-mediated strategy to yield a targeting vector in which, lox P-Frt-npt-lox P-Frt was placed in intron 1–2 and the third lox P, in the 3′-UTR (Fig. 1). Cre-mediated recombination between the remotest lox P sequences produced an allele that was missing exon 2 and a part of exon 3, which encode the amino acid residues 28 to 118 of the G2 subunit.
Immunoblot analysis
The brain and kidney were isolated and lysed in an extraction buffer containing 50 mM Tris-HCl (pH7.4), 1% sodium dodecyl sulphate (SDS) plus Complete proteinase inhibitors (Roche) and 1 mM phenylmethylsulfonyl fluoride. Protein concentration in the lysate was determined by the bicinchoninic acid colorimetric assay (Pierce). For the immunoblot analysis, 10 μg protein was loaded onto 5% to 20% SDS-polyacrylamide gel, electrophoresed, and immunoblotted as previously described19. Subunit G isoforms were detected by isoform-specific antibodies using the Pierce Western Blotting Substrate Plus system. Images were acquired and analyzed using the GE Healthcare LAS-4000 mini system37.
Animals
All animal experiments were approved by the institutional committee (ISIR, Osaka University) and carried out in accordance with the rules and regulations of the institutions and the government. C57Bl/6 mice were purchased from SLC Japan. The targeting construct was introduced into mouse embryonic stem cells40 (R1, kindly provided by Dr. A. Nagy) by electroporation and recombinants carrying the modified allele in the Atp6v1g2 locus were screened by PCR. The successful clones were injected into C57Bl/6 E3.5 blastocysts, to obtain chimeric mice, which were further crossed with C57Bl/6 mice. Cre-mediated recombination was performed in germ cells by crossing the chimeric mice with C57Bl/6 mice carrying an EIIa-promoter, Cre-recombinase transgene41.
Quantitative real-time PCR (qPCR) for G subunit isoforms
We prepared brains and kidneys by macroscopic dissection from 1.5–2 month old mice (n = 3 (Atp6v1g2+/− animals) or n = 4 (wild type and Atp6v1g2−/− animals)). The tissues were quick frozen in liquefied nitrogen and kept under −80 °C until use, then total RNA was extracted using Qiagen RNeasy mini kit. cDNA was obtained from the 1500 ng of RNA (determined by spectrophotometry) by reverse-transcription in 20 μl reaction using a SuperScript VILO cDNA synthesis kit (Life Technologies). Primer efficiencies were determined from standard curves obtained by a dilution series of mixed pool of cDNAs derived from brain and kidney of wild-type animals (Table 2). The cDNA (corresponding to 1.5 ng–0.015 pg RNA) was subjected to qPCR in 25 μl of 1× SYBR Premix Ex Taq II (Tli RNase H plus) (TaKaRa), 200 nM primers, and amplification was monitored in Life Technologies/Applied Biosystems Prism7000. The plots were analyzed with ABI Prism software (ver. 1.1) to obtain quantification cycles (Cq). The linear regression plots (Cq against log(RNA amount)) gave r2 > 0.99 in the range of 1.5 ng–0.15 pg of RNA, and the primer efficiencies (E) were determined from the slope according to a formula E = 10(−slope).
Table 2. PCR primers used for quantification of transcripts.
| Gene | primer | sequence | efficiency (%) | amplicon size (bp) | intron length (bp) |
|---|---|---|---|---|---|
| Atp6v1g1 | G1-S201 | 5′-ccctcagcaatggcgagtcagtc-3′ | 105.4 | 137 | 3585 |
| G1-A201 | 5′-tcagcctgggcttcttctttgg-3′ | ||||
| Atp6v1g2 | VG2-S201 | 5′-aagcgggcagcggagaaggtg-3′ | 88.5 | 140 | 201 |
| VG2-A202 | 5′-gccgcctgctgcttgctct-3′ | ||||
| Atp6v1g3 | VG3-S201 | 5′-agaaaagaaaaggaaagcgactga-3′ | 96.5 | 315 | 4020 |
| VG3-A201 | 5′-tctatgcctatggacttggatgtg-3′ | ||||
| Gapdh | GAPDH-S1 | 5′-tcccgtagacaaaatggtgaaggt-3′ | 106.2 | 184 | 1842 |
| GAPDH-A1 | 5′-tgtgccgttgaatttgccgtgagt-3′ |
The oligonucleotides used for specific amplification of G subunit isoform cDNA are shown. The upstream and downstream primers are designed to anneal separate exons, thus minimizing the amplification of genomic DNA.
We quantified transcripts of reference genes Gapdh, Actb, Tbp, Ppia and Ywhaz in the cDNAs from brains and kidneys of wild-type, Atp6v1g2+/−, Atp6v1g2−/− animals (Table 3). We then analyzed the amplification data by NormFinder algorithm42 in order to identify the best standard transcripts in the experiments (Table 4). Comparing expression levels in brain identified Gapdh as the most stable reference gene among the genotypes (stability value 0.066), following Tbp (0.133), Actb (0.134), Ywhaz (0.190), and Ppia (0.178). In kidney, Actb was the most stable (0.121), and Gapdh was the third (0.222). Although Actb appeared the best standard (0.193) if all the samples (both brain and kidney in three genotypes) were to be normalized, we chose Gapdh (0.233), the most stable standard reference in brain, as the normalizer in this experiment.
Table 3. PCR primers for the standard genes.
| gene | primer | sequence | efficiency (%) | references |
|---|---|---|---|---|
| Actb | ActB-S1: | 5′-gctgtattcccctccatcgtg-3′ | 106.4 | 43 |
| ActB-A1: | 5′-cacggttggccttagggttcag-3′ | |||
| Tbp | Tbp-S1: | 5′-ggcctctcagaagcatcacta-3′ | 119.4 | 43 |
| Tbp-A1: | 5′-gccaagccctgagcataa-3′ | |||
| Ppia | Cyc-S1: | 5′-agcactggagagaaaggatt-3′ | 103.2 | 44 |
| Cyc-A1: | 5′-agccattcagtcttggcagt-3′ | |||
| Ywhaz | Ywhaz-S1: | 5′-ttgagcagaagacggaaggt-3′ | 107.9 | 44 |
| Ywhaz-A1: | 5′-gaagcattggggatcaagaa-3′ |
The oligonucleotides used for amplification of candidate standard genes are described. Primer sequences were based on the references, modified to mouse sequence if necessary.
Table 4. Stability of the standard genes among genotypes.
| Gene |
Stability |
||
|---|---|---|---|
| Brain | Kidney | Brain & Kidney | |
| Gapdh | 0.066 | 0.222 | 0.233 |
| Actb | 0.134 | 0.121 | 0.193 |
| Tbp | 0.133 | 0.232 | 0.205 |
| Ppia | 0.178 | 0.305 | 0.283 |
| Ywhaz | 0.190 | 0.209 | 0.234 |
Stabilities of the candidate genes were examined. Cq was obtained in different samples, (brain and kidney, from the wild-type, Atp6v1g2+/−, and Atp6v1g2−/− animals) and transformed to linear scale quantities by dCq method, then analyzed by NormFinder algorithm to calculate stability index42.
The relative amounts of G isoform transcripts were determined by the qPCR. The cDNA corresponding to 75 pg of RNA was subjected to qPCR in 25 μl reaction as specified above. The profile of thermal cycle was 1 min initial denature at 96 ˚C, 40 cycles of 96 ˚C 5 sec/60 ˚C 35 sec, and then melt curves were obtained gradually raising the temperature from 65 ˚C to 90 ˚C. The amplification plots were analyzed by the ABI Prism software to obtain Cq. In each samples, the relative amounts of G isoform transcript was calculated as ratio = EGapdh(CqGapdh)/EGn isoform(CqGn). The changes in transcript levels among the genotypes were calculated as fold = EGn isoform, genotypes(Mean (CqGn, wild) − Mean (CqGn, genotypes))/EGapdh, genotypes(Mean (CqGapdh, wild) − Mean (CqGapdh, genotypes))25.
Additional Information
How to cite this article: Kawamura, N. et al. Loss of G2 subunit of vacuolar-type proton transporting ATPase leads to G1 subunit upregulation in the brain. Sci. Rep. 5, 14027; doi: 10.1038/srep14027 (2015).
Acknowledgments
We thank Dr. Andreas Nagy for ES cell line R1. We also thank Dr. Neal G. Copeland for the recombinogenic BAC engineering system. This study was supported in part by Individual Research Grants in Doshisha Women’s College of Liberal Arts to GHSW and Grants-in-Aid from MEXT, Japan.
Footnotes
The authors declare no competing financial interests.
Author Contributions N.K., G.H.S.W. and Y.W. performed the experiments. N.K., G.H.S.W. and Y.W. analyzed the data. Y.W. wrote the main manuscript. All the authors (N.K., G.H.S.W. and Y.W.) reviewed the manuscript.
References
- Sun-Wada G. H., Wada Y. & Futai M. Vacuolar H+ pumping ATPases in luminal acidic organelles and extracellular compartments: common rotational mechanism and diverse physiological roles. J Bioenerg Biomembr 35, 347–358. (2003). [DOI] [PubMed] [Google Scholar]
- Futai M. et al. Luminal acidification of diverse organelles by V-ATPase in animal cells. J Exp Biol 203 Pt 1, 107–116 (2000). [DOI] [PubMed] [Google Scholar]
- Sun-Wada G. H. et al. Acidic endomembrane organelles are required for mouse postimplantation development. Dev Biol 228, 315–325 (2000). [DOI] [PubMed] [Google Scholar]
- Sun-Wada G. H. et al. The a3 isoform of V-ATPase regulates insulin secretion from pancreatic β-cells. J Cell Sci 119, 4531–4540 (2006). [DOI] [PubMed] [Google Scholar]
- Pietrement C. et al. Distinct expression patterns of different subunit isoforms of the V-ATPase in the rat epididymis. Biol Reprod 74, 185–194 (2006). [DOI] [PubMed] [Google Scholar]
- Sun-Wada G. H., Tabata H., Kawamura N., Futai M. & Wada Y. Differential expression of a subunit isoforms of the vacuolar-type proton pump ATPase in mouse endocrine tissues. Cell Tissue Res 329, 239–248 (2007). [DOI] [PubMed] [Google Scholar]
- Trombetta E. S. & Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol 23, 975–1028 (2005). [DOI] [PubMed] [Google Scholar]
- Sun-Wada G. H., Tabata H., Kawamura N., Aoyama M. & Wada Y. Direct recruitment of H+-ATPase from lysosomes for phagosomal acidification. J Cell Sci 122, 2504–2513 (2009). [DOI] [PubMed] [Google Scholar]
- Li Y. P., Chen W., Liang Y., Li E. & Stashenko P. Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat Genet 23, 447–451 (1999). [DOI] [PubMed] [Google Scholar]
- Oka T. et al. a4, a unique kidney-specific isoform of mouse vacuolar H+-ATPase subunit a. J Biol Chem 276, 40050–40054 (2001). [DOI] [PubMed] [Google Scholar]
- Toyomura T. et al. From lysosomes to plasma membrane: Localization of vacuolar type H+-ATPase with the a3 isoform during osteoclast differentiation. J Biol Chem 278, 22023–22030 (2003). [DOI] [PubMed] [Google Scholar]
- Kikuta J. et al. Dynamic visualization of RANKL and Th17-mediated osteoclast function. J Clin Invest 123, 866–873 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton A., Bond S. & Forgac M. V-ATPase functions in normal and disease processes. Pflugers Arch 457, 589–598 (2009). [DOI] [PubMed] [Google Scholar]
- Oot R. A., Huang L. S., Berry E. A. & Wilkens S. Crystal structure of the yeast vacuolar ATPase heterotrimeric EGC(head) peripheral stalk complex. Structure 20, 1881–1892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane P. M. & Parra K. J. Assembly and regulation of the yeast vacuolar H+-ATPase. J Exp Biol 203, 81–87 (2000). [DOI] [PubMed] [Google Scholar]
- Charsky C. M., Schumann N. J. & Kane P. M. Mutational analysis of subunit G (Vma10p) of the yeast vacuolar H+-ATPase. J Biol Chem 275, 37232–37239 (2000). [DOI] [PubMed] [Google Scholar]
- Norgett E. E. et al. V1 and V0 domains of the human H+-ATPase are linked by an interaction between the G and a subunits. J Biol Chem 282, 14421–14427 (2007). [DOI] [PubMed] [Google Scholar]
- Sun-Wada G. H., Yoshimizu T., Imai-Senga Y., Wada Y. & Futai M. Diversity of mouse proton-translocating ATPase: presence of multiple isoforms of the C, d and G subunits. Gene 302, 147–153 (2003). [DOI] [PubMed] [Google Scholar]
- Murata Y. et al. Differential localization of the vacuolar H+ pump with G subunit isoforms (G1 and G2) in mouse neurons. J Biol Chem 277, 36296–36303 (2002). [DOI] [PubMed] [Google Scholar]
- Moriyama Y., Tsai H. L. & Futai M. Energy-dependent accumulation of neuron blockers causes selective inhibition of neurotransmitter uptake by brain synaptic vesicles. Arch Biochem Biophys 305, 278–281 (1993). [DOI] [PubMed] [Google Scholar]
- Takamori S. et al. Molecular anatomy of a trafficking organelle. Cell 127, 831–846 (2006). [DOI] [PubMed] [Google Scholar]
- Saw N. M. et al. Vacuolar H+-ATPase subunit Voa1 and Voa2 cooperatively regulate secretory vesicle acidification, transmitter uptake and storage. Mol Biol Cell 22, 3394–3409 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesinger P. R. et al. The V-ATPase Vo subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila. Cell 121, 607–620 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun-Wada G. H. et al. Mouse proton pump ATPase C subunit isoforms (C2-a and C2-b) specifically expressed in kidney and lung. J Biol Chem 278, 44843–44851 (2003). [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun-Wada G. H. & Wada Y. Vacuolar-type proton pump ATPases: roles of subunit isoforms in physiology and pathology. Histol Histopathol 25, 1611–1620 (2010). [DOI] [PubMed] [Google Scholar]
- Kane P. M. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev 70, 177–191 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunescu T. G. et al. Compensatory membrane expression of the V-ATPase B2 subunit isoform in renal medullary intercalated cells of B1-deficient mice. Am J Physiol Renal Physiol 293, F1915–1926 (2007). [DOI] [PubMed] [Google Scholar]
- Brown D., Paunescu T. G., Breton S. & Marshansky V. Regulation of the V-ATPase in kidney epithelial cells: dual role in acid-base homeostasis and vesicle trafficking. J Exp Biol 212, 1762–1772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. P., Krits I., Lu Y., Bai S. & Lee B. S. Regulation of enhanced vacuolar H+-ATPase expression in macrophages. J Biol Chem 277, 8827–8834 (2002). [DOI] [PubMed] [Google Scholar]
- Jeyaraj S., Dakhlallah D., Hill S. R. & Lee B. S. HuR stabilizes vacuolar H+-translocating ATPase mRNA during cellular energy depletion. J Biol Chem 280, 37957–37964 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J. et al. Actin binding activity of subunit B of vacuolar H+-ATPase is involved in its targeting to ruffled membranes of osteoclasts. J Bone Miner Res 21, 714–721 (2006). [DOI] [PubMed] [Google Scholar]
- Lu M., Ammar D., Ives H., Albrecht F. & Gluck S. L. Physical interaction between aldolase and vacuolar H+-ATPase is essential for the assembly and activity of the proton pump. J Biol Chem 282, 24495–24503 (2007). [DOI] [PubMed] [Google Scholar]
- Hurtado-Lorenzo A. et al. V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat Cell Biol 8, 124–136 (2006). [DOI] [PubMed] [Google Scholar]
- Merkulova M. et al. Aldolase directly interacts with ARNO and modulates cell morphology and acidic vesicle distribution. Am J Physiol Cell Physiol 300, C1442–1455 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M. et al. RILP regulates vacuolar ATPase through interaction with the V1G1 subunit. J Cell Sci 127, 2697–2708 (2014). [DOI] [PubMed] [Google Scholar]
- Sun-Wada G.-H. et al. Generation of chicken monoclonal antibodies against the a1, a2, and a3 subunit isoforms of vacuolar-type proton ATPase. Hybridoma 30, 199–203 (2011). [DOI] [PubMed] [Google Scholar]
- Lee E. C. et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73, 56–65. (2001). [DOI] [PubMed] [Google Scholar]
- Aoyama M., Agari K., Sun-Wada G. H., Futai M. & Wada Y. Simple and straightforward construction of a mouse gene targeting vector using in vitro transposition reactions. Nucleic Acids Res 33, e52 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Rossant J., Nagy R., Abramow-Newerly W. & Roder J. C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA 90, 8424–8428 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M. et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA 93, 5860–5865 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen C. L., Jensen J. L. & Orntoft T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64, 5245–5250 (2004). [DOI] [PubMed] [Google Scholar]
- Willems E. et al. Selection of reference genes in mouse embryos and in differentiating human and mouse ES cells. Int J Dev Biol 50, 627–635 (2006). [DOI] [PubMed] [Google Scholar]
- Bonefeld B. E., Elfving B. & Wegener G. Reference genes for normalization: a study of rat brain tissue. Synapse 62, 302–309 (2008). [DOI] [PubMed] [Google Scholar]