Abstract
The modification of intracellular proteins by monosaccharides of O-linked β-N-acetylglucosamine (O-GlcNAc) is an essential and dynamic post-translational modification of metazoans. The addition and removal of O-GlcNAc is catalyzed by the O-GlcNAc transferase (OGT) and O-GlcNAcase, respectively. One mechanism by which O-GlcNAc is thought to mediate proteins is by regulating phosphorylation. To provide insight into the pathways regulated by O-GlcNAc, we have utilized stable isotope labeling of amino acids in cell culture (SILAC)-based quantitative proteomics to carry out comparisons of site-specific phosphorylation in OGT wild-type (WT) and Null cells. Quantitation of the phosphoproteome demonstrated that out of 5,529 phosphoserine, phosphothreonine and phosphotyrosine sites, 232 phosphosites were upregulated and 133 downregulated in the absence of O-GlcNAc. Collectively, these data suggest that deletion of OGT has a profound effect on the phosphorylation of cell cycle and DNA damage response proteins. Key events were confirmed by biochemical analyses and demonstrate a increase in the activating autophosphorylation event on ATM (Ser1987) and on ATM’s downstream targets p53, H2AX and Chk2. Together, these data support widespread changes in the phosphoproteome upon removal of O-GlcNAc, suggesting that O-GlcNAc regulates processes such as the cell cycle, genomic stability, and lysosomal biogenesis.
Keywords: OGT, ATM, Signal Transduction, Glycobiology, Glycosylation
1. Introduction
The addition of the monosaccharide β-N-acetylglucosamine through an O-glycosidic bond (O-GlcNAc) to intracellular proteins is a ubiquitous post-translational modification that serves to mediate protein function and regulate cellular signaling. In contrast to other post-translational modifications cycled by hundreds of enzymes that mediate site specificity, O-GlcNAc is cycled by just two enzymes. The O-GlcNAc transferase (OGT) [1, 2] and the O-GlcNAcase (OGA) [3, 4], catalyze the addition and removal of O-GlcNAc from both serine and threonine residues, respectively. There are three splice variants of OGT termed nuclear/cytoplasmic OGT (ncOGT), mitochondrial OGT (mOGT) and short OGT (sOGT) [5]. All variants contain a C-terminal catalytic domain [1, 2] with varying numbers of N-terminal tetratricopeptide repeats, which regulate protein-protein interactions and may dictate substrate specificity [1, 2, 5]. O-GlcNAcase exists as two splice variants, one that contains an N-terminal hexosaminidase domain and a C-terminal domain that bares similarity to a histone acetyltransferase domain. The shorter variant contains the hexosaminidase domain and a short alternative C-terminus [3, 6].
Thousands of proteins are modified by O-GlcNAc, and the dynamic cycling of this modification is essential for cellular homeostasis. Knockout of OGT is embryonic lethal in mice [7], and Caenorhabditis elegans exhibit shortened lifespan and produce deficiencies in macronutrient storage and dauer larvae formation [8, 9]. In mice, the O-GlcNAcase knockout is perinatal lethal [10] and in C. elegans, presents with similar macronutrient deficiencies as the OGT Null, but demonstrate increased dauer larvae formation [11]. O-GlcNAc also appears essential for the response of cells and tissues to stress and injury. Studies in COS-7 cells have demonstrated elevated O-GlcNAc levels in response to oxidative, genotoxic, osmotic and heat stress, and a predisposition to cellular injury upon deletion of OGT [12]. Similarly, ischemic preconditioning produced a global increase of O-GlcNAc levels in mice [13]. This study also demonstrated that inhibition of O-GlcNAcase to raise O-GlcNAc levels prior to myocardial infarction reduced infarct size comparable to ischemic preconditioning. These data indicate that O-GlcNAc mediates a pro-survival response, although the mechanisms by which it achieves this remain unclear. One possible mechanism by which O-GlcNAc promotes survival and cellular homeostasis is through the regulation of phosphorylation events.
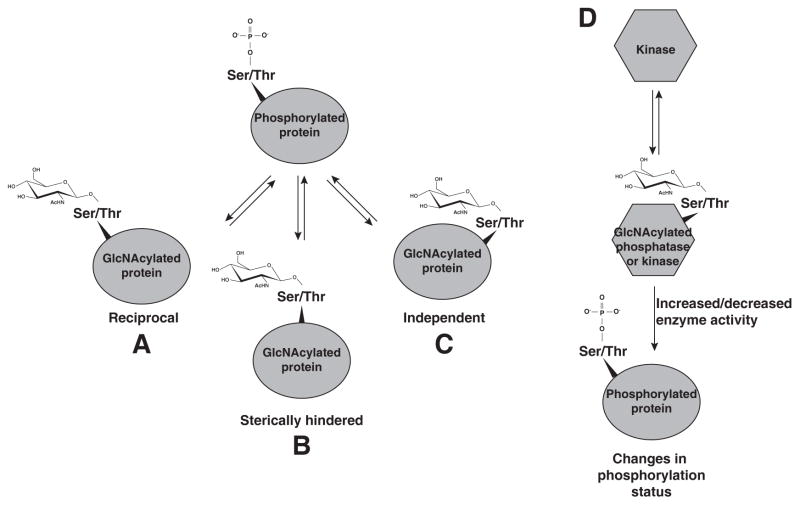
Due to the modification of the same serine and threonine residues by both O-GlcNAc and phosphorylation on proteins such as c-Myc and eNOS [14, 15], it has been proposed that crosstalk exists between these two modifications (Figure 1A). Several studies have shown that loss or inhibition of either OGT or O-GlcNAcase results in changes in serine and threonine phosphorylation profiles [11, 16, 17]. Similarly, inhibition of serine/threonine phosphatases 1, 2A, 2B with okadaic acid alters the profile of O-GlcNAc modified proteins [16]. Consistent with the idea that a subset of proteins can be rapidly cycled from phosphorylated to O-GlcNAcylated, both OGT and O-GlcNAcase are found in complexes with protein phosphatases and kinases [18, 19]. Many studies support a reciprocal regulation of serine and threonine residues by O-GlcNAc and O-phosphate [16, 20–22], which may depend on the active cycling of phosphorylation sites [16]. Indeed, a recent study identifying both O-GlcNAc modified proteins and phosphorylated proteins in the murine synapse showed that phosphorylated proteins are significantly more likely to be O-GlcNAc modified compared to non-phosphorylated proteins [23]. However, there did not appear to be an enrichment in sites that were reciprocally modified, indicating that O-GlcNAc may be modifying proteins at sites proximal to the phosphosite, possibly creating stearic hindrance for kinases and phosphatases (Figure 1B). An alternative mechanism by which OGT may regulate phosphorylation status is by modifying and regulating protein kinases and phosphatases (Figure 1D) [24, 25].
Figure 1. OGT regulates phosphorylation on serine, threonine and tyrosine residues of proteins.
Schematic representation of the interplay between O-phosphorylation and O-GlcNAcylation. A-C) Effect of O-GlcNAc modification on O-phosphate addition. (A) O-GlcNAc can occupy the same site as O-phosphate (reciprocal). (B) O-GlcNAc can occupy a nearby site that may cause steric hindrance to the addition of O-phosphate (sterically hindered). (C) O-GlcNAc may be added to a distinct site, separate from the phosphosite (independent). (D) O-GlcNAc can modify kinases and phosphatases that affect activity and thus phosphorylation of downstream targets.
Previous phosphoproteomic studies have focused on identifying changes in phosphorylation in cells with increased O-GlcNAc levels either by pharmacological inhibition of O-GlcNAcase or by overexpression of OGT [11, 20, 26]. We postulated that decreasing O-GlcNAcylation may provide more insight into the essential role of O-GlcNAc in cells, tissues, and organisms. Here, we applied a stable isotope labeling of amino acids in cell culture (SILAC)-based strategy to study phosphoproteomic changes induced by the deletion of OGT in cells [17, 27]. Using TiO2- and anti-phosphotyrosine antibody-based phosphopeptide enrichment, we identified 6,370 phosphosites on 2,070 phosphoproteins. We were able to quantify changes on 5,529 phosphosites by SILAC. Amongst them, a total of 364 phosphosites have been modulated by the deletion of OGT. We not only identified increased phosphorylation of vimentin, a known phosphoprotein regulated by O-GlcNAcylation, but also a number of novel proteins whose phosphorylation have not been reported to be regulated by OGT. These proteins include serine-protein kinase ATM, serine/threonine-protein kinase Chk1, mitogen-activated protein kinase 14 (MAPK14) and lysine-specific demethylase 2a. These proteins, and others identified in the screen, are predominantly involved in two different biological processes – cell cycle regulation and DNA damage response.
2. Material and methods
2.1 Reagents
Anti-ATM antibodies were purchased from Abcam (Cambridge, MA, USA), anti-ATM (pSer1987) antibodies from Calbiochem (Darmstadt, Germany), anti-OGT (DM17) and actin antibodies from Sigma (St. Louis, MO, USA), anti-γH2AX antibodies from Affinity Bioreagents (Golden, CO, USA), anti-H2AX and Stat3 from Santa Cruz Biotechnology (Dallas, TX, USA). Anti-p53, p-p53 (pSer15), Chk2, pChk2 (pThr68), AKT phosphosubstrate, pAKT (pSer473) and (pThr308), AKT, pStat3 (Tyr705) and anti-phosphotyrosine mouse mAb (pTyr-100) beads for immunoaffinity purification of phosphopeptides were purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-O-GlcNAc antibodies (CTD 110.6) were supplied by Core C4 (Johns Hopkins University School of Medicine, Baltimore, MD, USA). DMEM medium with and without Lysine and Arginine, fetal bovine serum (FBS), L-glutamine and antibiotics were from Invitrogen (Carlsbad, CA, USA). SILAC amino acids, 13C6, 15N2-Lysine and 13C6, 14N4-Arginine, were from Cambridge Isotope Laboratories (Andover, MA, USA). Titanspheres (TiO2, 5 μm beads) were from GL Sciences Inc. (Torrance, CA, USA). L-1-Tosylamide-2-phenylethyl chloromethyl ketone (TPCK) treated trypsin was from Worthington Biochemical Corp. (Lakewood, NJ, USA). All other reagents used in this study were from Fisher Scientific (Pittsburgh, PA, USA).
2.2 SILAC labeling and peptide preparation for mass spectrometry analysis
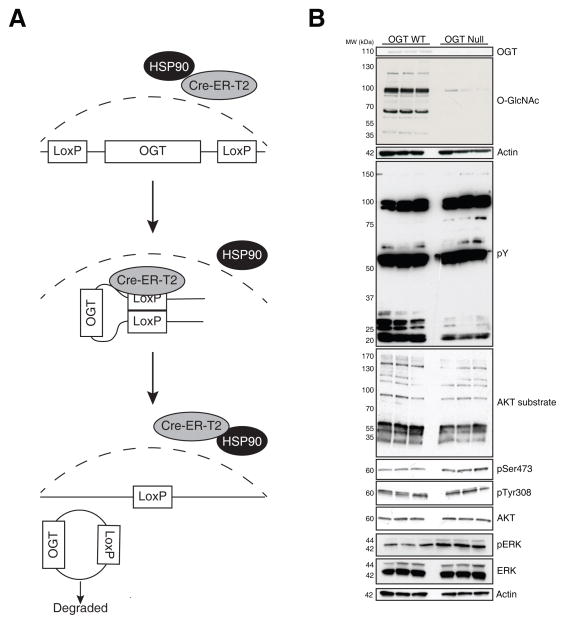
An immortalized mouse embryonic fibroblast (MEF) cell line in which OGT can be inducibly deleted was described previously [27]. Briefly, as shown in Figure 2A, MEFs in which the OGT allele is flanked by loxP recombination sites (OGTf/y) have been stably transduced with Cre-ErT2-GFP (OGTf/y Cre-ErT2-GFP). OGTf/y Cre-ErT2-GFP MEFs were maintained in DMEM medium supplemented with FBS, L-glutamine, penicillin, streptomycin at 37 °C in 5% CO2, and adapted to the SILAC media as described earlier [28]. MEFs grown in 13C6, 15N2-lysine/13C6, 15N4-arginine-containing media (heavy) and normal media (light) were plated at 5 × 105 cells per 100 mm plate. After 6 h, MEFs grown in heavy media were treated with 500 nM 4-hydroxytamoxifen (4-OHT) in ethanol for 12 h at 37 °C to induce deletion of OGT, and MEFs grown in light medium were treated with ethanol for 12 h at 37 °C as a control. 24 h prior to harvest, 4-OHT was removed and cells were fed complete medium.
Figure 2. Deletion of OGT in MEFs regulates phosphorylation events.
(A) Addition of 4-OHT can induce the recombination of Lox-P sites flanking the OGT allele, by promoting the translocation of mutated estrogen receptor-Cre-recombinase fusion protein into nucleus. (B) The effect of deleting OGT (36h) on protein phosphorylation was assessed by SDS-PAGE and western blot as indicated. As a control, the levels of OGT, O-GlcNAc and Actin were also assessed.
Peptides were prepared by an in-solution tryptic digestion protocol with modifications [29]. Briefly, OGT Null (4-OHT treated) or OGT WT (ethanol treated) MEFs (N=3) were lysed in lysis buffer (20 mM HEPES pH 8.0, 9 M urea, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 2 μM Thiamet-G), sonicated and cleared by centrifugation at 16,000 × g at 15 °C for 20 min. Equal amounts of protein (30 mg each) from the OGT WT and Null MEFs were mixed, reduced with 4.5 mM dithiothreitol and alkylated with 10 mM iodoacetamide. For tryptic digestion, protein extracts were diluted in 20 mM HEPES pH 8.0 to a final concentration of 2 M urea and incubated with TPCK-treated trypsin at 25 °C overnight. Protein digests were acidified by 1% v/v trifluoroacetic acid (TFA) and subjected to centrifugation at 2,000 × g at 25 °C for 5 min. The supernatant of the protein digests was loaded onto a Sep-Pak C18 column (Waters, Columbia, MD) equilibrated with 0.1% v/v TFA. Columns were washed with 6 ml of 0.1% v/v TFA twice and peptides were eluted in 2 ml of 40% v/v acetonitrile (ACN) with 0.1% v/v TFA three times. Eluted peptides were lyophilized and subjected to phosphopeptide enrichment.
2.3 Phosphopeptide enrichment strategy
The enrichment of phosphopeptides was carried out using the strategy described previously [30] that combines immunoaffinity purification (IAP) of phosphopeptides [29] with TiO2-based phosphopeptide enrichment following strong cation exchange chromatography fractionation (SCX) [31, 32]. Briefly, after lyophilization, 60 mg of peptides were split into two portions – 48 mg and 12 mg. 48 mg of peptide mixture was dissolved in 1.4 ml of IAP buffer (50 mM MOPS pH 7.2, 10 mM sodium phosphate, 50 mM NaCl) and subjected to centrifugation at 2,000 × g at room temperature for 5 min. Before IAP, pTyr-100 beads were washed with IAP buffer twice at 4 °C and the pH of the supernatant containing the peptides was adjusted to 7.2 by adding 1 M Tris base. For IAP, the supernatant was incubated with pTyr-100 beads (Cell Signaling Technology) at 4 °C for 30 min and the beads washed three times with IAP buffer and then twice with water. Peptides were eluted twice from beads by incubation with 0.15% TFA at room temperature.
The remaining 12 mg of lyophilized peptides were resuspended in 1 ml of solvent A (5 mM KH2PO4 pH 2.7, 30% v/v ACN) and fractionated by SCX on a PolySULPHOETHYL A (5 μm, 200 Å) column (200 × 9.4 mm; PolyLC Inc., Columbia, MD) using a linear gradient of solvent B (5 mM KH2PO4 pH 2.7, 30% v/v ACN, 350 mM KCl) on an Agilent 1100 LC system (Santa Clara, CA, USA). A total of 96 fractions were collected. For the whole proteomic analysis, 10% of the peptides from each fraction were pooled into 12 fractions and dried using a vacuum concentrator (Eppendorf, Hamburg, Germany). For the phosphoproteomic analysis, the remaining peptides (90%) in each fraction were pooled into 16 fractions, dried using a vacuum concentrator (Eppendorf) and subjected to TiO2-based phosphopeptide enrichment as described by Larsen et al. [31]. Briefly, TiO2 beads were pretreated by incubation with 2,5-dihydroxybenzoic acid (DHB) solution (80% v/v ACN, 1% v/v TFA, 3% w/v (DHB) for 2–4 hours at room temperature. Each fraction was resuspended in DHB and incubated with pretreated TiO2 beads. Phosphopeptide-bound TiO2 beads were washed three times with DHB solution and twice with 40% v/v ACN. Peptides were eluted three times with 40 μl of 2% v/v ammonia into 20 μl of 20% v/v TFA. The enriched phosphopeptides along with the fractionated peptides were cleaned up by C18 StageTip described previously [30].
2.4 Liquid chromatography tandem mass spectrometry
LC-MS/MS analysis of peptides and phosphopeptides was carried out using a reversed phase liquid chromatography system interfaced with an LTQ-Orbitrap Velos mass spectrometer. The mass spectrometer was operated in the “high-high” mode, where mass spectra of both precursor and product ions were acquired in the high resolution Orbitrap analyzer (Thermo Scientific). The peptides were loaded onto an analytical column (10 cm × 75 μm, Magic C18 AQ 5 μm, 120 Å) in 0.1% v/v formic acid and eluted using an ACN gradient (0–60% v/v) containing 0.1% v/v formic acid. The settings were: a) Precursor scans (FTMS) from 350–1,800 m/z at 60,000 resolution; and b) MS2 scan (FTMS) of HCD fragmentation of the 10 most intense ions (isolation width: 1.90 m/z; normalized collision energy: 35%; activation time=0.1 ms) at 7,500 resolution.
2.5 Mass spectrometry data analysis
The tandem mass spectra were searched using Andromeda algorithm [33] against a mouse UniProt database (released February 2014) through the MaxQuant platform (version 1.4.1.2) [34, 35]. The search parameters included: 2 SILAC states with Arg10 (13C6, 15N4-Arginine) and Lys8 (13C6, 15N2-Lysine); maximally-labeled amino acids of 3; a maximum of two missed cleavages; carbamidomethylation of cysteine as a fixed modification; oxidation of methionine; and phosphorylation of serine, threonine and tyrosine. The monoisotopic peptide tolerance was set to 10 ppm and the first search and main search for MS/MS were set to 20 ppm and 4.5 ppm, respectively. The maximum modifications per peptide was set to 6 and the maximum charge was set at 7. The reward type of the target-decoy analysis was chosen. The peptide-spectrum match (PSM) false discovery rate (FDR), protein FDR and the site decoy fraction were set to 0.01. The minimum peptide length was set to 7. The minimal scores for unmodified and modified peptides were 0 and 40, respectively. The minimal delta score for unmodified and modified peptides were 0 and 17, respectively. The minimum of unique and razor peptides for identification was set to 1. The ratio of heavy/light for each PSM was calculated using MaxQuant. The quantitation of identified proteins was carried out using at least 1 razor/unique non-phosphopeptide. The quantitation of each identified phosphosite was carried out using the least modified peptide and normalized. The probability of phosphorylation for each Ser/Thr/Tyr site on each peptide was calculated using Andromeda (MaxQuant).
2.6 Bioinformatics analysis
The Gene Ontology annotation of identified phosphoproteins was obtained from the PANTHER Classification System [36, 37] and the DAVID Bioinformatics Resource ver6.7 [38, 39]. The DNA damage response pathway map was constructed by manual curation of literatures [40–42] combined with the information from the DAVID Bioinformatics Resource.
2.7 Western blotting analysis
For western blot analysis, MEFs were grown as described above in DMEM (1 g/L glucose), 10% v/v FBS, Pen/Strep. As an alternative, U2OS cells were plated at 1×106 cells per 100 mm plate in DMEM (4.5 g/L glucose), 10% v/v FBS, Pen/Strep. All cells were maintained in a humidified water-jacketed incubator at 37 °C 5% CO2. Cells were extracted in NP-40 extraction buffer (1% v/v NP-40 in TBS, 2 mM EDTA, protease, phosphatase and glycosidase inhibitors) as described previously [27]. Total cell lysates were separated by SDS-PAGE, and proteins and phosphorylation events were detected by immunoblot as described previously [27]. Quantitation of western blots was performed using Image J.
2.8 Transfections and Transductions
O-GlcNAc levels were lowered in U2OS cells by viral transduction of O-GlcNAcase. Adenovirus expressing either GFP or O-GlcNAcase was placed on cells for 6 h at an multiplicity of infection of 50 [43]. For both transductions, reagents were removed at 6 h and cells were fed complete media. Media was changed at 24 h, and experiments initiated 18 h later.
2.9 Stress Treatments
U2OS and MEF cells in which O-GlcNAc levels had been suppressed (above) were subjected to 2 μM Doxorubicin (in DMSO) for the indicated lengths of time. Controls were treated with DMSO.
2.10 Nuclear and Cytoplasmic Extractions
Cells were washed in ice-cold PBS, pelleted and snap frozen. Nuclear and cytoplasmic extracts were performed as previously reported by Qing and co-workers [44]. The fractionation was assessed by immunoblotting for Tubulin (cytosolic) and Lamin-B (nuclear).
2.11 Statistical analysis
All statistical calculations were performed using GraphPad Prism 6 software® (San Diego, CA, USA). All data were assumed to be parametric. Student’s unpaired t-test was used to compare groups. Error bars represent the standard error of the mean. A p-value less than 0.05 was considered statistically significant.
3. Results
3.1 Inducible deletion of OGT results in complex changes to cellular phosphorylation
Previously, we have generated an immortalized MEF cell line in which deletion of OGT can be induced by addition of 4-OHT (Figure 2A). We have shown that this treatment results in significantly reduced OGT expression and thus O-GlcNAc levels at 36 h. Deletion of OGT is ultimately lethal in these cells. However, significant cell death does not occur until ~50 h [27]. Using these OGT WT and Null cells, we assessed changes in phosphorylation levels by immunoblotting (Figure 2B). Tyrosine phosphorylation displayed complex changes upon deletion of OGT, with some proteins exhibiting an increase in phosphorylation and others a decrease. O-GlcNAc has previously been shown to inhibit the AKT signaling pathway [45]. Using a substrate specific antibody, we probed the levels of AKT substrate phosphorylation. Our data suggest that deletion of OGT suppresses AKT signaling, which may reflect the enhanced sensitivity of these cells to injury. Next we assessed phosphorylation of AKT itself, focusing on phosphorylation of Thr308 and Ser473 which are thought to activate AKT. Phosphorylation of Thr308 was unaffected by deletion of OGT, whereas phosphorylation at Ser473 was paradoxically elevated. Finally, we probed for phosphorylation of ERK1/2, which was elevated in response to deletion of OGT. This is in contrast to previous findings that ERK1/2 phosphorylation is promoted by O-GlcNAcylation [46]. Together these data support a model in which there is a complex interplay between O-GlcNAc and phosphorylation that cannot simply be explained by competition between these two post-translational modifications for hydroxyl residues (Figure 1A). To provide insight into the phosphorylation events most affected by deletion of OGT, we performed a SILAC-based quantitative phosphoproteomic analysis of OGT Null MEFs.
3.2 A quantitative approach to identify changes in the phosphoproteome in response to OGT deletion
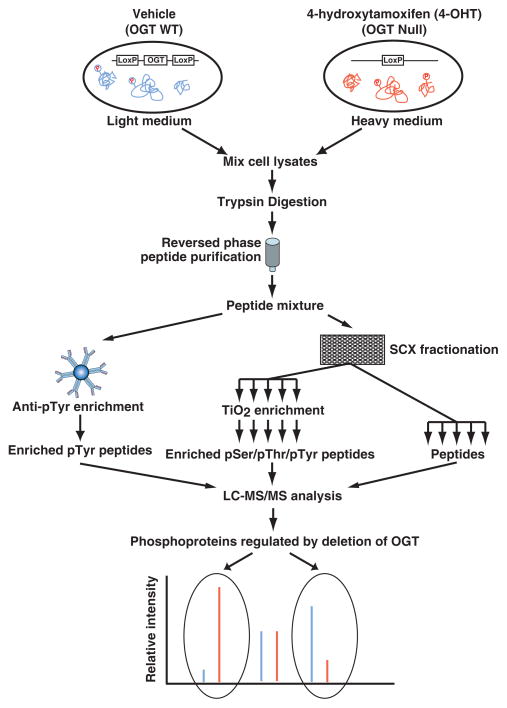
To systematically evaluate the effects of deleting OGT, and thus lowering O-GlcNAc levels on phosphorylation-based signaling pathways, we have used a SILAC-based approach outlined in Figure 3. Three separate biological replicates were assessed using this workflow. From each replicate, 80% of the peptides were first subjected to an anti-phosphotyrosine antibody-based phosphopeptide enrichment. The remaining peptides (20%) were fractionated into 96 fractions by SCX, 10% of which were merged into 12 fractions for quantifying changes in protein expression. The remaining 90% of peptides separated by SCX were merged into 16 fractions and subjected to a TiO2-based phosphopeptide enrichment. After the removal of salt by C18 StageTip cleanup, the fractionated peptides and the enriched phosphopeptides were analyzed by LC-MS/MS on an LTQ-Orbitrap Velos mass spectrometer.
Figure 3. The SILAC-based quantitative strategy to analyze changes in the phosphoproteome resulting from deletion of OGT in cells.
A schematic illustration of the SILAC-based quantitative whole proteomic/phosphoproteomic pipeline. Two populations of MEFs were cultured in ‘light’ or ‘heavy’ medium, separately. The cells grown in ‘light’ and ‘heavy’ medium were incubated in the presence of vehicle and 4-OHT, respectively. After lysis, the samples were mixed, digested with trypsin, desalted on a C18 column and subjected to lyophilization. 80% of the peptide mixture was subjected to anti-phosphotyrosine antibody-based phosphopeptide enrichment. The residual 20% of the peptide mixture was fractionated by SCX. The 90% of the SCX-fractionated peptides were merged and subjected to TiO2-based phosphopeptide enrichment. The enriched phosphopeptides from both enrichment methods were analyzed by LC-MS/MS. The residual 10% of SCX-fractionated peptides were also merged and analyzed by LC-MS/MS. The resulting high-resolution mass spectra reveal the changes of the phosphorylation status on each site and of the expression level of the protein by inducible deletion of OGT.
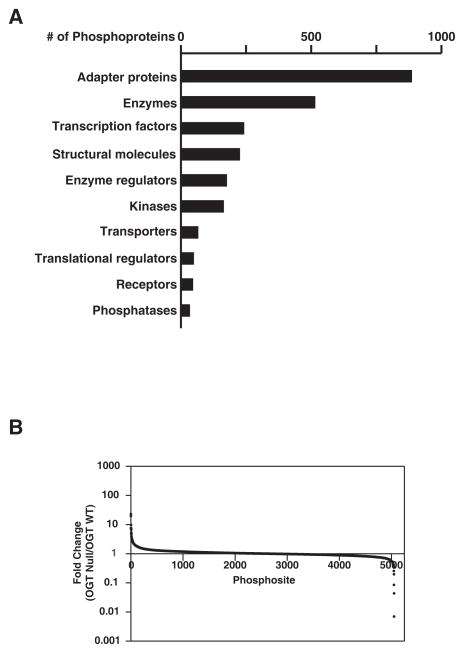
From three biological replicate experiments, 88 LC-MS/MS runs were carried out. A total 415,197 mass spectra were processed and searched against a Uniprot database using Andromeda [33] through MaxQuant (Version 1.4.1.2) [34, 35]. Using a FDR cut-off of 1% at both peptide and protein levels, a target-decoy analysis generated 152,700 PSM, of which 41,750 were phosphopeptide-spectrum-matches. After excluding reverse and contaminating matches, we identified a total of 3,516 protein groups and 19,623 peptides. Among these identified peptides, 5,603 were phosphopeptides that contained 6,370 phosphorylation sites (5,217 phosphoserine, 894 phosphothreonine and 259 phosphotyrosine sites). These phosphopeptides mapped to 2,070 phosphoproteins. Figure 4A shows the numbers of these phosphoproteins, categorized by molecular function based on the annotation by the PANTHER classification system. The majority of these proteins are adapter proteins and enzymes, including kinases and phosphatases, which are statistically overrepresented in our study.
Figure 4. Molecular function and relative distribution of changes in phosphoproteins regulated by OGT.
(A) The Gene Ontology annotation describing the numbers of identified phosphoproteins with different molecular functions as obtained from the PANTHER and DAVID bioinformatics tools. (B) Distribution of changes in phosphorylation levels on identified sites.
The quantitation of protein groups and phosphosites was carried out using the MaxQuant platform. The changes of identified proteins were carried out using the quantitation of non-phosphopeptides (Supplementary Table 1), while changes in phosphosites were calculated using the least-modified phosphopeptides and then normalized to the changes of the corresponding protein, if possible (Supplementary Table 2). We chose 1.5 fold-cutoff for increased protein phosphorylation and a 0.67 fold-cutoff for decreased phosphorylation. Figure 4B shows the distribution of the fold changes in phosphorylation events, among which the majority remain unchanged by the deletion of OGT. Of the phosphorylation sites identified and quantified in this study, phosphorylation of 232 serine, threonine or tyrosine residues was increased (Table 1, Supplementary Table 3), whereas, 131 serine, threonine or tyrosine residues exhibited decreased phosphorylation (Table 2, Supplementary Table 3). We also observed that the deletion of OGT altered the phosphorylation of a number of kinases and a phosphatase, including the serine/threonine kinase, ataxia telangiectasia mutated (ATM), checkpoint kinase 1 (Chk1) and the receptor tyrosine kinase c-Met (Figure 5; Table 3). Many of these enzymes are involved in the DNA damage response and regulation of the cell cycle, suggesting that OGT mediates signaling events within these pathways.
Table 1.
A partial list of phosphosites upregulated in OGT NULL MEFs compared with WT MEFs.
| Gene Symbol | Protein | Accession # | Phosphosite | Phosphopeptide Sequence | Fold Change |
|---|---|---|---|---|---|
| Arfgap2 | ADP-ribosylation factor GTPase-activating protein 2 | Q99K28 | S431 |
|
2.0 |
| Arhgef40 | Rho guanine nucleotide exchange factor 40 | Q3UPH7 | S959 |
|
1.7 |
| Atm | Serine-protein kinase ATM | Q62388 | S1987 |
|
2.5 |
| Bcr | Breakpoint cluster region protein | Q6PAJ1 | S113 |
|
1.7 |
| Cgn | Cingulin | P59242 | S129 |
|
1.8 |
| S131 |
|
1.7 | |||
| S437 |
|
1.9 | |||
| Chek1 | Serine/threonine-protein kinase Chk1 | O35280 | S317 |
|
2.2 |
| Cmtr1 | Cap-specific mRNA (nucleoside-2′-O-)-methyltransferase 1 | Q9DBC3 | S28, T29 |
|
1.7 |
| Eif4g1 | Eukaryotic translation initiation factor 4 gamma 1 | Q6NZJ6 | S1173 |
|
1.6 |
| Eif4g1 | Eukaryotic translation initiation factor 4 gamma 1 | Q6NZJ6 | S1197 |
|
1.9 |
| Ensa | Alpha-endosulfine | P60840 | S2 |
|
2.5 |
| Espl1 | Separin | P60330 | S239 |
|
2.0 |
| Hist1h1c; Hist1h1d | Histone H1.2;Histone H1.3 | P15864;P43277 | S37 |
|
2.3 |
| Hmga1 | High mobility group protein HMG-I/HMG-Y | P17095 | S6 |
|
2.7 |
| S9 |
|
2.2 | |||
| Hspb1 | Heat shock protein beta-1 | P14602 | S74 |
|
2.0 |
| Ifih1 | Interferon-induced helicase C domain- containing protein 1 | Q8R5F7 | S240 |
|
3.5 |
| S596, S599 |
|
3.3 | |||
| Kdm2a | Lysine-specific demethylase 2A | P59997 | S422 |
|
2.0 |
| Klc4 | Kinesin light chain 4 | Q9DBS5 | S590 |
|
1.7 |
| Maged2 | Melanoma-associated antigen D2 | Q9ER67 | T121 |
|
1.9 |
| Mapk14 | Mitogen-activated protein kinase 14 | P47811 | S2 |
|
2.5 |
| Mcm6 | DNA replication licensing factor MCM6 | P97311 | S13 |
|
2.1 |
| S704 |
|
2.0 | |||
| Micall1 | MICAL-like protein 1 | Q8BGT6 | S137 |
|
1.6 |
| Nbn | Nibrin | Q9R207 | S398 |
|
2.2 |
| Pafah1b2 | Platelet-activating factor acetylhydrolase IB subunit beta | Q61206 | S2 |
|
3.0 |
| Pdha1 | Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial | P35486 | S300 |
|
2.0 |
| Pkp4 | Plakophilin-4 | Q68FH0 | S220 |
|
9.8 |
| Plekha2 | Pleckstrin homology domain-containing family A member 2 | F8WIK5 | S314 |
|
1.7 |
| Pus7 | Pseudouridylate synthase 7 | Q91VU7 | S40 |
|
1.9 |
| Rab11fip1 | Rab11 family-interacting protein 1 | E9Q8L9 | S358 |
|
1.7 |
| Ranbp3 | Ran-binding protein 3 | Q9CT10 | S283 |
|
2.4 |
| T56 |
|
2.4 | |||
| Rnf213 | E3 ubiquitin-protein ligase RNF213 | E9Q555 | T46 |
|
3.3 |
| Rragc | Ras-related GTP-binding protein C | Q99K70 | S94 |
|
1.9 |
| T95 |
|
2.1 | |||
| Sept9 | Septin-9 | Q80UG5 | S30 |
|
1.8 |
| Snrnp70 | U1 small nuclear ribonucleoprotein 70 kDa | Q62376 | S408 |
|
7.6 |
| Trim28 | Transcription intermediary factor 1-beta | Q62318 | S471 |
|
1.8 |
| S473 |
|
1.9 | |||
| Ubap2 | Ubiquitin-associated protein 2 | Q91VX2 | S634 |
|
1.6 |
| Vim | Vimentin | P20152 | S430 |
|
2.3 |
| Yap1 | Yorkie homolog | P46938 | S94 |
|
1.7 |
Table 2.
A partial list of phosphosites downregulated in OGT NULL MEFs compared with WT MEFs.
| Gene Symbol | Protein | Accession # | Phosphosite | Phosphopeptide Sequence | Fold Change |
|---|---|---|---|---|---|
| Akap12 | A-kinase anchor protein 12 | Q9WTQ5 | S637 |
|
0.7 |
| Ice1 | Protein BC018507 | E9Q286 | S1820 |
|
0.5 |
| Met | Hepatocyte growth factor receptor | P16056 | Y1232 |
|
0.5 |
| Nrp1 | Neuropilin-1 | P97333 | Y920 |
|
0.6 |
| Ranbp10 | Ran-binding protein 10 | Q6VN19 | S365 |
|
0.6 |
| Tfeb | Transcription factor EB | Q9R210 | S108 |
|
0.4 |
| Ung | Uracil-DNA glycosylase | P97931 | S43 |
|
0.4 |
| Zcchc6 | Terminal uridylyltransferase 7 | E9PUA2 | S645 |
|
0.5 |
| Zdhhc5 | Palmitoyltransferase ZDHHC5 | Q8VDZ4 | S621 |
|
0.3 |
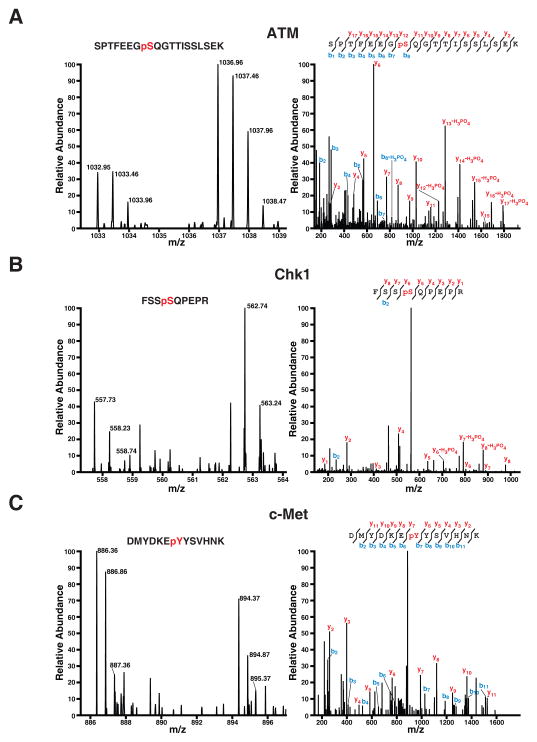
Figure 5. Phosphorylation of three kinases ATM, Chk1 and c-Met are changed by deletion of OGT.
MS spectra of one representative peptide each from ATM (A), Chk1 (B) and c-Met (C) are shown on the left. The panels on the right show the corresponding MS/MS spectra along with the peptide sequence.
Table 3.
A list of kinases and phosphatases regulated by OGT.
| Gene Symbol | Protein | Accession # | Phosphosite | Phosphopeptide Sequence | Fold Change |
|---|---|---|---|---|---|
| Atm | Serine-protein kinase ataxia telangiectasia mutated | Q62388 | S1987 |
|
2.5 |
| Chek1 | Serine/threonine-protein kinase Chk1 | G3UZ93 | T113 |
|
2.2 |
| S151 |
|
0.7 | |||
| Met | Hepatocyte growth factor receptor | P16056 | Y1232 |
|
0.5 |
| Mapk14 | Mitogen-activated protein kinase 14 | B2KF34 | S2 |
|
2.5 |
| Mapk3 | Mitogen-activated protein kinase 3 | Q63844 | T203 |
|
1.6 |
| Nek4 | Serine/threonine-protein kinase Nek4 | Q9Z1J2 | S430 |
|
1.8 |
| Pi4k2b | Phosphatidylinositol 4-kinase type 2-beta | Q8CBQ5 | S11 |
|
1.7 |
| Speg | Striated muscle-specific serine/threonine-protein kinase | Q62407 | S1281 |
|
1.5 |
| Sphk1 | Sphingosine kinase 1 | B1AT89 | S224 |
|
1.5 |
| Tk1 | Thymidine kinase, cytosolic | P04184 | S12 |
|
0.7 |
| Bcr | Breakpoint cluster region protein | Q6PAJ1 | S113 |
|
1.7 |
| Cdc25B | M-phase inducer phosphatase 2 | P30306 | S351 |
|
0.7 |
3.3 Inducible deletion of OGT changes phosphorylation on proteins involved in cell cycle and DNA damage response
Previous studies have characterized the phosphoproteome in cells in which O-GlcNAc levels have been enhanced either by inhibition of O-GlcNAcase or overexpression of OGT [16, 20]. In both mouse NIH3T3 cells [16] and Hela cells [20], elevation of O-GlcNAc appeared to positively and negatively regulate phosphorylation levels at distinct sites. We compared the phosphorylation sites regulated by deletion of OGT identified in our study with the phosphorylation sites regulated by elevating O-GlcNAc levels in mouse NIH3T3 cells. This comparison identified 13 phosphosites found in both studies (Supplementary Table 3). Among them, the phosphorylation of five sites that were upregulated by deletion of OGT were decreased by the inhibition of O-GlcNAcase. Similarly, the phosphorylation level of two sites that were downregulated by deletion of OGT were increased by the inhibition of O-GlcNAcase. The remaining six sites showed the same direction of changes in both studies. The results of this comparison suggest that increasing or decreasing O-GlcNAc levels may affect the cellular phosphoproteome at different points. Alternatively, these data may reflect the approaches used to modulate O-GlcNAc levels. In the OGT Null cells, there is no dynamic cycling of the O-GlcNAc modification, whereas in models where OGT has been overexpressed or O-GlcNAcase has been inhibited, cycling of the O-GlcNAc modification has been slowed. One other possibility is that modulation of OGT expression may affect cellular events in an O-GlcNAc independent manner, such as altering protein-protein interactions.
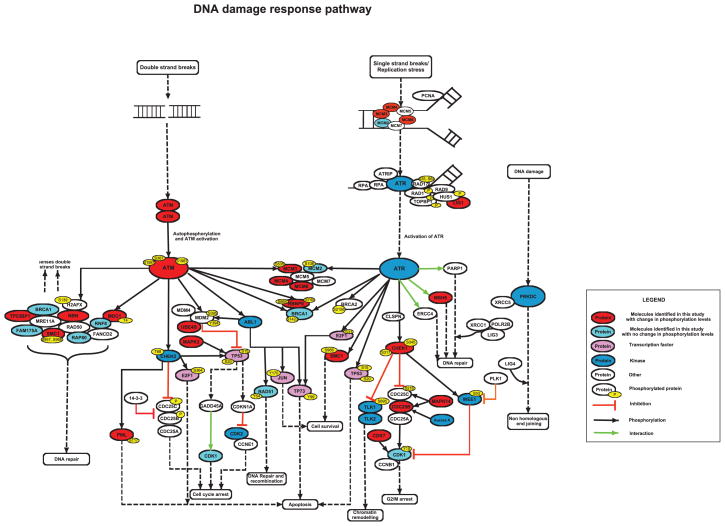
The DAVID bioinformatics resource (ver. 6.7) was used to search the biological process ontology terms of proteins with increased phosphorylation by inducible deletion of OGT (Figure 6). The proteins mapped into numerous pathways, although consistently these pathways were involved in regulating the cell cycle or DNA damage response. Several of these proteins are involved in homologous recombination, such as structural maintenance of chromosomes 1 and 3 (SMC1, SMC3) and ATM, or mismatch repair like mutS homolog 6 (MSH6). Due to incomplete annotation through DAVID, we sought to find whether other phosphoproteins quantified in our study are involved in DNA damage response by manual curation of the literature. The pathways and proteins outlined in Figure 6 demonstrate the phosphorylation events identified in our study in response to deletion of OGT, including those phosphoproteins identified that remain unchanged. These proteins appear to be most highly represented in double and single strand break repair pathways.
Figure 6. Phosphoproteins regulated by deletion of OGT are involved in DNA damage response pathway.
The phosphoproteins identified in our study were annotated in the DNA damage response pathway constructed through manual literature curation and DAVID bioinformatics resource. The legends are included in the inset. The data show that phosphoproteins involved in DNA damage response pathways are also upregulated by deletion of OGT.
3.4 The DNA Damage Kinase Ataxia Telangiectasia Mutated (ATM) and its Substrates are Basally Active in the OGT Null
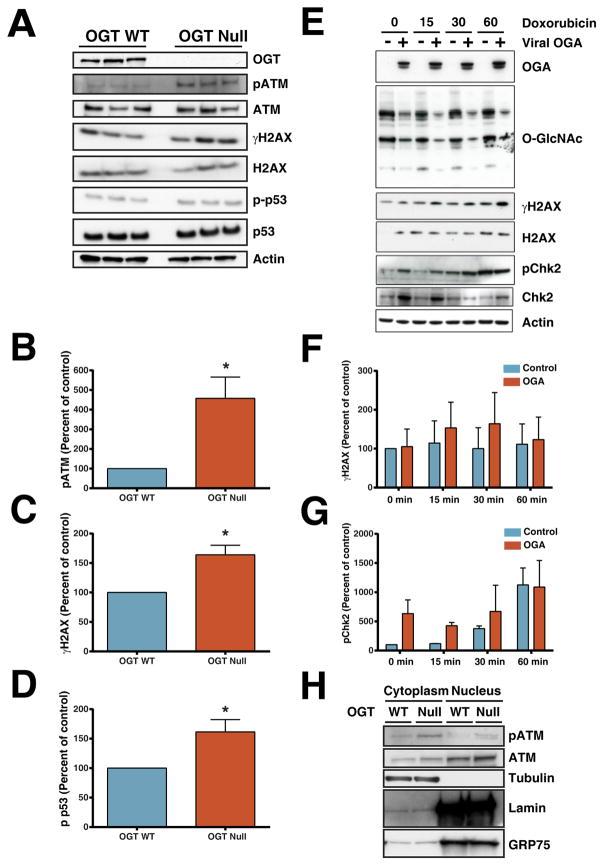
Because ATM regulates various cellular processes involved in the cellular stress response including, double strand break repair and cell cycle control [47], oxidative stress [48], and chromatin remodeling [49], we sought to further investigate the role of OGT in regulating ATM. ATM is a serine/threonine kinase belonging to the phosphatidylinositol 3-kinase related kinase (PIKK) family, and is activated by autophosphorylation of Ser1987 (Ser1981 in humans) [50]. Quantitation by MS analysis indicated that phosphorylation of this residue was increased 2.5 fold in OGT Null over WT, which was confirmed by western blot analysis (Figure 7A, B). Consistent with enhanced activation of ATM, phosphorylation of the ATM substrate p53 was elevated in response to OGT deletion (Figure 7A, D). Similarly, phosphorylation of the H2A histone family member X (H2AX), which is involved in DNA double strand break repair, was also elevated in OGT Null (Figure A, C). These data suggest that OGT regulates ATM, thus affecting downstream phosphorylation targets.
Figure 7. Deletion of OGT increases phosphorylation of ATM and its downstream signaling molecules.
(A) Deletion of OGT in MEFs increases phosphorylation of ATM at serine 1981 (pATM) and its downstream signaling molecules, H2AX and p53. The changes of the phosphorylation on ATM (B), H2AX (C), and p53 (D) were quantified by densitometry analysis from three independent experiments. (E) Overexpression of OGA in MEFs increases phosphorylation of the downstream targets of ATM, H2AX and Chk2. The changes in phosphorylation of H2AX (F), and Chk2 (G) were quantified by a densitometry analysis from three independent experiments. (H) Deletion of OGT can increase the phosphorylation of ATM in both nuclear and cytoplasmic fractions. pATM is present in both nuclear and cytoplasmic fractions in response to deletion of OGT.
To further evaluate the effects of decreasing O-GlcNAc levels on ATM, H2AX and serine/threonine-protein kinase Chk2 were examined in U2OS cells where O-GlcNAcase was overexpressed by viral transduction (Figure 7E, F, and G). Upon treatment of cells with the DNA intercalating agent, doxorubicin, γH2AX and pChk2 trended towards elevation in OGT Null compared to WT cells at 15 and 30 min.
Traditionally, induction of DNA double strand breaks causes ATM to become phosphorylated and translocate to the nucleus to participate in the repair process [50]. Recent studies suggest, however, that ATM is activated in response to other stressors [51]. ATM and pATM localization were evaluated in cytoplasmic and nuclear fractions. ATM was present in the cytoplasm and nucleus of both WT and OGT Null (Figure 7H). However, phosphorylated ATM is elevated in both the cytosolic and nuclear fractions of OGT Null cells. Thus, these data suggest that ATM phosphorylation may be regulated by O-GlcNAc in a manner independent of genomic instability.
3.5 Deletion of OGT alters the expression of a subset of proteins, including proteins involved in the biosynthesis of UDP-GlcNAc
We were able to compare the amount of 2,403 proteins from OGT WT and Null cells, of which 1,940 were quantified in at least two biological replicates. Surprisingly, only a modest number of proteins displayed increased expression or decreased expression (Table 4, Supplementary Table 4). We compared our data to a recent report in which O-GlcNAc levels were either elevated or suppressed using viral transduction of OGT or the O-GlcNAcase, and the effect on mitochondrial protein expression was quantified [52]. Comparison of these datasets with ours did not identify any corresponding proteins between the two studies or changes in individual protein expression levels. This is likely due to differences in experimental design, including choice of cell type (human SY5Y neuroblastoma cells vs. mouse embryonic fibroblasts) and the cellular fraction assessed (mitochondrial vs. nuclear and cytoplasmic).
Table 4.
A partial list of proteins regulated in OGT NULL MEFs compared with WT MEFs.
| Gene Symbol | Protein | Accession # | Fold Change |
|---|---|---|---|
| Atp6v1f | V-type proton ATPase subunit F | Q9D1K2 | 1.7 |
| Bst2 | Bone marrow stromal antigen 2 | Q8R2Q8 | 5.0 |
| Col5a2 | Collagen alpha-2(V) chain | Q3U962 | 0.5 |
| Fdps | Farnesyl pyrophosphate synthase | Q920E5 | 1.7 |
| Glg1 | Golgi apparatus protein 1 | Q61543 | 0.5 |
| Gng12 | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-12 | Q9DAS9 | 3.9 |
| Gnpda1 | Glucosamine-6-phosphate isomerase 1 | O88958 | 1.8 |
| Ifit1 | Interferon-induced protein with tetratricopeptide repeats 1 | Q64282 | 3.7 |
| Igtp | Protein Igtp | Q9DCE9 | 8.8 |
| Mt1 | Metallothionein-1 | P02802 | 1.8 |
| Parp12 | Poly [ADP-ribose] polymerase 12 | Q8BZ20 | 2.6 |
| Serpinc1 | Antithrombin-III | P32261 | 0.2 |
| Serpinf1 | Pigment epithelium-derived factor | P97298 | 0.2 |
| Snapc4 | snRNA-activating protein complex subunit 4 | Q8BP86 | 0.07 |
| Sprr1a | Cornifin-A | Q62266 | 2.3 |
| Timm8b | Mitochondrial import inner membrane translocase subunit Tim8 B | P62077 | 0.06 |
| Uap1l1 | UDP-N-acetylhexosamine pyrophosphorylase-like protein 1 | Q3TW96 | 1.8 |
| Ube2g1 | Ubiquitin-conjugating enzyme E2 G1 | P62254 | 1.7 |
| Ubxn7 | UBX domain-containing protein 7 | Q6P5G6 | 3.0 |
Many of the proteins identified in our study with changes in expression level appear to be involved in interferon based signaling pathways and ADP-ribosylation. In addition, several proteins involved in the synthesis of UDP-GlcNAc were upregulated. These proteins include UDP-N-acetylhexosamine pyrophosphorylase-1 (UAP1) and Glucosamine-6-phosphate isomerase 1 (Gnpda1). Previously, we and others have shown that the cell responds to pharmacological or genetic modulation of O-GlcNAc levels in an attempt to maintain basal O-GlcNAc levels [27, 43]. For instance, deletion of OGT results in suppression of expression of O-GlcNAcase. It’s possible that upregulation of enzymes within the hexosamine biosynthetic pathway is another mechanism used by the cell to maintain O-GlcNAc levels when the expression of OGT is suppressed.
4. Discussion
In this study we have evaluated and quantified the global phosphoproteome of OGT wild-type and Null cells using a SILAC-based methodology. In total, 5,529 phosphoserine, phosphothreonine and phosphotyrosine sites were quantified. Of those, 232 phosphosites were upregulated and 133 downregulated in the OGT Null compared with WT. In addition, from the 2,403 proteins quantified, 60 proteins had increased and 33 had decreased expression levels (Table 4, Supplementary Table 4). Previous studies in C. elegans have demonstrated that O-GlcNAc robustly regulates transcription and translation events involved in phosphatidylinositol 3,4,5 trisphosphate (PIP3) signaling, hexosamine biosynthetic pathway and lipid/carbohydrate metabolism [53]. While we also observed changes in the hexosamine biosynthetic pathway, widespread changes in protein levels were relatively modest, which may suggest that OGT is not a global regulator of transcription or translation in our model.
Comparison of our study with previous research examining changes in the phosphoproteome induced by elevating O-GlcNAc by inhibiting O-GlcNAcase identified 13 sites common to both studies, only 7 of which tracked with O-GlcNAc levels [20]. The remaining 6 sites were altered in the same direction in both studies, suggesting that particular pathways may attempt to compensate for changes in O-GlcNAc levels within the cell by utilizing similar pathways. Despite identification of 428 and 364 altered phosphorylation sites in response to elevated and decreased O-GlcNAc levels, respectively [16], we observed very little overlap in the sites that were identified. This may be indicative of differences in the chosen cell lines and experimental design or may suggest that the direction of the change of the O-GlcNAc level within the cell affects cellular processes differentially. These data may also reflect the approaches used to modulate O-GlcNAc levels. Inhibition of O-GlcNAcase increases cellular O-GlcNAc levels and reduces the cycling of the O-GlcNAc modification, whereas deletion of OGT decreases cellular O-GlcNAc level and terminates cycling.
We identified 11 kinases and 1 phosphatase with changed phosphorylation profiles in response to OGT deletion (Table 3). Of the 12 phosphorylation regulating enzymes identified, 7 are involved in the DNA damage response or regulation of the cell cycle and include c-Met, Mitogen activated protein kinase 3 (MAPK3; also known as ERK1), Mitogen activated protein kinase 14 (MAPK14; also known as p38α), Serine/threonine protein kinase Nek4 (Nek4), Serine/threonine protein kinase Chk1, Thymidine kinase 1 (TK1) and M-phase inducer phosphatase 2 (Cdc25B). For example, the phosphatase, Cdc25B, is phosphorylated by Aurora A kinase at Ser353 in human Hela cells [54]. This site is conserved in mouse, and was identified in our study. Phosphorylation of this residue results in co-localization with activated Aurora A kinase at the centrosome and induction of G-M phase transition via dephosphorylation of cyclins. As this site was found to have decreased phosphorylation levels, deletion of OGT may result in arrest at G-M phase. MAPK14 has also been shown to phosphorylate Cdc25B at Ser309 and Ser361, which results in binding to 14-3-3 proteins and initiation of the G-M phase checkpoint [55]. Furthermore, Chk1 phosphorylates Cdc25B in the non-catalytic N-terminus in response to DNA damage, inhibiting its phosphatase activity [56]. Together, these data may suggest that Cdc25B and its regulators may play an as yet unappreciated role in OGT regulation of DNA damage and the cell cycle.
Functional analysis by DAVID indicated that many of the phosphoproteins identified in our study are involved in DNA damage response (Figures 6 and 7). Manual literature curation, along with the functional annotation by DAVID, demonstrated an enrichment in proteins involved in both single and double strand DNA break repair (Figure 6). ATM, a key regulator of DNA damage repair, as well as additional cellular stress response pathways, was identified in our study (Figure 5). Induction of double strand breaks or other cellular stressors causes dissociation of inactive dimers to active monomers via ATM autophosphorylation [50]. Phosphorylation of Ser1987, which is the ATM activation site and is located in the FAT domain [50], was elevated 2.5 fold in OGT Null cells over WT (Figure 4). ATM is most noted for its role in DNA double strand break repair in which it has multiple roles. ATM participates in the MRN complex, composed of MRE11-RAD50-NBS1, which is recruited to the break site and acts as a bridge spanning the broken ends [57]. During the repair process, ATM interacts with MDC1 bound to histone H2AX, which allows for ATM phosphorylation of additional H2AX molecules, subsequently generating a positive feedback loop [58]. ATM activates and stabilizes p53 both directly through phosphorylation of p53 itself and indirectly through phosphorylation of ATM targets of p53, such as heterologous nuclear ribonucleoprotein K (HRNRPK) [59] and Chk2 [60]. Activated p53 then induces expression of cell cycle checkpoint and apoptotic genes [61].
We evaluated the role of ATM in DNA damage by addition of the DNA intercalating agent, doxorubicin, to USOS cells following overexpression of O-GlcNAcase. While not statistically significant, pChk2 trended towards elevation in OGT Null cells following overexpression of O-GlcNAcase. In addition, both pChk2 and γH2AX trended toward an increased response to doxorubicin treatment at 15 and 30 min time points (Figure 7E, F, G). This data suggests that ATM is activated via phosphorylation at Ser1987 in response to decreased O-GlcNAc levels, resulting in phosphorylation of downstream targets such as H2AX and Chk2 and possible activation of the DNA damage response. Previous studies have determined ATM to be O-GlcNAc modified [62]. We therefore assessed the glycosylation status of ATM in OGT WT MEFs. While we were able to immunoprecipitate ATM, we were unable to detect O-GlcNAcylation of ATM (data not shown), suggesting that OGT mediation of ATM is not likely through glycosylation of the enzyme. O-GlcNAc is required for precise cell cycle control, mitosis and cellular proliferation [10]. Thus it is possible that deletion of OGT induces genomic instability causing activation of ATM and its translocation to the nucleus. However, we have also observed that ATM is hyperphosphorylated in the cytosol suggesting a more complicated role for the regulation of ATM by OGT, possibly involving oxidative or other cellular stress processes. Indeed, previous studies have demonstrated ATM regulation of the pentose phosphate pathway by induction of the rate-limiting enzyme, glucose 6-phosphate dehydrogenase, which resulted in cellular protection from reactive oxygen species and an increase in the production of nucleotides [63]. Thus, OGT may negatively regulate ATM activation of the pentose phosphate pathway indirectly, in order to shunt glucose through the hexosamine biosynthetic pathway in an unstressed environment. Furthermore, O-GlcNAc modification of Ser529 of phosphofructokinase 1 (PFK1), the first committed step in glycolysis, inhibited the enzyme and redirected glucose flux through the pentose phosphate pathway [64]. Combined, the data from our group and others indicate that additional functional studies are necessary to evaluate the interplay between OGT and ATM in the regulation of the cellular stress response.
Here we have demonstrated that deletion of OGT results in global changes to both the proteome and phosphoproteome. The phosphoproteins identified in our study map primarily to DNA damage response and regulation of the cell cycle. Hence, future studies will be aimed at identifying the mechanisms by which OGT and O-GlcNAc regulate these pathways to maintain cellular homeostasis and the response to cellular stress.
Supplementary Material
Supplementary Table 1. A list of proteins quantified by SILAC.
Supplementary Table 2. A list of phosphorylated serine, threonine and tyrosine residues quantified by SILAC.
Supplementary Table 3. A list of phosphorylated serine, threonine and tyrosine residues regulated by an inducible deletion of OGT in MEFs.
Supplementary Table 4. A list of proteins regulated by an inducible deletion of OGT in MEFs.
Acknowledgments
The work was supported by a contract HHSN268201000032C from the National Heart Lung and Blood Institute (R.N.C. and A.P.) and by a grant S10RR023025 from the High End Instrumentation Program of the National Institutes of Health (A.P.) and an NIH Roadmap grant “Technology Center for Networks and Pathways” U54 RR 020839 (A.P.). The authors thank the National Heart, Lung, and Blood Institute PEG Core C4 for access to reagents and antibodies. Natasha E. Zachara, Ph.D., is funded by grants from the American Heart Association (SD0930162N) and the National Heart, Lung, and Blood Institute (R21-HL-108003 and P01-HL-107153). Marissa Martinez, Ph.D., receives funding as a GCF fellow from the National Heart, Lung and Blood Institute PEG Program (P01-HL-107153).
Abbreviations
- O-GlcNAc
O-Linked β-N-acetylglucosamine
- ATM
Ataxia telangiectasia mutated
- OGA
O-GlcNAcase
- OGT
O-GlcNAc transferase
- 4-OHT
4-hydroxytamoxifen
Footnotes
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [65] with the dataset identifier PXD001153.
Conflict of interest: Authors do not claim conflict of interest.
References
- 1.Lubas WA, Frank DW, Krause M, Hanover JA. O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. The Journal of biological chemistry. 1997;272:9316–9324. doi: 10.1074/jbc.272.14.9316. [DOI] [PubMed] [Google Scholar]
- 2.Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. The Journal of biological chemistry. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 3.Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. The Journal of biological chemistry. 2001;276:9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- 4.Dong DL, Hart GW. Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. The Journal of biological chemistry. 1994;269:19321–19330. [PubMed] [Google Scholar]
- 5.Hanover JA, Yu S, Lubas WB, Shin SH, et al. Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Archives of biochemistry and biophysics. 2003;409:287–297. doi: 10.1016/s0003-9861(02)00578-7. [DOI] [PubMed] [Google Scholar]
- 6.Wells L, Gao Y, Mahoney JA, Vosseller K, et al. Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic beta-N-acetylglucosaminidase, O-GlcNAcase. The Journal of biological chemistry. 2002;277:1755–1761. doi: 10.1074/jbc.m109656200. [DOI] [PubMed] [Google Scholar]
- 7.Shafi R, Iyer SP, Ellies LG, O’Donnell N, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahman MM, Stuchlick O, El-Karim EG, Stuart R, et al. Intracellular protein glycosylation modulates insulin mediated lifespan in C. elegans. Aging. 2010;2:678–690. doi: 10.18632/aging.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanover JA, Forsythe ME, Hennessey PT, Brodigan TM, et al. A Caenorhabditis elegans model of insulin resistance: altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci U S A. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang YR, Song M, Lee H, Jeon Y, et al. O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging cell. 2012;11:439–448. doi: 10.1111/j.1474-9726.2012.00801.x. [DOI] [PubMed] [Google Scholar]
- 11.Forsythe ME, Love DC, Lazarus BD, Kim EJ, et al. Caenorhabditis elegans ortholog of a diabetes susceptibility locus: oga-1 (O-GlcNAcase) knockout impacts O-GlcNAc cycling, metabolism, and dauer. Proc Natl Acad Sci U S A. 2006;103:11952–11957. doi: 10.1073/pnas.0601931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zachara NE, O’Donnell N, Cheung WD, Mercer JJ, et al. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. The Journal of biological chemistry. 2004;279:30133–30142. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]
- 13.Jones SP, Zachara NE, Ngoh GA, Hill BG, et al. Cardioprotection by N-acetylglucosamine linkage to cellular proteins. Circulation. 2008;117:1172–1182. doi: 10.1161/CIRCULATIONAHA.107.730515. [DOI] [PubMed] [Google Scholar]
- 14.Chou TY, Hart GW, Dang CV. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. The Journal of biological chemistry. 1995;270:18961–18965. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- 15.Musicki B, Kramer MF, Becker RE, Burnett AL. Inactivation of phosphorylated endothelial nitric oxide synthase (Ser-1177) by O-GlcNAc in diabetes-associated erectile dysfunction. Proc Natl Acad Sci U S A. 2005;102:11870–11875. doi: 10.1073/pnas.0502488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Gucek M, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc Natl Acad Sci U S A. 2008;105:13793–13798. doi: 10.1073/pnas.0806216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Molecular and cellular biology. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slawson C, Shafii S, Amburgey J, Potter R. Characterization of the O-GlcNAc protein modification in Xenopus laevis oocyte during oogenesis and progesterone-stimulated maturation. Biochimica et biophysica acta. 2002;1573:121–129. doi: 10.1016/s0304-4165(02)00369-0. [DOI] [PubMed] [Google Scholar]
- 19.Wells L, Vosseller K, Hart GW. Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science. 2001;291:2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Udeshi ND, Slawson C, Compton PD, et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc Natl Acad Sci U S A. 2009;106:8894–8899. doi: 10.1073/pnas.0900288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng X, Cole RN, Zaia J, Hart GW. Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor beta. Biochemistry. 2000;39:11609–11620. doi: 10.1021/bi000755i. [DOI] [PubMed] [Google Scholar]
- 23.Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, et al. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Molecular & cellular proteomics : MCP. 2012;11:215–229. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dias WB, Cheung WD, Hart GW. O-GlcNAcylation of kinases. Biochem Biophys Res Commun. 2012;422:224–228. doi: 10.1016/j.bbrc.2012.04.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dias WB, Cheung WD, Wang Z, Hart GW. Regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc modification. The Journal of biological chemistry. 2009;284:21327–21337. doi: 10.1074/jbc.M109.007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alfaro JF, Gong CX, Monroe ME, Aldrich JT, et al. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc Natl Acad Sci U S A. 2012;109:7280–7285. doi: 10.1073/pnas.1200425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazemi Z, Chang H, Haserodt S, McKen C, Zachara NE. O-linked beta-N-acetylglucosamine (O-GlcNAc) regulates stress-induced heat shock protein expression in a GSK-3beta-dependent manner. The Journal of biological chemistry. 2010;285:39096–39107. doi: 10.1074/jbc.M110.131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harsha HC, Molina H, Pandey A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat Protoc. 2008;3:505–516. doi: 10.1038/nprot.2008.2. [DOI] [PubMed] [Google Scholar]
- 29.Rush J, Moritz A, Lee KA, Guo A, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 30.Zhong J, Kim MS, Chaerkady R, Wu X, et al. TSLP signaling network revealed by SILAC-based phosphoproteomics. Molecular & cellular proteomics : MCP. 2012;11:M112 017764. doi: 10.1074/mcp.M112.017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Molecular & cellular proteomics : MCP. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox J, Neuhauser N, Michalski A, Scheltema RA, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. Journal of proteome research. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 34.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 35.Cox J, Matic I, Hilger M, Nagaraj N, et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nature protocols. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- 36.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas PD, Campbell MJ, Kejariwal A, Mi H, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 40.Yoo HY, Shevchenko A, Shevchenko A, Dunphy WG. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. The Journal of biological chemistry. 2004;279:53353–53364. doi: 10.1074/jbc.M408026200. [DOI] [PubMed] [Google Scholar]
- 41.Scully R, Chen J, Ochs RL, Keegan K, et al. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 42.Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci U S A. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slawson C, Zachara NE, Vosseller K, Cheung WD, et al. Perturbations in O-linked beta-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. The Journal of biological chemistry. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- 44.Qing G, Yan P, Xiao G. Hsp90 inhibition results in autophagy-mediated proteasome-independent degradation of IkappaB kinase (IKK) Cell research. 2006;16:895–901. doi: 10.1038/sj.cr.7310109. [DOI] [PubMed] [Google Scholar]
- 45.Vosseller K, Wells L, Lane MD, Hart GW. Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc Natl Acad Sci U S A. 2002;99:5313–5318. doi: 10.1073/pnas.072072399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tallent MK, Varghis N, Skorobogatko Y, Hernandez-Cuebas L, et al. In vivo modulation of O-GlcNAc levels regulates hippocampal synaptic plasticity through interplay with phosphorylation. The Journal of biological chemistry. 2009;284:174–181. doi: 10.1074/jbc.M807431200. [DOI] [PubMed] [Google Scholar]
- 47.Jazayeri A, Falck J, Lukas C, Bartek J, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nature cell biology. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 48.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 49.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 51.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nature cell biology. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 52.Tan EP, Villar MT, EL, Lu J, et al. Altering O-linked beta-N-Acetylglucosamine cycling disrupts mitochondrial function. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M113.525790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Love DC, Ghosh S, Mondoux MA, Fukushige T, et al. Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc Natl Acad Sci U S A. 2010;107:7413–7418. doi: 10.1073/pnas.0911857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dutertre S, Cazales M, Quaranta M, Froment C, et al. Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. Journal of cell science. 2004;117:2523–2531. doi: 10.1242/jcs.01108. [DOI] [PubMed] [Google Scholar]
- 55.Bulavin DV, Higashimoto Y, Popoff IJ, Gaarde WA, et al. Initiation of a G2/M checkpoint after ultraviolet radiation requires p38 kinase. Nature. 2001;411:102–107. doi: 10.1038/35075107. [DOI] [PubMed] [Google Scholar]
- 56.Comer FI, Hart GW. O-GlcNAc and the control of gene expression. Biochimica et biophysica acta. 1999;1473:161–171. doi: 10.1016/s0304-4165(99)00176-2. [DOI] [PubMed] [Google Scholar]
- 57.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 58.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 59.Moumen A, Magill C, Dry KL, Jackson SP. ATM-dependent phosphorylation of heterogeneous nuclear ribonucleoprotein K promotes p53 transcriptional activation in response to DNA damage. Cell cycle. 2013;12:698–704. doi: 10.4161/cc.23592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Advances in cancer research. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 61.Choi M, Shi J, Jung SH, Chen X, Cho KH. Attractor landscape analysis reveals feedback loops in the p53 network that control the cellular response to DNA damage. Sci Signal. 2012;5:ra83. doi: 10.1126/scisignal.2003363. [DOI] [PubMed] [Google Scholar]
- 62.Miura Y, Sakurai Y, Endo T. O-GlcNAc modification affects the ATM-mediated DNA damage response. Biochimica et biophysica acta. 2012;1820:1678–1685. doi: 10.1016/j.bbagen.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 63.Cosentino C, Grieco D, Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. The EMBO journal. 2011;30:546–555. doi: 10.1038/emboj.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yi W, Clark PM, Mason DE, Keenan MC, et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337:975–980. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vizcaino JA, Cote RG, Csordas A, Dianes JA, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. A list of proteins quantified by SILAC.
Supplementary Table 2. A list of phosphorylated serine, threonine and tyrosine residues quantified by SILAC.
Supplementary Table 3. A list of phosphorylated serine, threonine and tyrosine residues regulated by an inducible deletion of OGT in MEFs.
Supplementary Table 4. A list of proteins regulated by an inducible deletion of OGT in MEFs.