Abstract
Genetic factors contribute strongly to sex hormone levels, yet knowledge of the regulatory mechanisms remains incomplete. Genome-wide association studies (GWAS) have identified only a small number of loci associated with sex hormone levels, with several reproductive hormones yet to be assessed. The aim of the study was to identify novel genetic variants contributing to the regulation of sex hormones. We performed GWAS using genotypes imputed from the 1000 Genomes reference panel. The study used genotype and phenotype data from a UK twin register. We included 2913 individuals (up to 294 males) from the Twins UK study, excluding individuals receiving hormone treatment. Phenotypes were standardised for age, sex, BMI, stage of menstrual cycle and menopausal status. We tested 7 879 351 autosomal SNPs for association with levels of dehydroepiandrosterone sulphate (DHEAS), oestradiol, free androgen index (FAI), follicle-stimulating hormone (FSH), luteinizing hormone (LH), prolactin, progesterone, sex hormone-binding globulin and testosterone. Eight independent genetic variants reached genome-wide significance (P<5 × 10−8), with minor allele frequencies of 1.3–23.9%. Novel signals included variants for progesterone (P=7.68 × 10−12), oestradiol (P=1.63 × 10−8) and FAI (P=1.50 × 10−8). A genetic variant near the FSHB gene was identified which influenced both FSH (P=1.74 × 10−8) and LH (P=3.94 × 10−9) levels. A separate locus on chromosome 7 was associated with both DHEAS (P=1.82 × 10−14) and progesterone (P=6.09 × 10−14). This study highlights loci that are relevant to reproductive function and suggests overlap in the genetic basis of hormone regulation.
Introduction
Studies have suggested that genetic factors contribute significantly to population variance in sex hormone levels, however, few associated genetic variants and genes have been identified to date.1 As well as playing an important role in reproduction, variations in sex hormone levels can have wider implications for health and disease. Reproductive functions include control of the menstrual cycle, spermatogenesis, steroidogenesis and lactation, and sex hormone levels have been implicated in breast cancer, cardiovascular disease, osteoporosis, type 2 diabetes and ageing.2, 3, 4, 5 Circulating levels of sex hormones are limited by sex hormone-binding globulin (SHBG), which is a glycoprotein that binds and transports oestradiol, testosterone and dehydroepiandrosterone (DHEA) to a lesser extent.6
GWAS have been performed for dehydroepiandrosterone sulphate (DHEAS), SHBG, follicle-stimulating hormone (FSH), luteinizing hormone (LH), oestradiol and testosterone.2, 4, 7, 8, 9, 10 The largest of these GWAS was in over 28 000 males and females and identified 12 loci associated with differences in SHBG levels, including four loci with sex-specific genetic effects and considerable allelic heterogeneity at the SHBG gene locus.7 A recent study in 3495 Chinese men has identified a novel locus associated with oestradiol and FSH levels, and a further novel locus for oestradiol.10 A GWAS of total testosterone in males identified three loci, including two in the SHBG gene, that were also associated with SHBG levels.9 In an analysis of males and females combined, eight loci associated with DHEAS were identified, of which several were associated with changes in gene expression levels in pathways linked to ageing.4 GWAS studies of sex hormone-related phenotypes have explained less than 10% of variance in oestradiol and SHBG, and less than 5% of variance in testosterone, DHEAS and FSH.4, 7, 9, 10
In this study, we performed a 1000 Genomes imputed GWAS to identify novel genetic variants in sex hormone-related phenotypes where either GWAS has not yet been performed or has not been performed at 1000-Genomes-density variant coverage.
Materials and Methods
Study population
The study included up to 2913 individuals of European ancestry from the Twins UK study with genotype and phenotype data.11 Twins UK is a supported access resource with all data access requests overseen by the Twins UK Resource Executive Committee. All studies have ethical approval from the Guy's and St Thomas' Ethics Committee (for further information, see http://www.twinsuk.ac.uk/data-access/). The Twins UK cohort is 51% monozygotic and 49% dizygotic.11 Individuals included in the analysis were mostly females, however, a small number of males (maximum of 294) were also included (Supplementary Table 1). Individuals who were pregnant or currently receiving hormone replacement therapy or oral contraceptive treatments were excluded from the analysis. Twins UK samples have been included in previous GWAS of DHEAS and SHBG.4, 7
Phenotypes
Plasma levels of DHEAS, FSH, LH, oestradiol, progesterone, prolactin, SHBG and testosterone were measured by commercial ElectroChemiLuminescent immunoassays on a Modular Analytics E170 analyser (Roche Diagnostics GmbH, Mannheim, Germany) using the prescribed assay calibrators and performed according to the manufacturer's protocol. The specific assays used were: DHEA-S (03000087; CalSet 03000095), FSH (11775863; CalSet II 03032680), LH (11732234; CalSet II 03561097), Estradiol II (03000079; CalSet II 03064921), Progesterone II (12145383; CalSet 12145391), Prolactin II (03203093; CalSet 03277356), SHBG (03052001: CalSet 03052028) and Testosterone II (05200067; CalSet II 05202230). Details of the immunoassays are provided in the Supplementary Information. Free androgen index (FAI) was calculated as (testosterone/sex hormone-binding globulin) × 100.12 Individual sex hormone measures were fitted in a regression model against age, sex, BMI, phase of menstrual cycle (for females, as a categorical variable), menopausal status, after which the residuals were transformed to approximate a normal distribution (either through log, square-root or inverse rank normal transformation) and outliers more than four standard deviations from the mean were removed. Single nucleotide polymorphism (SNP) beta estimate effect sizes are quoted as a per-allele standard deviation change in the covariate-adjusted transformed residuals. The number of individuals included in the analysis of each hormone was 2899 for DHEAS, 2906 for oestradiol, 2699 for FAI, 2885 for FSH, 2881 for LH, 2865 for prolactin, 2689 for progesterone, 2913 for SHBG and 2657 for testosterone (differences in the numbers for FAI and testosterone are accounted for by removal of outliers prior to inclusion in the GWAS).
Genotypes
Genotyping of the TwinsUK dataset was done with HumanHap300, HumanHap610Q, HumanHap1M Duo and HumanHap1.2M Duo 1M arrays. Imputation was done in two datasets (n=2040 from the HumanHap300 array; n=3614 from the HumanHap610Q, HumanHap1M Duo and 1.2M Duo 1M arrays) which were then merged with GTOOL. We performed imputation for Twins UK study subjects based on 1000 Genomes data as described previously.13 This involved estimating the phase of contiguous variants in the subjects using the haplotypes calculated from the 1000 Genomes Project consisting of 1094 individuals and 2188 haplotypes and the program MACH 1.0.16. The variants in the build-37 November 2010 release of 1000 Genomes (Phase 1-α interim) were imputed into the phased haplotypes using MINIMAC. This resulted in 37 426 733 imputed SNPs. We excluded SNPs that were imputed with an r2imp<0.5. This left 10 879 115 SNPs and after filtering for minor allele frequency (MAF)>0.01 the number fell to 7 879 351. We used a multi-ethnic reference panel that included 381 Europeans (including 98 Tuscans), 181 Americans, 246 Africans and 286 Asians to improve the quality of imputation, particularly at lower frequency variants.14
Statistical analysis
We performed a linear mixed-model GWAS analysis for each of the hormones using the program GEMMA,15 which is capable of accounting for relatedness of the study subjects when applicable, as well as population stratification and cryptic relatedness. Association statistics using the score test were calculated for 7 879 351 autosomal SNPs passing a MAF filter of 0.01 and an imputation quality score of 0.5.
The Bonferroni-corrected P-value for the number of SNPs tested across nine traits was P<7 × 10−10; however, this is likely to be conservative given that many SNPs are unlikely to be independent and there are Bayesian arguments for less conservative P-values.13 Hence, we considered independent significant SNPs to be those with P<5 × 10−8 and more than 1 Mb away from another significant SNP. The UCSC Genome Browser and Locus Zoom were used to identify genes in the regions where significant SNPs were identified.16, 17 SNAP, HaploReg v2, Locus Zoom and Ensembl Biomart were used to identify HapMap proxies for the 1000 Genomes signals, with linkage disequilibrium evaluated in the 1000 Genomes Phase I CEU population.17, 18, 19, 20 We analysed expression quantitative trait loci data to identify associations between SNPs associated with variation in hormone levels and expression levels of nearby genes in the Multiple Tissue Human Expression Resource (MuTHER).21 Functional annotation of SNPs in strong linkage disequilibrium with the significant signals (r2>0.8) was performed using wANNOVAR, GWAVA and HaploReg v2.20, 22, 23
Results
Hormone phenotypes are correlated
There were strong correlations between three groups of phenotypes included in our study (Table 1): (i) FAI, SHBG and testosterone; (ii) progesterone, DHEAS and testosterone; and, (iii) FSH and LH. FAI was positively correlated with testosterone (r=0.69) and negatively correlated with SHBG (r=−0.61), as would be expected because FAI is a calculated index of the amount of androgen not bound by SHBG. Testosterone and SHBG were not correlated (r=0.04). Progesterone was positively correlated with DHEAS (r=0.60) and, to a lesser extent, testosterone (r=0.44). As a result of the correlation with testosterone, progesterone was also correlated with FAI (r=0.39). DHEAS was also positively correlated with testosterone (r=0.55) and, as a result, FAI (r=0.52). There was a strong positive correlation between FSH and LH (r=0.63). Though the other correlations were smaller, oestradiol was positively correlated with testosterone (r=0.22) and was negatively correlated with FSH (r=−0.24).
Table 1. Correlation coefficients between the sex hormone-related phenotypes.
| Oestradiol | Prolactin | DHEAS | SHBG | Progesterone | LH | FSH | Testosterone | ||
|---|---|---|---|---|---|---|---|---|---|
| log10 (Oestradiol residuals) | log10 (Prolactin residuals) | square root (DHEAS residuals) | ln (SHBG residuals) | log10 (Progesterone residuals) | LH residuals | inverse normal (FSH residuals) | Testosterone residuals | ||
| Oestradiol | log10 (Oestradiol residuals) | 1 | — | — | — | — | — | — | — |
| Prolactin | log10 (Prolactin residuals) | 0.09 | 1 | — | — | — | — | — | — |
| DHEAS | square root (DHEAS residuals) | 0.09 | −0.01 | 1 | — | — | — | — | — |
| SHBG | ln (SHBG residuals) | 0.08 | 0.06 | −0.15 | 1 | — | — | — | — |
| Progesterone | log10 (Progesterone residuals) | 0.17 | 0.04 | 0.60 | −0.06 | 1 | — | — | — |
| LH | LH residuals | 0.00 | 0.14 | 0.03 | 0.03 | 0.04 | 1 | — | — |
| FSH | inverse rank normal transformed (FSH residuals) | −0.24 | 0.08 | 0.01 | −0.01 | 0.00 | 0.63 | 1 | — |
| Testosterone | Testosterone residuals | 0.22 | 0.03 | 0.55 | 0.04 | 0.44 | 0.06 | −0.01 | 1 |
| FAI | FAI residuals | 0.11 | 0.00 | 0.52 | −0.61 | 0.39 | 0.02 | −0.01 | 0.69 |
Note: Values in bold are significant at P<0.05.
Three novel association signals
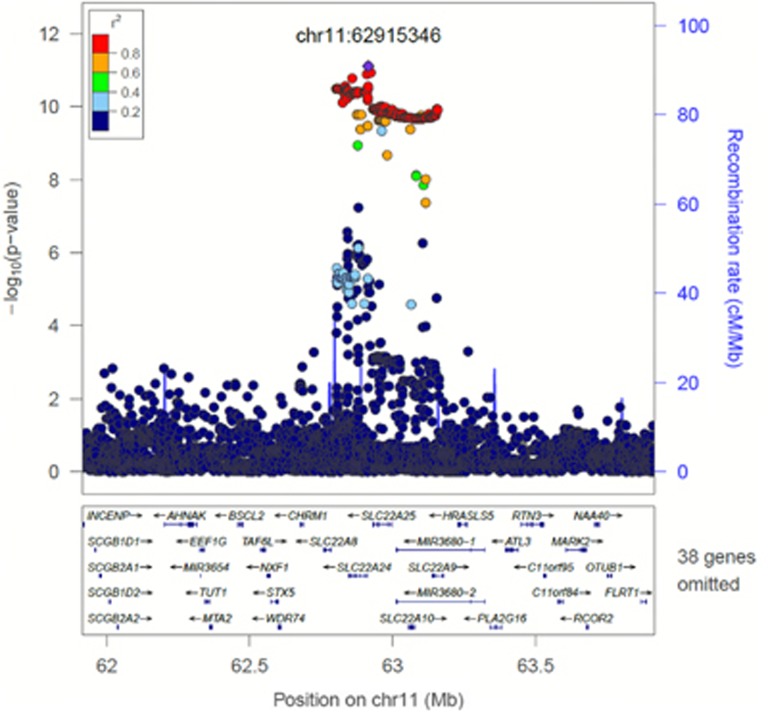
We identified new signals for progesterone, oestradiol and FAI (Table 2). The signal for progesterone (rs112295236, P=7.68 × 10−12, MAF=0.06) was identified in an intergenic region of chromosome 11 (Figure 1). We searched for associations of the progesterone SNP with other traits that might be influenced by progesterone levels, but we found no evidence for association with other phenotypes (P>0.05 in published GWAS of age at menopause, early menopause, age at menarche, BMI, height, type 2 diabetes and glycaemic traits, endometriosis, and birth weight (maternal and foetal genotype)) or in a GWAS of pre-term delivery (five gestational age/pre-term delivery traits for mother's and child's genotype, unpublished data). The signals for FAI and oestradiol were located at 16q12.2 (rs117145500, near LOC643714, P=1.50 × 10−8, MAF=0.06) and 12p13.31 (rs117585797, in ANO2, P=1.63 × 10−8, MAF=0.01), respectively, and demonstrated no association with other tested complex traits.
Table 2. Variants significantly associated with hormone levels (P<5 × 10−8).
| Hormonea | Chr-positionb | SNP id | Imputation quality | Minor allele frequency | Minor allele effect (% of sd)c | P-value | Location (gene) | Best candidate (distance) | Comments |
|---|---|---|---|---|---|---|---|---|---|
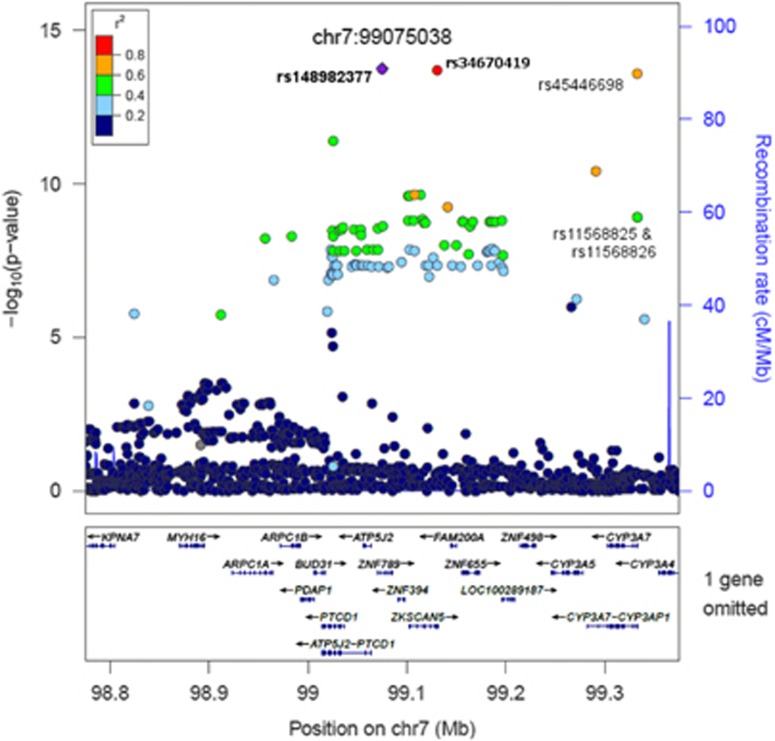
| DHEAS | chr7.hg19:g.99075038 T>C | rs148982377T>C | 0.927 | 0.038 | −53.1 | 1.82 × 10−14 | ZNF789 (intron) | CYP3A7 (228 kb) | Published signal for DHEAS.d Same signal as rs34670419 (progesterone)e |
| FAI | chr16.hg19:g.52947630 A>C | rs117145500A>C | 0.959 | 0.063 | −35.9 | 1.50 × 10−8 | intergenic | LOC643714 (307 kb) | New signal |
| FSH | chr11.hg19:g.30226356 T>C | rs11031005T>C | 0.978 | 0.129 | −23.2 | 1.74 × 10−8 | intergenic | FSHB (26 kb) | New signal. Overlaps with LH signal (rs11031002)e |
| LH | chr11.hg19:g.30215261 T>A | rs11031002T>A | 0.971 | 0.121 | 25.2 | 3.94 × 10−9 | intergenic | FSHB (37 kb) | New signal. Overlaps with FSH signal (rs11031005)e |
| Oestradiol | chr12.hg19:g.6011490C>A | rs117585797C>A | 0.572 | 0.013 | 87.1 | 1.63 × 10−8 | ANO2 (intron) | ANO2 (intronic) | New signal |
| Progesterone | chr7.hg19:g.99130834G>T | rs34670419G>T | 0.927 | 0.037 | −55.6 | 6.09 × 10−14 | ZKSCAN5 (3' UTR) | CYP3A4 (224 kb) or CYP3A7 (172 kb) | Published signal for DHEAS.d Same signal as rs148982377 (DHEAS)e |
| Progesterone | chr11.hg19:g.62915346C>G | rs112295236C>G | 0.962 | 0.062 | 41.0 | 7.68 × 10−12 | intergenic | SLC22A9 (222 kb) | New signal |
| SHBG | chr17.hg19:g.7574775C>T | rs1641549C>T | 0.895 | 0.239 | −28.0 | 1.21 × 10−15 | TP53 (intron) | SHBG (38 kb) | Published signal for testosteroned |
Abbreviations: Chr, chromosome; DHEAS, dehydroepiandrosterone sulphate; FAI, free androgen index; FSH, follicle-stimulating hormone; LH, luteinizing hormone; sd, standard deviation; SHBG, sex hormone binding globulin; UTR, untranslated region.
Results are for square root of the DHEAS residuals and FAI residuals; inverse rank normal transformed FSH residuals and LH residuals; log10 of the oestradiol residuals and progesterone residuals; and ln of the SHBG residuals.
Details of the reference sequence on which the variant descriptions are based is given in the Supplementary information.
Minor allele effect sizes are quoted as a per-allele change expressed as a percentage of a standard deviation in the covariate-adjusted transformed residuals.
Further details regarding the published genetic variants associated with reproductive hormones are given in Supplementary Table 2.
Effect sizes and P-values for each signal in each hormone phenotype are given in Supplementary Table 3.
Figure 1.
SNPs within 1 Mb of the significant signal for progesterone on chromosome 11 (rs112295236; chr11.hg19:g.62915346C>G). Note: Not all genes are shown. Linkage disequilibrium is based on 1000 Genomes Nov 2010 EUR.
Five signals in known regions
We identified two signals that replicated previous associations, and a further three that have been reported previously, but for other phenotypes (Table 2). The signals for SHBG (rs1641549, near SHBG gene, P=1.21 × 10−15, MAF=0.24) and DHEAS (rs148982377, in ZNF789, P=1.82 × 10−14, MAF=0.04) have both been reported previously4, 7, 9 (Supplementary Table 2). The significant associations for FSH (rs11031005; P=1.74 × 10−8, MAF=0.13) and LH (rs11031002; P=3.94 × 10−9, MAF=0.12) are highly correlated SNPs (r2=0.79), residing in an intergenic region near the FSHB gene, and are in linkage disequilibrium with a published variant for menopause age (rs12294104) (r2=0.37 for FSH (rs11031005) and r2=0.45 for LH (rs11031002)).24 The strongest association for progesterone (rs34670419; P=6.09 × 10−14, MAF=0.04) was located on chromosome 7 in the 3' untranslated region of ZKSCAN5 (Table 2; Supplementary Figure 1), a locus previously reported as associated with DHEAS levels.
Two pairs of phenotypes have common signals
We identified overlaps between signals for FSH/LH and progesterone/DHEAS (association results for significant signals in all phenotypes are in Supplementary Table 3). The signals for FSH and LH were in linkage disequilibrium (r2=0.79), and the most significant SNP for FSH also reached genome-wide significance for LH; however, the direction of effects was the opposite of that expected by the phenotypic correlation (ie, levels of FSH and LH are positively correlated, though the minor allele decreased FSH and increased LH (Supplementary Table 3). The strongest signal for progesterone on chromosome 7 (rs34670419) was in linkage disequilibrium with the signal for DHEAS (r2=1), with allelic effects consistent with the expected phenotypic correlation (Supplementary Table 3).
Overlap between DHEAS and progesterone variants
To investigate the genetic overlap of DHEAS and progesterone further, we tested whether five published variants for DHEAS (identified prior to conditional analysis) were associated with progesterone levels in our data.4 One of these variants reached genome-wide significance in our progesterone data (rs11761528, P=3.34 × 10−8) (Supplementary Table 4), and was at the same locus as our strongest progesterone signal (rs34670419, chr7:99130834). The published signal was not the strongest signal in our analysis though it was 12 kb from and in moderate linkage disequilibrium with our top chromosome 7 progesterone signal (r2=0.49). However, four of the five published variants were consistent in direction of effect (P=0.19) and two were nominally significant after Bonferroni correction (Supplementary Table 4). In addition, we investigated whether our two progesterone signals were significant in other published DHEAS meta-analysis data by looking up our two progesterone signals in data from Zhai et al.4 A proxy for our progesterone signal on chromosome 7 (r2=0.58) was strongly associated with DHEAS (P=2.34 × 10−34) and our progesterone signal on chromosome 11 showed weak evidence of association (P=1.53 × 10−4) (Supplementary Table 5).4
FSH and LH signals overlap with a menopause locus
There was evidence of overlap between the signals for FSH and LH with a variant for menopause age on chromosome 11 (Supplementary Tables 6–9). There was moderate linkage disequilibrium between the signals for FSH and LH and a published variant for menopause age (rs12294104), with r2=0.37 for FSH (rs11031005) and r2=0.45 for LH (rs11031002), P= 3.02 × 10−7 for FSH and P=6.25 × 10−7 for LH.24, 25 None of the other published menopause or menarche variants were associated with FSH or LH at Bonferroni-corrected P<0.05.24, 26
Identification of potentially causal candidate genes
The signal for SHBG (rs1641549) was 38 kb from the protein-coding gene SHBG. Four other signals were in linkage disequilibrium with known polymorphisms that have functional consequences: The signals for DHEAS (rs148982377) and progesterone (rs34670419) were in linkage disequilibrium with three SNPs (rs45446698, rs11568825 and rs11568826; r2>0.4 for all) that are part of a polymorphism in the promoter of CYP3A7, which is known to be associated with lower DHEAS levels (Figure 2). All three of these SNPs were genome-wide significant for DHEAS (P=2.49 × 10−14 for rs45446698; P=1.18 × 10−9 for rs11568825 and rs11568826) and one was genome-wide significant for progesterone (rs45446698, P=7.7 × 10−11), with the other two almost reaching significance (P<5 × 10−7). The top signals for LH and FSH were within 38 kb of, and in moderate linkage disequilibrium with, a known polymorphism (rs10835638) in the promoter of FSHB (rs11031005, r2=0.62; rs11031002, r2=0.74) (Supplementary Figure 2).27 Although the published FSHB promoter polymorphism was not the strongest signal in this region, it was genome-wide significant for association with LH in our data (P=4.84 × 10−9), and nearly significant for FSH (P=2.31 × 10−7) (Supplementary Table 11). Other candidate genes were identified by a search of a 300 kb region around each signal and are listed in Supplementary Table 10. Functional annotation of the signals with wANNOVAR did not reveal any additional likely causative variants.
Figure 2.
SNPs within 300 kb of the significant signal for DHEAS on chromosome 7 (rs148982377; chr7.hg19:g.99075038 T>C). SNPs indicated are the strongest progesterone signal (rs34670419 (chr7.hg19:g.99130834G>T)) and those in the CYP3A7 promoter polymorphism that were identified in this analysis (rs45446698 (chr7.hg19:g.99332948 T>G), rs11568825 (chr7.hg19:g.99332986 A>C) and rs11568826 (chr7.hg19:g.99332978 A>T)). Note: Not all genes are shown. Linkage disequilibrium is based on 1000 Genomes Nov 2010 EUR.
Discussion
In this study of nine sex hormone-related phenotypes, we identified three new signals and two pairs of phenotypes with a common signal. Four of the eight significant signals reached a conservative significance level of P<7 × 10−10, including the new signal for progesterone. This is the first published GWAS for the hormones progesterone, prolactin and the hormone measure FAI, and we identified genetic associations for all except prolactin. As we are not aware of any other genotyped cohorts with measurements for progesterone and FAI, we have been unable to replicate these findings. The hormones DHEAS, FSH, LH, SHBG, testosterone and oestradiol have been included in previously published GWAS, and we have compared our results with these existing data.2, 4, 7, 8, 9, 10 This study is, to our knowledge, one of the first published GWAS of hormones using 1000 Genomes Phase I imputed data. Three of the signals we identified were low frequency (less than 5%) and had large effect sizes (more than 50% change relative to standard deviation). In addition to identifying novel signals, we also observed two previously identified signals, demonstrating that true signals can be identified for these traits even in modest sample sizes.
The progesterone signal (rs112295236) that we identified on chromosome 11 was upstream of SLC22A9, which codes for an organic anion transporter OAT7 found in the liver. OAT7 is involved with the transport of DHEAS and oestrone-3-sulphate in exchange for butyrate, and is thought to be important for the release of oestrogen-3-sulphate into the blood.28 We did not find evidence to support this hypothesis in expression data, however, such data are only currently available for a limited range of tissues (skin, lymphoblastoid cell lines and adipose), not including ovary, which is the main site of progesterone synthesis. However, there was evidence of an association between this progesterone signal and DHEAS levels in data from a published GWAS, albeit at sub-genome-wide significance levels,4 and DHEAS and progesterone were both strongly correlated in our study (r=0.60). We postulated that the progesterone variant may be associated with phenotypic outcomes, such as offspring birth weight, or age at menopause, but we found no associations in data from other studies. Of course the effect size of our variant was relatively small and we may be underpowered to detect additional phenotypic associations.
The variant associated with decreased FAI in our analyses (rs117145500) showed some evidence of association with increased SHBG and decreased DHEAS, consistent with the role of DHEA in testosterone synthesis and the effect of SHBG on the amount of free androgens. This was supported by a negative correlation of FAI with SHBG in our data (r=−0.61) and positive correlation with DHEAS (r=0.55).
We identified a locus showing borderline significant association with oestradiol that requires further replication. The genetic variant associated with oestradiol (rs117585797) is a low frequency variant (MAF=0.013) with a large effect size, and is in an intron of the ANO2 gene on chromosome 12. A previous GWAS of oestradiol levels in postmenopausal women did not identify any genetic variants reaching genome-wide significance in this region, though this may have been underpowered to detect this signal.2 Two other genes are present within the same chromosomal region (vWF and CD9), though there is not strong evidence to support one as a more likely candidate over the other. VWF (47 kb away) codes for the von Willebrand factor (vWF) protein which is involved in haemostasis, aiding platelet adhesion and preventing factor VIII degradation. Oestradiol has been shown to increase vWF production by endothelial cells in vitro,29 and in post-menopausal women, oral oestrogen treatment has been shown to increase vWF (though transdermal treatment did not show this effect).30 CD9 (298 kb away) is a widely expressed cell surface molecule that has been shown to be required for sperm–egg fusion in mice.31
We provide evidence for overlap in the genetic regulation of two pairs of hormones whose levels are strongly correlated: FSH and LH (r=0.63); and progesterone and DHEAS (r=0.6). The FSH/LH variants were in linkage disequilibrium with a functional polymorphism (-211 G→T) in a progesterone response element of the promoter of the FSHB gene, which codes for the beta polypeptide of FSH. In females, FSH receptors are reported in endometrium32 and in granulosa cells. In granulosa cells, stimulation with FSH augments the expression of LH receptors.33 In vitro studies have demonstrated that the allele in linkage disequilibrium with the effect alleles in our study reduces levels of FSHB expression.34, 35 Previous studies provide conflicting data regarding the direction of effect of this polymorphism on FSH and LH levels.27, 36, 37 In our study, despite a positive overall correlation between FSH and LH levels, the genetic variants were negatively associated with FSH and positively associated with LH. Although this appears counter-intuitive, a similar situation is seen for other traits, for example, genetic variants that increase fasting glucose are not always risk factors for type 2 diabetes.38 Thus, the relationship between FSH and LH is complex and will involve additional genetic and non-genetic factors. A previous GWAS including FSH and LH did not find this signal, though this study was in Chinese men10 compared with our study of mainly female Europeans. Our study sheds light on part of the biological interaction between FSH and LH, but further variants need to be identified to understand this more fully.
The second pair of hormones with overlap in genetic regulation was DHEAS and progesterone. We identified a signal for progesterone on chromosome 7 in linkage disequilibrium with a known signal for DHEAS, plus evidence for association of progesterone levels with six of eight known DHEAS SNPs and of DHEAS levels with our newly identified chromosome 11 progesterone SNP. Both progesterone and DHEAS are steroid hormones that have a common precursor in their synthesis pathways (pregnenolone), but neither are directly synthesised from each other.3 These hormones were positively correlated in our data (r=0.60). The top signal for DHEAS in our study was rs148982377, which tagged a polymorphism in the promoter of CYP3A7, which was also in linkage disequilibrium with the top progesterone signal. CYP3A7 has a progesterone response element that is thought to regulate expression during pregnancy.39, 40, 41 In the same region is the CYP3A4 gene, which codes for a cytochrome P450 enzyme that metabolises progesterone, DHEAS, oestrone and testosterone.42
The main limitation of our study is the absence of suitable replication cohorts for the hormone measures progesterone, oestradiol and FAI. Half of the signals reached a conservative, Bonferonni-adjusted significance level of P<7 × 10−10, giving us confidence in the findings for progesterone, DHEAS and SHBG. Although the signals for FAI, FSH, LH and oestradiol reached a less conservative significance threshold, there are strong arguments for the validity of less stringent P-values in 1000 Genomes imputed GWAS.13 Further large studies are required to enable validation of our results. Such studies should also allow detection of new signals because the power of our study was limited by sample size and by the relatedness of the study individuals. Approximately 10% of our cohort consisted of males, and thus we will have been more likely to detect genetic effects in females than males, though our ability to detect genetic variants affecting both sexes should not have been affected. The cohort used in our study was of European ancestry and as such our findings will need to be replicated in other ethnic populations. Further GWAS studies of these hormones may benefit from an increase in power by implementing newly emerging multi-variate methods which would take account of correlations between the hormones.43 Any additional studies should also consider the need to directly quantify the effect of the variants, as it is difficult to relate the adjusted and transformed hormone measure that we used for our analysis to actual physiological changes. Further studies are required to address these issues.
In this GWAS of nine sex hormone-related phenotypes, we were able to detect three new signals (oestradiol, FAI and progesterone traits), two pairs of signals overlapping with other traits (FSH/LH and progesterone/DHEAS) and two signals seen before (DHEAS and SHBG traits). We have demonstrated potential overlap in the genetics of hormone regulation, as might be expected from common pathways in hormone synthesis. As well as the overlap in the top signals for DHEAS and progesterone, and FSH and LH, there were other variants associated with more than one hormone at lower significance levels, suggesting further commonality in hormone regulation. We identified novel genetic variants and potential overlap in the genetic basis of hormone regulation that will inform future studies, not only of hormones but also of common diseases, ageing and reproductive lifespan that include sex related hormones on the aetiological pathway.
Acknowledgments
We thank Roche Diagnostics Australia Pty Limited, Castle Hill, Australia, who provided support for the analysis of the hormones. We thank the volunteer twins for their participation in the study. Twins UK received funding support from NIHR Biomedical Research Centre (grant to Guys' and St Thomas' Hospitals and King's College London); the Chronic Disease Research Foundation; Canadian Institutes of Health Research, the Canadian Foundation for Innovation, the Fonds de la Recherche en Santé Québec, The Lady Davis Institute, the Jewish General Hospital and Ministère du Développement économique, de l'Innovation et de l'Exportation du Quebec. The Australian National Health and Medical Research Council (NHMRC project grants 1010494, 1048216), and Sir Charles Gairdner Hospital Research (grant PP2009/028). This work was supported by funding from the Wellcome Trust (092447/Z/10/Z) and Medical Research Council (MC_U106179472).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- 1Vandenput L, Ohlsson C: Genome-wide association studies on serum sex steroid levels. Mol Cell Endocrinol 2013; 382: 758–766. [DOI] [PubMed] [Google Scholar]
- 2Prescott J, Thompson DJ, Kraft P et al: Genome-wide association study of circulating estradiol, testosterone, and sex hormone-binding globulin in postmenopausal women. PLoS One 2012; 7: e37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Nussey S, Whitehead S: Endocrinology: An Integrated Approach. Oxford: BIOS Scientific Publishers, 2001. [PubMed] [Google Scholar]
- 4Zhai G, Teumer A, Stolk L et al: Eight common genetic variants associated with serum DHEAS levels suggest a key role in ageing mechanisms. PLoS Genet 2011; 7: e1002025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Perry JRB, Weedon MN, Langenberg C et al: Genetic evidence that raised sex hormone binding globulin (SHBG) levels reduce the risk of type 2 diabetes. Hum Mol Genet 2010; 19: 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Hammond GL, Bocchinfuso WP: Sex hormone-binding globulin: gene organization and structure/function analyses. Horm Res 1996; 45: 197–201. [DOI] [PubMed] [Google Scholar]
- 7Coviello AD, Haring R, Wellons M et al: A genome-wide association meta-analysis of circulating sex hormone-binding globulin reveals multiple Loci implicated in sex steroid hormone regulation. PLoS Genet 2012; 8: e1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Jin G, Sun J, Kim ST et al: Genome-wide association study identifies a new locus JMJD1C at 10q21 that may influence serum androgen levels in men. Hum Mol Genet 2012; 21: 5222–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Ohlsson C, Wallaschofski H, Lunetta KL et al: Genetic determinants of serum testosterone concentrations in men. PLoS Genet 2011; 7: e1002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Chen Z, Tao S, Gao Y et al: Genome-wide association study of sex hormones, gonadotropins and sex hormone-binding protein in Chinese men. J Med Genet 2013; 50: 794–801. [DOI] [PubMed] [Google Scholar]
- 11Moayyeri A, Hammond CJ, Hart DJ, Spector TD: The UK Adult Twin Registry (TwinsUK Resource). Twin Res Hum Genet 2013; 16: 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Christ-Crain M, Meier C, Huber P, Zimmerli L, Trummler M, Muller B: Comparison of different methods for the measurement of serum testosterone in the aging male. Swiss Med Wkly 2004; 134: 193–197. [DOI] [PubMed] [Google Scholar]
- 13Wood AR, Perry JRB, Tanaka T et al: Imputation of variants from the 1000 Genomes Project modestly improves known associations and can identify low-frequency variant - phenotype associations undetected by HapMap based imputation. PLoS One 2013; 8: e64343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Marchini J, Howie B: Genotype imputation for genome-wide association studies. Nat Rev Genet 2010; 11: 499–511. [DOI] [PubMed] [Google Scholar]
- 15Zhou X, Stephens M: Genome-wide efficient mixed-model analysis for association studies. Nat Genet 2012; 44: 821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Meyer LR, Zweig AS, Hinrichs AS et al: The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 2013; 41: D64–D69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Pruim RJ, Welch RP, Sanna S et al: LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010; 26: 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Flicek P, Amode MR, Barrell D et al: Ensembl 2012. Nucleic Acids Research 2012; 40: D84–D90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PIW: SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008; 24: 2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Ward LD, Kellis M: HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012; 40: D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Grundberg E, Small KS, Hedman AK et al: Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet 2012; 44: 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Chang X, Wang K: wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet 2012; 49: 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Ritchie GR, Dunham I, Zeggini E, Flicek P: Functional annotation of noncoding sequence variants. Nat Methods 2014; 11: 294–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Stolk L, Perry JR, Chasman DI et al: Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat Genet 2012; 44: 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Stolk L, Zhai G, van Meurs JBJ et al: Loci at chromosomes 13, 19 and 20 influence age at natural menopause. Nat Genet 2009; 41: 645–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Elks CE, Perry JR, Sulem P et al: Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet 2010; 42: 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Schuring AN, Busch AS, Bogdanova N, Gromoll J, Tuttelmann F: Effects of the FSH-beta-subunit promoter polymorphism -211G->T on the hypothalamic-pituitary-ovarian axis in normally cycling women indicate a gender-specific regulation of gonadotropin secretion. J Clin Endocrinol Metab 2013; 98: E82–E86. [DOI] [PubMed] [Google Scholar]
- 28Koepsell H: The SLC22 family with transporters of organic cations, anions and zwitterions. Mol Aspects Med 2013; 34: 413–435. [DOI] [PubMed] [Google Scholar]
- 29Harrison RL, McKee PA: Estrogen stimulates von Willebrand factor production by cultured endothelial cells. Blood 1984; 63: 657–664. [PubMed] [Google Scholar]
- 30Rabbani LE, Seminario NA, Sciacca RR, Chen HJ, Giardina EG: Oral conjugated equine estrogen increases plasma von Willebrand factor in postmenopausal women. J Am Coll Cardiol 2002; 40: 1991–1999. [DOI] [PubMed] [Google Scholar]
- 31Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C: Severely reduced female fertility in CD9-deficient mice. Science 2000; 287: 319–321. [DOI] [PubMed] [Google Scholar]
- 32La Marca A, Carducci Artenisio A, Stabile G, Rivasi F, Volpe A: Evidence for cycle-dependent expression of follicle-stimulating hormone receptor in human endometrium. Gynecol Endocrinol 2005; 21: 303–306. [DOI] [PubMed] [Google Scholar]
- 33Hunzicker-Dunn M, Maizels ET: FSH signaling pathways in immature granulosa cells that regulate target gene expression: branching out from protein kinase A. Cell Signal 2006; 18: 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Benson CA, Kurz TL, Thackray VG: A human FSHB promoter SNP associated with low FSH levels in men impairs LHX3 binding and basal FSHB transcription. Endocrinology 2013; 154: 3016–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Hoogendoorn B, Coleman SL, Guy CA et al: Functional analysis of human promoter polymorphisms. Hum Mol Genet 2003; 12: 2249–2254. [DOI] [PubMed] [Google Scholar]
- 36Grigorova M, Punab M, Ausmees K, Laan M: FSHB promoter polymorphism within evolutionary conserved element is associated with serum FSH level in men. Hum Reprod 2008; 23: 2160–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Grigorova M, Punab M, Ẑilaitienė B et al: Genetically determined dosage of follicle-stimulating hormone (FSH) affects male reproductive parameters. J Clin Endocrinol Metab 2011; 96: E1534–E1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Scott RA, Lagou V, Welch RP et al: Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 2012; 44: 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Burk O, Tegude H, Koch I et al: Molecular mechanisms of polymorphic CYP3A7 expression in adult human liver and intestine. J Biol Chem 2002; 277: 24280–24288. [DOI] [PubMed] [Google Scholar]
- 40Itoh S, Yanagimoto T, Tagawa S et al: Genomic organization of human fetal specific P-450IIIA7(cytochrome P-450HFLa)-related gene(s) and interaction of transcriptional regulatory factor with its DNA element in the 5′ flanking region. Biochim Biophys Acta 1992; 1130: 133–138. [DOI] [PubMed] [Google Scholar]
- 41Smit P, van Schaik RH, van der Werf M et al: A common polymorphism in the CYP3A7 gene is associated with a nearly 50% reduction in serum dehydroepiandrosterone sulfate levels. J Clin Endocrinol Metab 2005; 90: 5313–5316. [DOI] [PubMed] [Google Scholar]
- 42Kliewer SA, Lehmann JM, Milburn MV, Willson TM: The PPARs and PXRs: nuclear xenobiotic receptors that define novel hormone signaling pathways. Recent Prog Horm Res 1999; 54: 345–367. [PubMed] [Google Scholar]
- 43Galesloot TE, van Steen K, Kiemeney LA, Janss LL, Vermeulen SH: A comparison of multivariate genome-wide association methods. PLoS One 2014; 9: e95923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.