Abstract
Background
Altered expression of S100A16 has been reported in human cancers, but its biological role in tumorigenesis is not fully understood. This study aimed to investigate the clinical significance and functional role of S100A16 in oral squamous cell carcinoma (OSCC) suppression.
Methods
S100A16 mRNA and/or protein levels were examined by quantitative RT-PCR and immunohistochemistry in whole- and laser microdissected-specimens of normal human oral mucosa (NHOM, n = 65), oral dysplastic lesions (ODL, n = 21), OSCCs (n = 132) and positive cervical nodes (n = 17). S100A16 protein expression in OSCC was examined for correlations with clinicopathological variables and patient survival. S100A16 was over-expressed and knocked-down in OSCC-derived (CaLH3 and H357) cells by employing retroviral constructs to investigate its effects on cell proliferation, sphere formation and three dimensional (3D)-organotypic invasive abilities in vitro and tumorigenesis in a mouse xenograft model.
Results
Both S100A16 mRNA and protein levels were found to be progressively down-regulated from NHOM to ODL and OSCC. Low S100A16 protein levels in OSCC significantly correlated with reduced 10-year overall survival and poor tumor differentiation. Analysis of two external OSCC microarray datasets showed a positive correlation between the mRNA expression levels of S100A16 and keratinocyte differentiation markers. CaLH3 and H357 cell fractions enriched for differentiated cells either by lack of adherence to collagen IV or FACS sorting for low p75NTR expression expressed significantly higher S100A16 mRNA levels than the subpopulations enriched for less differentiated cells. Corroborating these findings, retroviral mediated S100A16 over-expression and knock-down in CaLH3 and H357 cells led to respective up- and down-regulation of differentiation markers. In vitro functional studies showed significant reduction in cell proliferation, sphere formation and 3D-invasive abilities of CaLH3 and H357 cells upon S100A16 over-expression. These functional effects were associated with concomitant down-regulation of self-renewal (Bmi-1 and Oct 4A) and invasion related (MMP1 and MMP9) molecules. S100A16 over-expression also suppressed tumorigenesis of H357 cells in a mouse xenograft model and the resulting tumor xenografts displayed features/expression of increased differentiation and reduced proliferation/self-renewal.
Conclusions
These results indicate that S100A16 is a differentiation promoting protein and might function as a tumor suppressor in OSCC.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-015-1622-1) contains supplementary material, which is available to authorized users.
Background
Oral squamous cell carcinoma (OSCC) is an aggressive neoplasm which is highly invasive and frequently metastasizes to cervical lymph nodes leading to a severely reduced patient survival. Despite recent advances in diagnosis and treatment modalities, less than 50 % of OSCC patients survive for 5 years [1]. Among the molecular and cellular changes occurring during OSCC development, a significant disturbance in cellular differentiation and maturation process has been reported to be a common event in oral carcinogenesis [2–4]. Nevertheless, the precise molecular mechanism regulating differentiation and its contribution to OSCC progression is not fully understood.
The S100 protein family is a multifunctional group of EF-hand calcium binding proteins. This family consists of small acidic proteins (10–12 kDa) that are expressed only in vertebrates in a cell and tissue specific manner. To date, 25 S100 protein members have been described in humans [5, 6]. Genes encoding several of the members of this family are clustered in the epidermal differentiation complex (EDC) on chromosome 1q21 [7–9], and many of the S100 members have been reported to be involved in cellular differentiation and differentiation-related pathologies [10, 11]. In addition, S100 proteins have recently been implicated in the regulation of epithelial-mesenchymal transition, cancer stem cells and tumor heterogeneity in human malignancies [12–14].
S100A16 is a recent addition to the S100 protein family [15]. Although it has been reported to be widely expressed in human tissues [15], its precise biological functions are not fully understood. In a recent study, S100A16 has been suggested to be related with cell invasion and poor prognosis in human breast cancer [16]. We have identified S100A16 to be an interaction partner of S100A14, a proliferation and invasion-related protein in OSCC [17–19]. These observations indicate that S100A16 might be related with OSCC progression. Nevertheless, functional roles and prognostic significance of this protein are currently unknown in OSCC. In the current study, we demonstrate that down-regulation of S100A16 expression in OSCC specimens was associated with poor prognosis and poor differentiation grade. Experimentally, S100A16 was found to promote malignant keratinocyte differentiation and to suppress aggressive tumor phenotype such as proliferation, sphere formation and 3D-organotypic invasive abilities of OSCC-derived cells in vitro and tumorigenesis in a mouse xenograft model.
Methods
Human tissue specimens
All tissue samples were collected from Haukeland University Hospital after informed written patient consent. This study was approved by the Committee for Medical and Health Research Ethics in West Norway (2011/1244 REK vest, 2010/481 REK vest). A total number of 75 normal human oral mucosa [NHOM, 31 formalin fixed-paraffin embedded (FFPE) and 44 frozen], 21 oral dysplastic lesion (ODL, all FFPE), 132 OSCC (82 FFPE and 50 frozen) and 17 positive cervical lymph nodes (all FFPE) were used in the current study for the expression analysis of S100A16 by immunohistochemistry (IHC) and/or quantitative RT-PCR (qRT-PCR). All OSCC patients included in the study were newly diagnosed cases, and had no history of chemo- or radiotherapy prior to surgery. All NHOM specimens were donated by patients undertaking wisdom tooth extraction. For S100A16 IHC, FFPE specimens of NHOM (n = 21), ODL (n = 11; 1 carcinoma in situ, 1 severe, 7 moderate and 2 mild dysplastic lesions), OSCCs (n = 65), and positive cervical lymph nodes (n = 17) were used. Details of the clinicopathological information of these OSCC cases are reported in Table 1. FFPE specimens of NHOM (n = 10), ODL (n = 10) and OSCC (n = 17) were laser microdissected and used for quantification of S100A16 mRNA by qRT-PCR. In OSCC specimens, paratumor (dysplastic) epithelium, tumor center/core and the corresponding invading front/island were microdissected. Detailed methodology for laser microdissection is reported in Additional file 1. S100A16 mRNA expression was examined in frozen tissues of normal human oral mucosa (NHOM, n = 44) and OSCCs (n = 50). These tissues were stored at −80 °C till RNA extraction.
Table 1.
S100A16 expression (PLI score) and clinicopathological variables of the OSCC patients
| PLI score at invading fronts/islandsa | |||
|---|---|---|---|
| Variables | Low, n (%) | High, n (%) | P |
| Ageb (years) | |||
| ≤64 | 18 (60.0) | 12 (40.0) | 0.108 |
| >64 | 14 (60.0) | 21 (60.0) | |
| Gender | |||
| Female | 10 (47.6) | 11 (52.4) | 0.857 |
| Male | 22 (50.0) | 22 (50.0) | |
| Location | |||
| Tongue | 14 (45.2) | 17 (54.8) | 0.566 |
| Gingiva, buccal mucosa & oral lip | 11 (47.8) | 12 (52.2) | |
| Floor of mouth & oro-pharynx | 7 (49.2) | 4 (36.4) | |
| Differentiation | |||
| Poor and moderate | 22 (62.9) | 13 (37.1) | 0.018 |
| Well | 10 (33.3) | 20 (66.7) | |
| Lymph node involvement | |||
| Negative (N0) | 15 (39.5) | 23 (60.5) | 0.062 |
| Positive (N1 & N2) | 17 (63.0) | 10 (37.0) | |
| Tumor size | |||
| T1 & T2 | 19 (52.8) | 17 (47.2) | 0.638 |
| T3 & T4 | 13 (44.8) | 16 (55.2) | |
| Recurrence | |||
| No | 20 (43.5) | 26 (56.5) | 0.149 |
| Yes | 12 (63.2) | 7 (36.8) | |
| Tumor stage | |||
| Early (1 & 2) | 8 (38.1) | 13 (61.9) | 0.215 |
| Late (3 & 4) | 24 (54.5) | 20 (45.5) | |
aOSCCs were stratified into high and low S100A16 expression groups by using median S100A16 PLI score as a cut-off
bpatients were categorized into low- and high-age groups based on the median age
External microarray databases
Eight external microarray datasets, four for OSCC and head and neck SCC (mainly consisting of OSCC) [20–23], and one each for esophageal squamous cell carcinoma (ESCC) [24], colorectal carcinoma (CRC) [25], prostate cancer [26] and ovarian cancer [27] were used either i) to validate the down-regulation of S100A16 in OSCC or in the above mentioned malignancies or ii) for the correlation analyses of S100A16 and differentiation related molecules.
IHC
S100A16 IHC was performed in FFPE tissue specimens of NHOM, ODL, OSCCs, and positive cervical lymph nodes as described previously [19]. Briefly, antigen retrieval was done by microwave treatment in Tris-EDTA buffer, pH 9.0 (DAKO). After blocking with 10 % goat serum, rabbit polyclonal anti-human S100A16 primary antibody (11456-1-AP, Proteintech, Chicago, IL, USA, 1:100 dilutions) was applied. After wash, anti-rabbit secondary antibody conjugated with horseradish peroxidase labeled polymer (EnVision System, DAKO) was applied. Presence of antigen was visualized by staining with 3, 3′-diaminobenzidine (DAKO), counterstained with hematoxylin (DAKO) and mounted with EuKit mounting medium. Sections incubated with 3 % BSA instead of primary antibody served as negative controls. FFPE tissues from mouse tumor xenografts were stained with anti-S100A16, anti-involucrin, anti-Ki67, and anti-Bmi-1. For detailed methodology of IHC and the antibody used, see Additional file 1.
IHC evaluation
Blinded for the clinical information, IHC evaluation of all specimens was done at 400× (40× objective lens) using Leica DMLB microscope (Leica Microsystems). Inter-observer variation was controlled by calibrating the evaluation done by three investigators (DS, TAO and HP). Afterwards, all specimens were evaluated by one investigator (DS). Expression pattern of S100A16 was evaluated semiquantitatively by scoring three consecutive fields (>500 cells/field, whenever possible) on the surface epithelium of NHOM and ODL, and at the invading tumor islands of lymph nodes. For OSCCs, the evaluation was done both at the central and the invading front (the deepest part of an invasive tumor, >3–4 cell layers thick). When it was not possible to identify clear invasive fronts, deepest invading tumor islands consisting of >50 cells were used for quantification. A composite scoring system combining the number of S100A16 positive cells (P score), cellular localization (membranous or cytoplasmic or both, L score) and intensity (I score) was used for S100A16 scoring. The final (PLI) score was calculated by multiplying the individual P, L and I scores and averaging PLI scores of the three evaluated fields. For details of the PLI scoring system, see Additional file 1.
The evaluation of Ki67 staining in the tumor xenografts was done only at the invading fronts (5–6 cell layers). Positive and negative tumor cell nuclei were manually counted (at least 300 cells were counted in 3–6 representative areas, at 40× objective lens) and the fraction of the positive cells were calculated. Bmi-1, S100A16 and involucrin staining in the tumor xenografts were evaluated qualitatively only.
Cell culture, construction of expression vector and transfection
The oral squamous cell carcinoma-derived cell-lines CaLH3 [28] and H357 [29] were cultured as described elsewhere [17]. S100A16 expression and shRNA vectors were constructed as described previously [17, 19]. For details of the expression and shRNA vector construction, see Additional file 1. CaLH3 and H357 cells infected with retrovirus with S100A16 insert and retrovirus without S100A16 insert are referred to as ‘S100A16-CaLH3 and S100A16-H357’, and ‘control-CaLH3 and control-H357’ cells, respectively.
Tissue engineering (3D-models) and evaluation of carcinoma cell invasion
Primary carcinoma associated fibroblasts isolated from a patient with OSCC were embedded in collagen type I biomatrix (BD Biosciences), and seeded on top with control or S100A16 over-expressing CaLH3 cells, as previously described [30]. 3D constructs were harvested, formalin-fixed and paraffin-embedded. Depth of invasion was measured on 5-μm sections stained with hematoxylin and eosin using Olympus DP.Soft 5.0 software. For the measurement of carcinoma cell invasion, each 3D-organotypic section was divided into fifths. The central and the two outer fifths were excluded from measurements, depth of invasion being assessed in the remaining two fifths only. For this, a horizontal line was drawn (using the software Olympus DP.Soft 5.0) through the uppermost remnants of the collagen gel to visualize the basement membrane zone; depth of invasion was determined every 100 μm along this horizontal line as the vertical distance from this line to the limit of invading epithelial cells (Fig. 5f).
Fig. 5.

Retroviral mediated S100A16 over-expression inhibited proliferation, sphere formation and 3D-invasion abilities of OSCC cells in vitro. S100A16 was over-expressed in CaLH3 and H357 cell-lines and subsequent effect on malignant phenotype was examined. a Six thousands control or S100A16 over-expressing CaLH3 or H357 cells were seeded in duplicates in the microtiter E-plates and cell proliferation was measured in real time for 72 h with the xCELLigence system. S100A16 over-expressing cells proliferated significantly slower as compared to the control cells as demonstrated by the normalized cell index. Similarly, sphere formation abilities of S100A16-CaLH3 (b and d) and S100A16-H357 (c and e) cells were significantly reduced as compared to the corresponding controls. Error bars represent SEM of 6 replicates for each experiment. Student’s-t test was performed for statistical analysis. Expt., Experiment. S100A16 over-expression led to significant reduction in the invasive potential of CaLH3 cells in 3D-organotypic cultures (f, invasion quantified in g). Yellow dotted line represents the imaginary basement membrane. Black vertical lines with double arrowheads represent the depth of invasion of malignant keratinocytes. h–j S100A16 mediated functional effects on malignant phenotype were associated with concomitant down-regulation of proliferation/self-renewal markers (Bmi-1 and Oct4A) (h) and invasion-promoting molecules (MMP1 and MMP9 mRNA levels) in CaLH3 and H357 cell-lines (i and j). Error bars in (g) represent SEM of 4 repeated experiments where as in (i and j) represent 3 repeated experiments. Student’s-t test was performed for statistical analyses
RNA extraction, cDNA synthesis and qRT-PCR
RNA was extracted from frozen specimens (NHOM and OSCC), laser microdissected FFPE tissues (NHOM, ODL and OSCC) and OSCC-derived cell-lines respectively using Dynabeads mRNA Direct kit (Invitrogen), RNeasy FFPE Kit (#73504, Qiagen) and RNeasy fibrous tissue mini kit (cat no: 74704, Qiagen Inc.). See Additional file 1: Supplementary methods and Table S2 for details of the cDNA synthesis and qRT-PCR.
Immunoblotting
Twenty to 30 μg of cell lysates were resolved in NuPAGE® Novex 4–12 % Bis-TrisTris gel (NP0329, Life technologies, NY, USA) and immunoblotted with antibodies as described in Additional file 1: Table S3.
Real time cell proliferation assay (xCELLigence system)
The xCELLigence DP device from Roche Diagnostics (Mannheim, Germany) was used to quantitatively and dynamically monitor cell proliferation in real-time. Six thousands control or S100A16 over-expressing CaLH3 and H357 cells were seeded in duplicates in the electronic microtiter E-plates (Cat. No: 5469830001; Roche Diagnostic) and proliferation was measured in real time for 72 h. Data acquisition and analysis was performed with the RTCA software (version 1.2.1.1002, Roche Diagnostics).
In vitro sphere formation assay
Inner surface of each well of 48 well-plate was coated evenly with a 12 mg/mL solution of polyHEMA (sigma, P3932) in 95 % ethyl alcohol and sterilized under UV overnight. Afterwards, 490 μL of cell culture medium with 1 mg/mL methylcellulose was added in each well. One thousand cells suspended in 10 μL medium was then added in each well and evenly mixed with the medium. Sphere formation was quantified on 14th day by counting the number of spheres (>50 cells) at 4× objective under Nikon ECLIPSE TS100 fluorescent microscope. Each experiment was repeated thrice in 6 replicates.
Adherence to collagen IV
Previous studies have shown that rapid adherence of keratinocytes to collagen IV is a robust method to enrich cells for stem cell properties [31, 32]. According to this method, cells adhering most rapidly to collagen IV are considered to be enriched for cells with a less differentiated phenotype (stem cell properties); whereas the late adherent cell population contains relatively fewer cells with stem cell properties and the non-adherent cell population consists of cells with a more differentiated phenotype. This assay was performed as described previously [33, 34]. Briefly, cell suspension was allowed to attach to culture dishes coated with collagen IV (10 μg/mL) (BD Biosciences, USA) in the cell incubator for 10 min. Cells attached to the dishes were collected and referred to as rapid adherent cells (RAC). The unattached cells within the first 10 min were then transferred to a new collagen IV-coated dish for an additional 30 min in incubator. Cells that adhered within this period were referred to as middle adherent cells (MAC). Remaining unattached cells were collected as late adherent cells (LAC).
Fluorescent activated cell sorting (FACS) for p75NTR and cytokeratin 13
p75NTR, a member of tumor necrosis factor receptor superfamily, is a low affinity neurotrophin receptor. Accumulated evidences suggest that p75NTR is a putative stem cell marker both in the normal oral and esophageal tissues [35–37] as well as in the malignancies including OSCC [37–40]. Accordingly, cells with p75NTR high expression are considered to be enriched for cells with a less differentiated phenotype (stem cell properties), whereas the cells with low P75NTR expression are enriched for cells with a more differentiated phenotype. Unfixed oral cancer cells were stained with anti-p75NTR antibody (Sigma Aldrich, 1:250 dilutions) whereas methanol fixed cells were stained with anti-cytokeratin 13 antibody (Novacastra, 1:350 dilutions). For detailed methodology of FACS, see Additional file 1.
In vivo tumorigenesis assay
Protocols for all animal studies were approved by the Norwegian Animal Research Authority (Project ID: 20124236). Twelve nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice were randomly divided into two groups (n = 6, each group). One thousand S100A16-H357 or control-H357 cells suspended in 50 μL of Matrigel (BD Biosciences) were injected in the tongue of each mouse. Tumor development was monitored regularly under inhalation anesthesia. Length and breadth of the formed tumors were measured by Vernier caliper and tumor volume was calculated using the following formula-(length × breadth2)/2. Tumor formation was confirmed histologically.
Statistics
Statistical analysis was done using SPSS 21 and/or GraphPad prism 5. Difference in means between two groups was analyzed by using unpaired t-tests, whereas comparison between more than two groups was done by using ANOVA test with Bonferroni Post-Hoc. Median PLI scores both at the tumor center and at the invading front/island were used as cut-off values to stratify OSCCs into high- and low-S100A16 expression groups. According to the differentiation status, OSCCs were categorized into two groups: highly differentiated and moderately-poorly differentiated. Association between the expression status of S100A16 and other binary variables was done using Chi-square Test. Survival analysis was performed using the Kaplan-Meier analysis (log-rank test). Cox proportional hazard model was used to examine the effect of S100A16 expression on 10-year overall survival. Level of significance was set at 5 %.
Results
S100A16 was progressively down-regulated from normal tissue to dysplasia and OSCC; and low S100A16 expression at the invading front/islands correlated with reduced survival and poor tumor differentiation
To examine the expression and localization of S100A16, IHC was performed on archived FFPE specimens of NHOM (n = 21), ODL (n = 11), OSCC (n = 65) and positive cervical lymph nodes (n = 17). A strong membranous expression of S100A16 was found in the supra-basal (committed/differentiating) epithelial cell layers of all NHOM tissues (Fig. 1a). Negative or weak cytoplasmic staining was found in the basal cell layer (stem cell compartment) in most of NHOM samples (Fig. 1a and A1). The expression pattern of S100A16 in ODL was similar to that found in NHOM (Fig. 1b). The superficial and central areas of OSCC specimens demonstrated similar staining pattern to that found in NHOM, whereas very weak or negative expression was observed at the invading front/island of tumor cells with concomitant membrane to cytoplasmic translocation in majority of the cases (Fig. 1 C1 and C2). Nevertheless, S100A16 staining was relatively strong with membranous localization at the invading front/island of well-differentiated OSCCs (Additional file 2: Figure S1A). S100A16 staining was very weak or absent in the infiltrating tumor islands of positive cervical lymph nodes (Additional file 2: Figure S1B).
Fig. 1.

S100A16 protein was progressively down-regulated from NHOM to ODL and OSCC and low S100A16 protein expression correlated with poor OSCC prognosis. a Representative NHOM specimen showed strong, predominantly membranous S100A16 expression in the epithelial compartment. Basal cell layer (arrowheads), however, was mostly negative for S100A16 expression (A1). b Expression pattern of S100A16 in ODL was similar to that of NHOM. However, the expression intensity was weaker than that in NHOM (c) Representative OSCC lesion showing a gradient of S100A16 expression: central area (C1) showed a strong, membranous staining in contrast to a very weak, mostly cytoplasmic staining in the invading front area (C2). d Graphic illustration of S100A16 PLI score demonstrated gradual down-regulation of S100A16 from NHOM to ODL, OSCC and positive cervical nodes. ANOVA test with Bonferroni Post-Hoc was used for the statistical analysis. P-value: ***, <0.001; ns, not significant. e Kaplan-Meier curves showing reduced 10-year survival probabilities for patients with low S100A16 PLI score. Log-Rank test was used for statistical analysis
Quantification of S100A16 staining showed that S100A16 PLI score was gradually decreased during the transition from NHOM to ODL and OSCC (Fig. 1d). Of note, PLI score was found to be lower at the invading front/island as compared to the central areas in OSCCs (Fig. 1d). Examination of possible correlation between S100A16 expression and clinical parameters showed that low S100A16 PLI score at the invading front/island was associated with reduced 10-year overall survival (Log-Rank test, P = 0.017) (Fig. 1e), moderate-poorly differentiated OSCCs (P = 0.018) and lymph node involvement (P = 0.062) (Table 1). Multivariate Cox regression analysis demonstrated that S100A16 expression was a significant prognostic factor (HR = 0.483, CI = 0.24–0.95, P = 0.037) for the survival of OSCC patients (Table 2). However, no significant correlations were observed between the PLI score at the tumor center and clinicopathological variables (Additional file 1: Table S1). A trend for better survival probabilities was found for well differentiated and early stage tumors, but the results were not statistically significant (data not shown).
Table 2.
Results of a multivariate Cox regression analysis for predicting the overall survival of OSCC cases
| Variables | Assigned score | Hazard ratio | 95 % CI | P-value |
|---|---|---|---|---|
| Age | ||||
| ≤64 | 0 | 1.51 | 0.82–2.94 | 0.169 |
| >64 | 1 | |||
| Sex | ||||
| Female | 0 | 1.18 | 0.60–2,33 | 0.623 |
| Male | 1 | |||
| Differentiation | ||||
| Well | 0 | 1.09 | 0.53–2.94 | 0.803 |
| Moderate & poor | 1 | |||
| T-stage | ||||
| T1 & T2 | 0 | 0.871 | 0.39–1.90 | 0.728 |
| T3 & T4 | 1 | |||
| Clinical stage | ||||
| Early (1 & 2) | 0 | 1.371 | 0.55–3.33 | 0.494 |
| Late (3 & 4) | 1 | |||
| S100A16 | ||||
| High | 0 | 0.483 | 0.24–0.95 | 0.037 |
| Low | 1 | |||
CI, Confidence interval
S100A16 mRNA level was progressively down-regulated from NHOM to ODL and OSCC
Expression levels of S100A16 mRNA were quantitatively examined in an independent cohort of frozen specimens of NHOM (n = 44) and OSCC (n = 50) by qRT-PCR. The mean expression of S100A16 mRNA was found to be significantly down-regulated in OSCC compared to NHOM (P < 0.0001) (Fig. 2a). Down-regulation of S100A16 mRNA levels was verified in three independent microarray datasets for OSCC (Fig. 2b–d). To validate the progressive down-regulation of S100A16 mRNA expression during OSCC progression, FFPE specimens of NHOM, ODL and OSCC were laser dissected and mRNA levels were quantitatively examined. Parallel to the IHC findings, mRNA expression level was progressively down-regulated in the oral keratinocytes during the transition from NHOM to ODL, including paratumor epithelium, and OSCC (Fig. 2e).
Fig. 2.

S100A16 mRNA level was progressively down-regulated from NHOM to ODL and OSCC. a S100A16 mRNA expression was examined in frozen specimens of NHOM (n = 44) and OSCCs (n = 50) by using qRT-PCR. Mean S100A16 mRNA was found to be significantly down-regulated in OSCCs (P < 0.0001). S100A16 mRNA expression levels were normalized to GAPDH mRNA expression. Error bars represent SEM. Student’s-t test was performed for statistical analysis. b–d Down-regulation of S100A16 mRNA levels in OSCC was verified in three independent microarray datasets. Error bars represent SEM. Student’s-t test was performed for statistical analysis. e Gradual down-regulation of S100A16 mRNA during the transition from NHOM to ODL and OSCC was validated in laser dissected specimens of NHOM, ODL, paratumor (dysplastic) epithelium, tumor center and invading front by qRT-PCR. qRT-PCR was done in duplicates and S100A16 mRNA level was normalized to GAPDH and ACTB mRNA levels. Error bars represent SEM. ANOVA test with Bonferroni Post-Hoc was used for the statistical analysis. P-value: ***, <0.001; **, 0.001–0.01
S100A16 mRNA level was down-regulated during tumor progression of several other human malignancies
To investigate whether S100A16 down-regulation is a common event during tumor progression of other carcinomas as well, the expression levels of S100A16 mRNA were examined in external microarray datasets of other human malignancies and tumor progression model systems. Similar to OSCC, S100A16 mRNA level was found to be significantly down-regulated in ESCC and CRC as compared to the corresponding control specimens (Additional file 3: Figure S2A-B). Moreover, progressive down-regulation was observed during various stages of tumor progression in prostate cancer and in ovarian cancer model systems (Additional file 3: Figure S2C-D).
S100A16 mRNA expression was positively correlated with differentiation markers in OSCC specimens and in cell fractions enriched for differentiated cells
Positive correlation between the expression of S100A16 as examined by IHC and the differentiation status found in the OSCC specimens prompted us to further examine the correlation between S100A16 and differentiation markers in OSCC specimens in vivo, and in the differentiated cell fractions in vitro. S100A16 mRNA levels were positively correlated with mRNA levels of several of the differentiation markers (IVL, KRT13, TGM1 and FLG) in two independent microarray datasets [20, 21] (Fig. 3a–d and Additional file 4: Figure S3). In parallel, similar correlation was also found in the LAC and p75NTRlow cell fractions (enriched for differentiated cells) compared to the RAC/MAC and p75NTRhigh fractions (enriched for less differentiated cells) (Fig. 3e–g).
Fig. 3.

S100A16 mRNA expression was positively correlated with differentiation markers in OSCC specimens and in the cell fractions enriched for more differentiated cells. a–d S100A16, IVL, KRT13, TGM1 and FLG mRNA levels were obtained from external microarray dataset (Rickmen) and their correlation was examined using Pearson analysis. e–g Cell fractions were enriched for differentiated cells either by using lack of adherence to collagen IV or by FACS sorting for low p75NTR expression and mRNA expression levels of S100A16 and IVL and KRT10 were examined by qRT-PCR. e Significantly higher mRNA levels of S100A16, IVL and KRT10 were found in LAC cell fractions (enriched for more differentiated cells) as compared to RAC/MAC (enriched for less differentiated cells). Error bars represent SEM of 3 repeated experiments. ANOVA test with Bonferroni Post-Hoc was used for statistical analysis. P-value: ***, <0.001. f and g Fractions enriched for differentiated cell (p75NTRlow) expressed significantly higher expression of S100A16, IVL and KRT10 as compared to p75NTRhigh fractions in CaLH3 (f) and H357 (g) cells. Expression levels were normalized to GAPDH mRNA expression. Error bars represent SEM of 3 repeated experiments. Student’s-t test was performed for statistical analysis
S100A16 modulated expression of differentiation-related markers in OSCC-derived cells
The in vivo and in vitro association of S100A16 with a more differentiated phenotype led us to investigate whether S100A16 can induce expression of differentiation-related markers in OSCC-derived cells. Retroviral mediated over-expression of S100A16 resulted in up-regulation of involucrin, cytokeratin 13 and transglutaminase 1 in CaLH3 cells (expression of filaggrin could not be detected in both control and S100A16-CaLH3 cells) (Fig. 4a). In H357 cells, over-expression of S100A16 was associated with up-regulation of involucrin, cytokeratin 10 and filaggrin (expression of transglutaminase 1 and cytokeratin 13 could not be detected in both control and S100A16-H357 cells (Fig. 4a). FACS analysis further confirmed the up-regulation of cytokeratin 13 upon S100A16 over-expression (Fig. 4b–d). Confirming the above results, shRNA mediated knock-down of S100A16 resulted in down-regulation of involucrin and cytokeratin 13 in CaLH3 cells (Fig. 4e). The total p38 or phospho-p38 expression levels were not affected by S100A16 over-expression (Fig. 4a).
Fig. 4.

S100A16 over-expression modulated differentiation-related markers in OSCC cell-lines. S100A16 was over-expressed and knocked-down in OSCC-derived cells by retroviral vectors and concomitant modulation of differentiation markers was examined. a Western blot analysis showed up-regulation of several of the differentiation markers with S100A16 over-expression. ** anti human-cytokeratin 13 (sc-58721, Santa Cruz); *anti human-cytokeratin 13 (NCL-CK13, Novacastra). b Up-regulation of cytokeratin 13 in S100A16-CaLH3 was further verified by FACS analysis (b–d). Error bars in (c and d) represent SEM of 3 repeated experiments. Student’s-t test was performed for statistical analysis. d In parallel with over-expression, S100A16 knock-down led to down-regulation of involucrin and cytokeratin 13 in CaLH3 cells
S100A16 over-expression reduced cell proliferation, sphere formation ability and 3D-invasive potential of OSCC-derived cells in vitro
The functional role of S100A16 in OSCC tumorigenesis was next examined by performing a number of established functional assays. Proliferation rates (as measured by normalized cell index) of CaLH3 and H357 cell-lines were found to be significantly reduced upon S100A16 over-expression (Fig. 5a). More importantly, over-expression of S100A16 led to significant reduction in the sphere formation abilities (in vitro surrogate for the in vivo tumorigenesis assay) of both CaLH3 and H357 cell-lines as compared to the corresponding control cells (Fig. 5b–e) (P < 0.05). Suppression in sphere formation abilities correlated with a simultaneous down-regulation of self-renewal markers (Oct 4A and Bmi-1) in S100A16-CaLH3 and S100A16-H357 cells (Fig. 5h). Furthermore, S100A16 over-expression led to significant reduction of the invasive potential of CaLH3 cells in 3D-organotypic cultures (Fig. 5f, quantified in g). In parallel, S100A16 over-expression led to significant down-regulation of MMP9 mRNA levels in both CaLH3 and H357 cells-lines (Fig. 5j). MMP1 mRNA expression, however, was significantly down-regulated only in H357 cells (Fig. 5i).
S100A16 over-expression decreased tumor formation ability of H357 cells in NOD/SCID mice and the resulting tumor xenografts exhibited a more differentiated and less proliferative phenotype
The effect of S100A16 on the in vivo tumor formation ability was examined by injecting S100A16 over-expressing (S100A16-H357) or control (control-H357) H357 cells in the tongue of NOD/SCID mice. When 1000 cells/mouse were injected, control-H357 cells formed tongue tumors in all of the NOD/SCID mice (6/6, 100 % tumors) whereas S100A16-H357 cells formed tumors in 5 of the mice (5/6, 83.4 % tumors). More importantly, tongue tumors formed by the control-H357 cells were significantly larger (at 33 days, P = 0.04) compared to that of S100A16-H357 cells (Fig. 6a). In addition, lag phase for S100A16-H357 cells to form tongue tumors was longer than that of control-H357 cells (Fig. 6b). We next examined whether the phenotype of S100A16-H357 tumor xenografts would correlate with the expression of differentiation and proliferation/self-renewal markers. As expected, S100A16-H357 xenografts demonstrated features of well differentiation (presence of keratin pearls, Fig. 6c and d) with higher expression of the terminal differentiation marker involucrin as compared to the control-H357 xenografts (Fig. 6e and f) Additionally, S100A16-H357 xenografts expressed lower levels of Ki67 (Fig. 6g and h) and Bmi-1 (Fig. 6i and j) as compared to the control-H357 xenografts.
Fig. 6.
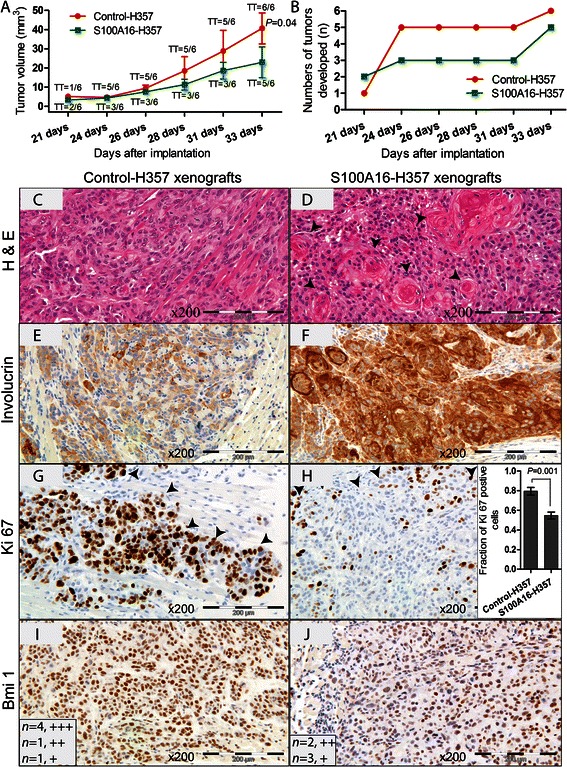
S100A16 over-expression reduced tumorigenesis in vivo and the resulting tumor xenografts exhibited features of increased differentiation. a One thousand control or S100A16 over-expressing H357 cells were injected in the tongue of each mouse (NOD/SCID) and tumor development was monitored. All mice injected with control cells formed tumors (Tumor Take, TT = 6/6) whereas S100A16-H357 cells formed tumors in 5 of the mice (TT = 5/6). In addition, tumors formed by control cells were significantly larger as compared to that of the S100A16-H357 cells (P = 0.04, at 33 days). Error bars represent SEM. Student’s-t test was performed for statistical analysis. b S100A16-H357 cells took longer time (lag phase) to form tumors as compared to the control cells. Tumor xenografts were harvested, formalin fixed, paraffin embedded and examined for histomorphology, IHC expression of differentiation, proliferation and self-renewal markers. Representative images demonstrating highly differentiated phenotype of S100A16-H357 xenografts with formation of keratin pearls (arrowheads) and higher expression of involucrin (c–f). On the other hand, S100A16-H357 xenografts demonstrated lower expression levels of Ki67 (g and h) and Bmi-1 (i and j) as compared to the control-H357 xenografts. Arrowheads in (g) and (h) mark the invading front. Error bars in (h) represent SEM. Student’s-t test was performed for statistical analysis in (f). +++, strong; ++, moderate; +, weak staining
Discussion
In the current study, assessment of whole tissue specimens of NHOM, ODL, OSCC and positive lymph node showed progressive down-regulation of both S100A16 protein and mRNA levels during OSCC progression (Figs. 1 and 2). These data were confirmed by analyzing S100A16 mRNA levels in laser captured microdissected specimens and in three independent OSCC microarray datasets (Fig. 2). These findings indicate that the reduced level of S100A16 might be related to OSCC progression. Given the high probability of chromosomal rearrangement in 1q21 region (where S100A16 is located) in human cancers, one of the mechanisms for S100A16 down-regulation in OSCC could be the deletion of S100A16 locus in these lesions. Indeed, a recent work reported a loss in the S100A16 locus in OSCC specimens from India and Sri Lanka [41]. The clinicopathological analysis of the current study showed a significant correlation between low S100A16 protein (at the invading front/island) levels and reduced 10-year overall survival probabilities for OSCC patients (Fig. 1e), poor tumor differentiation and positive cervical nodes. These data suggest a prognostic value for S100A16 in OSCC. Of note, S100A16 protein expression at the tumor center did not reveal any association with clinicopathological variables. These findings are in agreement with the concept that tumor invading fronts/islands are the more active areas of a malignant lesion and molecular/morphological changes at these areas are better prognosticators than those at the central/superficial region of the tumor [42, 43].
Positive correlation between S100A16 protein level and tumor differentiation, as found in the current study, pointed to a functional role of S100A16 in the regulation of keratinocyte differentiation. In agreement, mRNA levels of S100A16 and differentiation markers were positively correlated in the OSCC specimens in vivo in two independent microarray datasets (Fig. 3a–d, Additional file 4: Figure S3) and in the cell fractions enriched for differentiated cells in vitro (Fig. 3e–g). Furthermore, modulation of earlier and terminal differentiation markers in OSCC-derived cells by over-expression and knock-down of S100A16 provided direct evidence that S100A16 functions as a differentiation promoting protein in OSCC (Fig. 4). This corroborates well with the observation that other members of S100 proteins are involved in the regulation of cellular differentiation [10, 11]. Indeed, a gradual increase in S100A16 expression has been described previously during the differentiation of preadipocytes to adipocytes [44]. However, in malignant oral keratinocytes this function seems to be independent of the p38 MAP kinase pathway, as no change was observed in the current study on the p38 phosphorylation status with S100A16 over-expression (Fig. 4a). This warrants investigation of p38 independent mechanisms possibly involved in S100A16 mediated modulation of differentiation markers in oral cancer cells.
Poorly differentiated phenotypes with excessive cellular proliferation and invasive abilities are considered to be characteristics of aggressive tumors. For several tumor types, lesions with more differentiated phenotype have been shown to have a less aggressive behavior and better clinical outcome [45, 46], indicating that molecular regulators that promote cellular differentiation might have tumor suppressive functions [47–49]. Several observations in the current study suggested that loss of S100A16 might contribute to the acquisition of aggressive OSCC phenotype. Firstly, significantly reduced S100A16 expression at the invading front/island as compared to the tumor center and, severely down-regulated expression in the positive lymph nodes indicated that loss of S100A16 might be necessary for the tumor cells to acquire an invasive phenotype (Figs. 1 and 2e). Further, correlation between reduced S100A16 expression level and reduced OSCC patient survival pointed towards a role for S100A16 in the maintenance of less aggressive tumor phenotype. Indeed, retroviral mediated S100A16 over-expression significantly suppressed several aspects of aggressive tumor phenotype; such as proliferation, sphere formation and 3D-organotypic invasive abilities of OSCC-derived cells in vitro (Fig. 5a–g). In parallel, S100A16 over-expression reduced tumorigenic abilities (tumor incidence and tumor volume) of H357 cells in NOD/SCID mice (Fig. 6a and b). These tumor suppressive functions were paralleled at the molecular level by concomitant down-regulation of self-renewal (Bmi-1and Oct4A) and invasion related (MMP1 and MMP9) molecules in vitro (Fig. 5h–j). Likewise, S100A16-H357 xenografts were found to be more differentiated and less proliferative, both histologically and at the molecular level as evidenced by the higher involucrin expression and lower expression of Ki67 and Bmi-1 (Fig. 6c–j). Taken together, these findings suggested a role for S100A16 as a tumor suppressor in OSCC and indicated that progressive loss of S100A16 might be related with aggressive tumor growth and invasion leading to reduced patient survival. Similar to our results, IRF6 (INF regulatory factor 6), a pro-differentiating factor which shares similar expression pattern to that of S100A16, has been shown to have a tumor suppressive activity in squamous cell carcinoma by promoting keratinocyte differentiation [47, 49]. Additionally, progressive down-regulation of S100A16 as found in CRC, prostate and ovarian cancers (Additional file 3: Figure S2) demonstrates a broader relevance for S100A16 in the process of tumorigenesis of other human malignancies and warrants further investigation in these tumor types.
Conclusion
Our results indicate a novel role for S100A16 in the regulation of OSCC differentiation and tumor suppression. Further molecular characterization of S100A16 mediated tumor suppressive functions might contribute to the better understanding of OSCC carcinogenesis and provide opportunity for S100A16 based better prognostication and management of OSCC.
Acknowledgments
The authors would like to thank Gunnvor Øijordsbakken (Department of Clinical Medicine, The Gade Laboratory for Pathology) for assistance with immunohistochemistry and Prof. Olav Dahl (Department of Clinical Science, Section of Oncology) for allowing us to use the facilities in the Mohn Cancer Research Laboratory. This study was co-funded by post-doctoral fund (DS), Bergen Medical Research Foundation (DEC), Norwegian Cancer Society (DEC), Helse Vest (DEC) and Center for Cancer Biomarkers (ACJ, University of Bergen).
Abbreviations
- NHOM
Normal human oral mucosa
- ODL
Oral dysplastic lesion
- OSCC
Oral squamous cell carcinoma
- IHC
Immunohistochemistry
- qRT-PCR
Quantitative reverse transcription-polymerase chain reaction
- ESCC
Esophageal squamous cell carcinoma
- CRC
Colorectal cancer
Additional files
Supplementary Methods and Tables. Table S1. S100A16 expression and clinicopathological variables of the OSCC patients. Table S2. Details of the TaqMan assays used for qRT-PCR. Table S3. Details of the antibodies used for immunoblotting. (DOCX 31 kb)
(A) S100A16 staining was strong with membranous localization at the invading front/island of well-differentiated OSCC. (B) S100A16 staining was very weak or absent in the infiltrating tumor islands of positive cervical lymph nodes. (TIFF 2613 kb)
Down-regulation of S100A16 mRNA levels in tumor tissues as compared to the normal controls in independent microarray datasets. (A) ESCC, (B) CRC, (C) prostate cancer and (D) in vitro progression model of ovarian cancer (MOSE-E, non-tumorigenic; MOSE-I, intermediate and MOSE-L, aggressive malignant phenotype). For statistical analysis, paired-t test was performed in (A) and one way ANOVA test with Bonferroni Post-Hoc in (B–D). Error bars represent SEM. **, 0.001–0.01. ns, not significant. (TIFF 275 kb)
Positive correlation between S100A16 mRNA expression and differentiation markers in OSCC specimens. Expression data were obtained from external microarray dataset [20]. (TIFF 337 kb)
Footnotes
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
DS, ACJ and DEC conceived and designed research; DS, OB, HP, TAO, M-T T, ACJ and DEC performed the experiments, analyzed the data and reviewed the manuscript. All authors read and approved the manuscript.
Contributor Information
Dipak Sapkota, Phone: +47 55973231, Email: Dipak.Sapkota@k1.uib.no.
Ove Bruland, Email: ove.bruland@helse-bergen.no.
Himalaya Parajuli, Email: Himalaya.Parajuli@k1.uib.no.
Tarig A. Osman, Email: Tarig.Osman@k1.uib.no
Muy-Teck Teh, Email: m.t.teh@qmul.ac.uk.
Anne C. Johannessen, Email: Anne.Johannessen@k1.uib.no
Daniela Elena Costea, Email: Daniela.Costea@k1.uib.no.
References
- 1.Silverman SJ. Demographics and occurrence of oral and pharyngeal cancers: the outcomes, the trends, the challenge. J Am Dent Assoc. 2001;132(suppl_1):7S–11S. doi: 10.14219/jada.archive.2001.0382. [DOI] [PubMed] [Google Scholar]
- 2.Leethanakul C, Patel V, Gillespie J, Pallente M, Ensley JF, Koontongkaew S, Liotta LA, Emmert-Buck M, Gutkind JS. Distinct pattern of expression of differentiation and growth-related genes in squamous cell carcinomas of the head and neck revealed by the use of laser capture microdissection and cDNA arrays. Oncogene. 2000;19(28):3220–3224. doi: 10.1038/sj.onc.1203703. [DOI] [PubMed] [Google Scholar]
- 3.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakamoto K, Aragaki T, Morita K-i, Kawachi H, Kayamori K, Nakanishi S, Omura K, Miki Y, Okada N, Katsube K-i, et al. Down-regulation of keratin 4 and keratin 13 expression in oral squamous cell carcinoma and epithelial dysplasia: a clue for histopathogenesis. Histopathology. 2011;58(4):531–542. doi: 10.1111/j.1365-2559.2011.03759.x. [DOI] [PubMed] [Google Scholar]
- 5.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochem Biophys Res Commun. 2004;322(4):1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 6.Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33(7):637–668. doi: 10.1016/S1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- 7.Schäfer BW, Wicki R, Engelkamp D, Mattei MG, Heizmann CW. Isolation of a YAC clone covering a cluster of nine S100 genes on human chromosome 1q21: rationale for a new nomenclature of the S100 calcium-binding protein family. Genomics. 1995;25(3):638–643. doi: 10.1016/0888-7543(95)80005-7. [DOI] [PubMed] [Google Scholar]
- 8.Pietas A, Schlüns K, Marenholz I, Schäfer BW, Heizmann CW, Petersen I. Molecular cloning and characterization of the human S100A14 gene encoding a novel member of the S100 family. Genomics. 2002;79(4):513–522. doi: 10.1006/geno.2002.6744. [DOI] [PubMed] [Google Scholar]
- 9.Volz A, Korge BP, Compton JG, Ziegler A, Steinert PM, Mischke D. Physical mapping of a functional cluster of epidermal differentiation genes on chromosome 1q21. Genomics. 1993;18(1):92–99. doi: 10.1006/geno.1993.1430. [DOI] [PubMed] [Google Scholar]
- 10.Martinsson H, Yhr M, Enerbäck C. Expression patterns of S100A7 (psoriasin) and S100A9 (calgranulin-B) in keratinocyte differentiation. Exp Dermatol. 2005;14(3):161–168. doi: 10.1111/j.0906-6705.2005.00239.x. [DOI] [PubMed] [Google Scholar]
- 11.Wolf R, Lewerenz V, Büchau AS, Walz M, Ruzicka T. Human S100A15 splice variants are differentially expressed in inflammatory skin diseases and regulated through Th1 cytokines and calcium. Exp Dermatol. 2007;16(8):685–691. doi: 10.1111/j.1600-0625.2007.00587.x. [DOI] [PubMed] [Google Scholar]
- 12.Lo J-F, Yu C-C, Chiou S-H, Huang C-Y, Jan C-I, Lin S-C, Liu C-J, Hu W-Y, Yu Y-H. The epithelial-mesenchymal transition mediator S100A4 maintains cancer-initiating cells in head and neck cancers. Cancer Res. 2011;71(5):1912–1923. doi: 10.1158/0008-5472.CAN-10-2350. [DOI] [PubMed] [Google Scholar]
- 13.Rasanen K, Sriswasdi S, Valiga A, Tang H-Y, Zhang G, Perego M, Somasundaram R, Li L, Speicher K, Klein-Szanto A, et al. Comparative secretome analysis of epithelial and mesenchymal subpopulations of head and neck squamous cell carcinoma identifies S100A4 as a potential therapeutic target. Mol Cell Proteomics. 2013;12(12):3778–92. doi: 10.1074/mcp.M113.029587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein U, Arlt F, Walther W, Smith J, Waldman T, Harris ED, Mertins SD, Heizmann CW, Allard D, Birchmeier W, et al. The metastasis-associated gene S100A4 is a novel target of β-catenin/T-cell factor signaling in colon cancer. Gastroenterology. 2006;131(5):1486–1500. doi: 10.1053/j.gastro.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 15.Marenholz I, Heizmann CW. S100A16, a ubiquitously expressed EF-hand protein which is up-regulated in tumors. Biochem Biophys Res Commun. 2004;313(2):237–244. doi: 10.1016/j.bbrc.2003.11.115. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka M, Ichikawa-Tomikawa N, Shishito N, Nishiura K, Miura T, Hozumi A, Chiba H, Yoshida S, Ohtake T, Sugino T. Co-expression of S100A14 and S100A16 correlates with a poor prognosis in human breast cancer and promotes cancer cell invasion. BMC Cancer. 2015;15:53. doi: 10.1186/s12885-015-1059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sapkota D, Bruland O, Costea DE, Haugen H, Vasstrand EN, Ibrahim SO. S100A14 regulates the invasive potential of oral squamous cell carcinoma derived cell-lines in vitro by modulating expression of matrix metalloproteinases, MMP1 and MMP9. Eur J Cancer. 2011;47(4):600–610. doi: 10.1016/j.ejca.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Sapkota D, Costea DE, Blø M, Bruland O, Lorens JB, Vasstrand EN, Ibrahim SO. S100A14 inhibits proliferation of oral carcinoma derived cells through G1-arrest. Oral Oncol. 2012;48(3):219–225. doi: 10.1016/j.oraloncology.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Sapkota D, Costea DE, Ibrahim SO, Johannessen AC, Bruland O. S100A14 interacts with S100A16 and regulates its expression in human cancer cells. PLoS One. 2013;8(9):e76058. doi: 10.1371/journal.pone.0076058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thurlow JK, Peña Murillo CL, Hunter KD, Buffa FM, Patiar S, Betts G, West CML, Harris AL, Parkinson EK, Harrison PR, et al. Spectral clustering of microarray data elucidates the roles of microenvironment remodeling and immune responses in survival of head and neck squamous cell carcinoma. J Clin Oncol. 2010;28(17):2881–2888. doi: 10.1200/JCO.2009.24.8724. [DOI] [PubMed] [Google Scholar]
- 21.Rickman DS, Millon R, De Reynies A, Thomas E, Wasylyk C, Muller D, Abecassis J, Wasylyk B. Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene. 2008;27(51):6607–6622. doi: 10.1038/onc.2008.251. [DOI] [PubMed] [Google Scholar]
- 22.Rentoft M, Coates PJ, Laurell G, Nylander K. Transcriptional profiling of formalin fixed paraffin embedded tissue: pitfalls and recommendations for identifying biologically relevant changes. PLoS ONE. 2012;7(4):e35276. doi: 10.1371/journal.pone.0035276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng C-H, Liao C-T, Peng S-C, Chen Y-J, Cheng A-J, Juang J-L, Tsai C-Y, Chen T-C, Chuang Y-J, Tang C-Y, et al. A novel molecular signature identified by systems genetics approach predicts prognosis in oral squamous cell carcinoma. PLoS ONE. 2011;6(8):e23452. doi: 10.1371/journal.pone.0023452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su H, Hu N, Yang HH, Wang C, Takikita M, Wang Q-H, Giffen C, Clifford R, Hewitt SM, Shou J-Z, et al. Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin Cancer Res. 2011;17(9):2955–2966. doi: 10.1158/1078-0432.CCR-10-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanz-Pamplona R, Berenguer A, Cordero D, Mollevi D, Crous-Bou M, Sole X, Pare-Brunet L, Guino E, Salazar R, Santos C, et al. Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer. Mol Cancer. 2014;13(1):46. doi: 10.1186/1476-4598-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22(14):2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 27.Creekmore AL, Silkworth WT, Cimini D, Jensen RV, Roberts PC, Schmelz EM. Changes in gene expression and cellular architecture in an ovarian cancer progression model. PLoS ONE. 2011;6(3):e17676. doi: 10.1371/journal.pone.0017676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harper LJ, Piper K, Common J, Fortune F, Mackenzie IC. Stem cell patterns in cell lines derived from head and neck squamous cell carcinoma. J Oral Pathol Med. 2007;36(10):594–603. doi: 10.1111/j.1600-0714.2007.00617.x. [DOI] [PubMed] [Google Scholar]
- 29.Prime SS, Nixon SV, Crane IJ, Stone A, Matthews JB, Maitland NJ, Remnant L, Powell SK, Game SM, Scully C. The behaviour of human oral squamous cell carcinoma in cell culture. J Pathol. 1990;160(3):259–269. doi: 10.1002/path.1711600313. [DOI] [PubMed] [Google Scholar]
- 30.Costea DE, Kulasekara K, Neppelberg E, Johannessen AC, Vintermyr OK. Species-specific fibroblasts required for triggering invasiveness of partially transformed oral keratinocytes. Am J Pathol. 2006;168(6):1889–1897. doi: 10.2353/ajpath.2006.050843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones PH, Harper S, Watt FM. Stem cell patterning and fate in human epidermis. Cell. 1995;80(1):83–93. doi: 10.1016/0092-8674(95)90453-0. [DOI] [PubMed] [Google Scholar]
- 32.Li D-Q, Chen Z, Song XJ, de Paiva CS, Kim H-S, Pflugfelder SC. Partial enrichment of a population of human limbal epithelial cells with putative stem cell properties based on collagen type IV adhesiveness. Exp Eye Res. 2005;80(4):581–590. doi: 10.1016/j.exer.2004.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang X, Graham KA, Johannessen AC, Costea DE, Labeed FH. Human oral cancer cells with increasing tumorigenic abilities exhibit higher effective membrane capacitance. Integr Biol. 2014;6(5):545–554. doi: 10.1039/C3IB40255J. [DOI] [PubMed] [Google Scholar]
- 34.Liang X, Osman TA-H, Sapkota D, Neppelberg E, Lybak S, Liavaag PG, Johannessen AC, Jacobsen HK, Enger PØ, Costea DE, et al. Rapid adherence to collagen IV enriches for tumour initiating cells in oral cancer. Eur J Cancer. 2014;50(18):3262–3270. doi: 10.1016/j.ejca.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura T, Endo K-i, Kinoshita S. Identification of human oral keratinocyte stem/progenitor cells by neurotrophin receptor p75 and the role of neurotrophin/p75 signaling. Stem Cells. 2007;25(3):628–638. doi: 10.1634/stemcells.2006-0494. [DOI] [PubMed] [Google Scholar]
- 36.Okumura T, Shimada Y, Imamura M, Yasumoto S. Neurotrophin receptor p75NTR characterizes human esophageal keratinocyte stem cells in vitro. Oncogene. 2003;22(26):4017–4026. doi: 10.1038/sj.onc.1206525. [DOI] [PubMed] [Google Scholar]
- 37.Tomellini E, Lagadec C, Polakowska R, Le Bourhis X. Role of p75 neurotrophin receptor in stem cell biology: more than just a marker. Cell Mol Life Sci. 2014;1–15. [DOI] [PMC free article] [PubMed]
- 38.Huang S-D, Yuan Y, Liu X-H, Gong D-J, Bai C-G, Wang F, Luo J-H, Xu Z-Y. Self-renewal and chemotherapy resistance of p75NTR positive cells in esophageal squamous cell carcinomas. BMC Cancer. 2009;9(1):9. doi: 10.1186/1471-2407-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Civenni G, Walter A, Kobert N, Mihic-Probst D, Zipser M, Belloni B, Seifert B, Moch H, Dummer R, van den Broek M, et al. Human CD271-positive melanoma stem cells associated with metastasis establish tumor heterogeneity and long-term growth. Cancer Res. 2011;71(8):3098–3109. doi: 10.1158/0008-5472.CAN-10-3997. [DOI] [PubMed] [Google Scholar]
- 40.Osman TA, Parajuli H, Sapkota D, Ahmed IAH, Johannessen AC, Costea DE. The low-affinity nerve growth factor receptor p75NTR identifies a transient stem cell-like state in oral squamous cell carcinoma cells. J Oral Pathol Med. 2014. doi:10.1111/jop.12251. [DOI] [PubMed]
- 41.Lunde MLS, Roman E, Warnakulasurya S, Mehrotra R, Laranne J, Vasstrand EN, Ibrahim SO. Profiling of chromosomal changes in potentially malignant and malignant oral mucosal lesions from South and South-East Asia using array-comparative genomic hybridization. Cancer Genomics Proteomics. 2014;11(3):127–140. [PubMed] [Google Scholar]
- 42.Bryne M. Is the invasive front of an oral carcinoma the most important area for prognostication? Oral Dis. 1998;4(2):70–77. doi: 10.1111/j.1601-0825.1998.tb00260.x. [DOI] [PubMed] [Google Scholar]
- 43.Bryne M, Jenssen N, Boysen M. Histological grading in the deep invasive front of T1 and T2 glottic squamous cell carcinomas has high prognostic value. Virchows Arch. 1995;427(3):277–281. doi: 10.1007/BF00203395. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Zhang R, Xin J, Sun Y, Li J, Wei D, Zhao AZ. Identification of S100A16 as a novel adipogenesis promoting factor in 3T3-L1 cells. Endocrinology. 2011;152(3):903–911. doi: 10.1210/en.2010-1059. [DOI] [PubMed] [Google Scholar]
- 45.Kademani D, Bell RB, Bagheri S, Holmgren E, Dierks E, Potter B, Homer L. Prognostic factors in intraoral squamous cell carcinoma: the influence of histologic grade. J Oral Maxillofac Surg. 2005;63(11):1599–1605. doi: 10.1016/j.joms.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 46.Arduino PG, Carrozzo M, Chiecchio A, Broccoletti R, Tirone F, Borra E, Bertolusso G, Gandolfo S. Clinical and histopathologic independent prognostic factors in oral squamous cell carcinoma: a retrospective study of 334 cases. J Oral Maxillofac Surgery. 2008;66(8):1570–1579. doi: 10.1016/j.joms.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 47.Botti E, Spallone G, Moretti F, Marinari B, Pinetti V, Galanti S, De Meo PD, De Nicola F, Ganci F, Castrignano T, et al. Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proc Natl Acad Sci U S A. 2011;108(33):13710–13715. doi: 10.1073/pnas.1110931108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X, Cao W, Wang X, Zhang J, Lv Z, Qin X, Wu Y, Chen W. TGM3, a candidate tumor suppressor gene, contributes to human head and neck cancer. Mol Cancer. 2013;12(1):151. doi: 10.1186/1476-4598-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Restivo G, Nguyen B-C, Dziunycz P, Ristorcelli E, Ryan RJH, Özuysal ÖY, Di Piazza M, Radtke F, Dixon MJ, Hofbauer GFL, et al. IRF6 is a mediator of Notch pro-differentiation and tumour suppressive function in keratinocytes. EMBO J. 2011;30(22):4571–4585. doi: 10.1038/emboj.2011.325. [DOI] [PMC free article] [PubMed] [Google Scholar]


