Abstract
Progeroid disorders overlapping with De Barsy syndrome (DBS) are collectively denoted as autosomal-recessive cutis laxa type 3 (ARCL3). They are caused by biallelic mutations in PYCR1 or ALDH18A1, encoding pyrroline-5-carboxylate reductase 1 and pyrroline-5-carboxylate synthase (P5CS), respectively, which both operate in the mitochondrial proline cycle. We report here on eight unrelated individuals born to non-consanguineous families clinically diagnosed with DBS or wrinkly skin syndrome. We found three heterozygous mutations in ALDH18A1 leading to amino acid substitutions of the same highly conserved residue, Arg138 in P5CS. A de novo origin was confirmed in all six probands for whom parental DNA was available. Using fibroblasts from affected individuals and heterologous overexpression, we found that the P5CS-p.Arg138Trp protein was stable and able to interact with wild-type P5CS but showed an altered sub-mitochondrial distribution. A reduced size upon native gel electrophoresis indicated an alteration of the structure or composition of P5CS mutant complex. Furthermore, we found that the mutant cells had a reduced P5CS enzymatic activity leading to a delayed proline accumulation. In summary, recurrent de novo mutations, affecting the highly conserved residue Arg138 of P5CS, cause an autosomal-dominant form of cutis laxa with progeroid features. Our data provide insights into the etiology of cutis laxa diseases and will have immediate impact on diagnostics and genetic counseling.
Keywords: ALDH18A1, De Barsy syndrome, cutis laxa, mitochondria, proline metabolism, pyrroline-5-carboxylate synthase, P5CS, PYCR1, progeroid syndrome
Main Text
Syndromes with cutis laxa (CL) exhibit autosomal-dominant, autosomal-recessive, and X-linked modes of inheritance.1 The currently known autosomal-dominant forms are exclusively due to mutations in genes encoding proteins of the extracellular matrix (ECM). Beside the cutis laxa phenotype, affected individuals with mutations in ELN (MIM: 130160), encoding elastin (ADCL1 [MIM: 123700]), show severe cardiovascular and pulmonary symptoms.2,3 Additionally, mutations in FBLN5 (MIM: 604580) (ADCL2 [MIM: 614434]) were shown to cause an overlapping condition with more pronounced skeletal involvement.4
The autosomal-recessive forms of cutis laxa are clinically and genetically most variable.1,5 They can be subdivided into three distinct groups based on the clinical presentation and the localization of the affected gene products. ARCL type 1 (ARCL1 [MIM: 2191009]) is the most severe form with manifestations in the cardiovascular, pulmonary, genitourinary, and gastrointestinal systems. They are due to mutations in EFEMP2 (MIM: 604633), FBLN5, or LTBP4 (MIM: 604710) encoding components or modifiers of the ECM.6–9 In one case, biallelic mutations in ELN also were detected.10 An overlapping form of ARCL, which mainly shows arterial tortuosity and only mild cutis laxa, is due to mutations affecting the facilitated glucose transporter GLUT10 (SLC2A10 [MIM: 606145]).11,12
Autosomal-recessive cutis laxa type 2A (ARCL2A [MIM: 219200]) is caused by mutations in ATP6V0A2 (MIM: 611716), which encodes the a2 subunit of the V-type H+-ATPase mainly localized to the Golgi apparatus. ARCL2A is further characterized by a typical facial Gestalt, skeletal involvement, and variable intellectual disability. The presence of a glycosylation defect classifies this disorder also as a congenital disorder of glycosylation.13–15 Macrocephaly, alopecia, cutis laxa, and scoliosis (MACS [MIM: 613075]) syndrome is another disorder belonging to the ARCL disease spectrum, which is due to mutations in RIN2 (MIM: 600222), encoding an effector of the small GTPase Rab5. A mild hypoglycosylation of serum proteins can also be observed in RIN2-related cutis laxa.16,17 Gerodermia osteodysplastica (GO [MIM: 231070]) is caused by mutations in GORAB (MIM: 607983) encoding an effector of the small GTPases RAB6 and ARF5.18 Individuals affected by GO have a progeroid aspect, pronounced osteoporosis with fractures in early infancy, and usually no intellectual disability.19,20 ARCL2B (MIM: 612940) partially resembles GO but can be distinguished by a typical facial Gestalt and intellectual disability in most of the affected individuals. It is caused by mutations in PYCR1 (MIM: 179035) encoding pyrroline-5-carboxylate reductase 1, an enzyme involved in the de novo synthesis of proline from pyrroline-5-carboxylate (P5C) residing in mitochondria.21–25
ARCL type 3 (ARCL3A [MIM: 219150]; ARCL3B [MIM: 614438]) comprises De Barsy syndrome,26,27 which can be regarded as an extreme form of the clinical spectrum of ARCL2B with more pronounced progeroid appearance, cataract or corneal clouding, and profound intellectual disability. Most De Barsy-like (ARCL3)-affected persons carry mutations in PYCR1.21,28 Interestingly, mutations in PYCR2 (MIM: 616406) encoding the closely related enzyme pyrroline-5-carboxylate reductase 2 cause a phenotype with microcephaly and hypomyelination, but without skin manifestations.29 Mutations in ALDH18A1 (MIM: 138250), which encodes the mitochondrial enzyme pyrroline-5-carboxylate synthase (P5CS) responsible for the conversion of glutamic acid to P5C, have been found in the most severely affected ARCL3 individuals.25,30–35 PYCR1/2 and the enzyme proline dehydrogenase (PRODH) catalyzing the reverse reaction are key components of the proline cycle, which shuttles redox equivalents in and out mitochondria.36 Furthermore, this pathway is tightly connected to the urea cycle via ornithine aminotransferase (OAT) and to the TCA cycle via glutamate dehydrogenase (GLDH).37 P5CS and its counterpart pyrroline-5-carboxylate dehydrogenase (P5CDH) can also be regarded as integral parts of this proline cycle, because transporters for both end products, proline and glutamate, are present in mitochondria.
In the present study we describe eight affected individuals originating from Jordan, Finland, the Netherlands, Mexico, Arab Emirates, and the United States presenting with a progeroid, De Barsy-like cutis laxa phenotype. The family histories contained no information on other subjects with cutis laxa or other connective tissue disorders, miscarriage, early abortion, or consanguinity (Table 1, Figure 1A). All affected individuals showed a typical facial Gestalt with a triangular face, a prominent and broad forehead, cataracts or corneal clouding, and prominent low-set ears (Figure 1B). Furthermore, all probands presented with prenatal, and seven also with postnatal, growth restriction. All individuals had lax and thin skin with visible veins and joint hyperlaxity, six presented with cataracts, five with adducted thumbs, and four with cranial vessel tortuosity (Figure 1B, Table 1). Psychomotor development was delayed in all probands. Joint hyperlaxity and muscle hypotonia might have contributed to the motor delay as noted in the previously published case report for the affected individual 2-II:1.38 Furthermore, at least three probands presented with brisk peripheral reflexes. In addition, proband 3-II:1 was diagnosed with an autism spectrum disorder and in the affected individuals 2-II:1 and 3-II:1, a foramen magnum stenosis was observed. Based on the clinical features, the initial diagnosis of De Barsy syndrome was made in all affected individuals, except 1-II:4, 4-II:1, and 7-II:1 (Table 1). Because of a clinical overlap with progeroid Ehlers-Danlos syndrome, collagen type 1 and 3 production was assessed in proband 2-II:1 and showed reduced levels for the age range.38 In individuals 3-II:1 and 4-II:1, plasma amino acid profiling revealed no abnormalities. However, in the affected individual 7-II:1, the plasma amino acid levels of ornithine, arginine, and proline were decreased and in proband 8-II:2, ornithine, citrulline, and proline were reduced.
Table 1.
Summary of Clinical and Molecular Findings in Individuals with De Novo ALDH18A1 Mutations
| Family 1 | Family 2 | Family 3 | Family 4 | Family 5 | Family 6 | Family 7 | Family 8 | Freq. | |
|---|---|---|---|---|---|---|---|---|---|
| Clinical Findings | |||||||||
| Origin | Jordan | Finland | Netherlands | USA | Mexico | UAE | USA | USA | |
| Age at Last Evaluation | 2.5 years | 2.9 years | 2 years | 6 years | 4 years | 13 years | 3 years | 3 years | |
| IUGR | + | + | + | + | + | + | + | + | 100% |
| Postnatal growth delay | + | + | − | + | + | + | + | + | 88% |
| Dysmorphic features | − | + | + | + | + | + | + | + | 88% |
| Lax, wrinkled skin | + | + | + | + | + | + | + | + | 100% |
| Thin, translucent skin | + | + | + | + | + | + | + | + | 100% |
| Joint hyperlaxity | + | + | + | + | + | + | + | + | 100% |
| Hernias | + | + | + | + | − | + | − | + | 100% |
| Hip dislocation | ND | + | − | − | + | + | + | + | 71% |
| Adducted thumb | + | + | − | − | + | + | − | + | 63% |
| Club foot | − | + | − | − | + | + | − | − | 38% |
| Abnormal spine curvature | − | + | − | − | + | + | − | − | 38% |
| Osteopenia | − | + | − | − | + | − | ND | + | 43% |
| Late fontanel closure | − | + | + | − | + | ND | − | + | 50% |
| Wormian bones | ND | + | + | − | − | − | − | − | 29% |
| Microcephaly | + | − | − | + | + | + | + | − | 63% |
| Brain anomalies | − | + | − | − | − | ND | − | − | 14% |
| Cranial vessel tortuosity | − | − | + | + | − | − | + | + | 50% |
| Psychomotor retardation | + | + | + | + | + | + | + | + | 100% |
| Autism | ND | ND | + | − | ND | ND | ND | − | 33% |
| Feeding difficulties | − | + | + | + | ND | ND | + | + | 83% |
| Hypotonia | + | + | + | + | + | ND | + | + | 100% |
| Brisk reflexes | − | + | + | ND | + | ND | − | − | 50% |
| Cataract | − | + | + | + | + | + | + | + | 88% |
| Strabismus | − | − | + | − | − | − | − | + | 25% |
| Additional clinical findings | − | foramen magnum stenosis | foramen magnum stenosis, shallow sella turcica | os odontoideum, disharmonic bone age | − | − | − | − | |
| Plasma amino acid levels | ND | ND | normal | normal | ND | ND | ornithine ↓, arginine ↓, proline ↓ | ornithine ↓, citrulline ↓, proline ↓ | |
| Initial diagnosis | WSS | DBS | DBS | connective tissue disorder | DBS | DBS | cutis laxa | DBS | |
| Molecular Findings | |||||||||
| cDNA | c.412C>T | c.412C>T | c.412C>T | c.413G>T | c.413G>T | c.413G>A | c.413G>A | c.413G>A | |
| Parental origin | paternal | ND | paternal | ND | ND | ND | paternal | ND | |
| Protein | p.Arg138Trp | p.Arg138Trp | p.Arg138Trp | p.Arg138Leu | p.Arg138Leu | p.Arg138Gln | p.Arg138Gln | p.Arg138Gln | |
| MutationTaster score | 0.9999 (DC) | 0.9999 (DC) | 0.9999 (DC) | ||||||
| PolyPhen2 score | 1.000 (PD) | 1.000 (PD) | 1.000 (PD) | ||||||
| SIFT score | 0 (D) | 0.04 (D) | 0.04 (D) | ||||||
Abbreviations and symbols are as follows: +, present; −, absent; ND, not determined; DC, disease causing; PD, probably damaging; D, damaging; Freq, frequency of feature.
Figure 1.
Pedigrees and Clinical Phenotype of Eight De Barsy-like Affected Individuals
(A) Pedigrees of families 1–8 with De Barsy-like affected individuals.
(B) Clinical phenotype of probands 1-II:4, 3-II:1, 4-II:1, 5-II:3, and 6-II:1 at various ages between 5 months and 13 years. Ages are given in the lower right corner. Note triangular face, wrinkly and translucent skin (especially visible in 2-II:2 and 5-II:3), adducted thumbs, and abnormal spine curvature in the oldest proband 6-II:1. Muscular hypotonia is visible in proband 4-II:1. Written consent for publication of photographs from affected individuals was given by the probands’ legal representatives.
To identify the molecular genetic basis of the observed De Barsy-like phenotype, we investigated the genes ALDH18A1 (GenBank: NM_002860.3) and PYCR1 (GenBank: NM_001282280.1) by conventional Sanger sequencing in families 1–4 and 6–8 as described previously.31,34 In proband 5-II:3 we used a gene panel approach capturing the most relevant genes linked to connective tissue disorders (Figure S1A). In all affected individuals, sequencing of PYCR1, the most frequently affected gene in ARCL3, revealed no pathogenic sequence alteration.13 However, in the affected individuals 1-II:4, 2-II:1, and 3-II:1, a heterozygous change c.412C>T (p.Arg138Trp) and in 4-II:1 and 5-II:3 the transition c.413G>T (p.Arg138Leu) in ALDH18A1 were found (Figure 2A, Table 1). In the affected individuals 6-II:1, 7-II:1, and 8-II:2, we found the heterozygous substitution c.413G>A (p.Arg138Gln) (Figure 2A, Table 1). In none of the case subjects was a second mutation or pathogenic rare variant in the ALDH18A1 coding region detectable. Next, we excluded an additional exonic deletion in the ALDH18A1 locus in individuals 2-II:1, 5-II:3, and 6-II:1 by quantitative PCR and by cDNA expression analysis in probands 1-II:4 and 3-II:1 (data not shown). Because no second hit in ALDH18A1 was detectable, we sequenced DNA from the parents of families 1–3, 5, 7, and 8 to investigate whether the mutations were inherited. To our surprise, in all six families analyzed, a de novo occurrence of the mutation was proven. Unfortunately, no DNA was available for the parents from family 6 and only the maternal DNA from family 4 was available. In three case subjects, nearby SNPs allowed us to elucidate a paternal origin of the mutated allele (Figures S1B and S1C). Mutations affecting the residue Arg138 have not been described previously and are not mentioned in the ExAC, EVS, or the 1000 Genomes Project datasets. These mutations were predicted to be pathogenic according to MutationTaster, PolyPhen-2, and SIFT (Table 1). The residue Arg138 is phylogenetically highly conserved among all eukaryotes (Figure 2B). However, a 19 amino acid stretch containing Arg138 is absent in the orthologous E. coli enzyme γ-glutamyl kinase.
Figure 2.
Heterozygous De Novo Mutations Affecting Arg138, a Highly Conserved Residue of P5CS
(A) Sequence traces of ALDH18A1 exon 4 from affected individuals and parents demonstrating a de novo origin of the c.412C>T, c.413G>T, and c.413G>A mutations leading to the amino acid substitutions p.Arg138Trp, p.Arg138Leu, and p.Arg138Gln, respectively, in P5CS. Consents for molecular studies were obtained from all legal representatives of the affected infants. The Charité-Universitaetsmedizin Berlin ethics committee and the Institutional Review Board from the National University of Singapore approved the study.
(B) Alignment of P5CS protein sequence from different species around mutated human residue Arg138. Shading indicates degree of conservation. Note that E. coli does not contain any residue orthologous to Arg138, although it is conserved in all other species.
(C) Domains and alterations of the P5CS protein. The substitutions described in this study are given in red; all known alterations are given in black below the drawing. Cys612 is a catalytically important residue. Abbreviations are as follows: MTS, mitochondrial targeting sequence; GK, γ-glutamyl kinase; GPR, glutamyl-phosphate reductase.
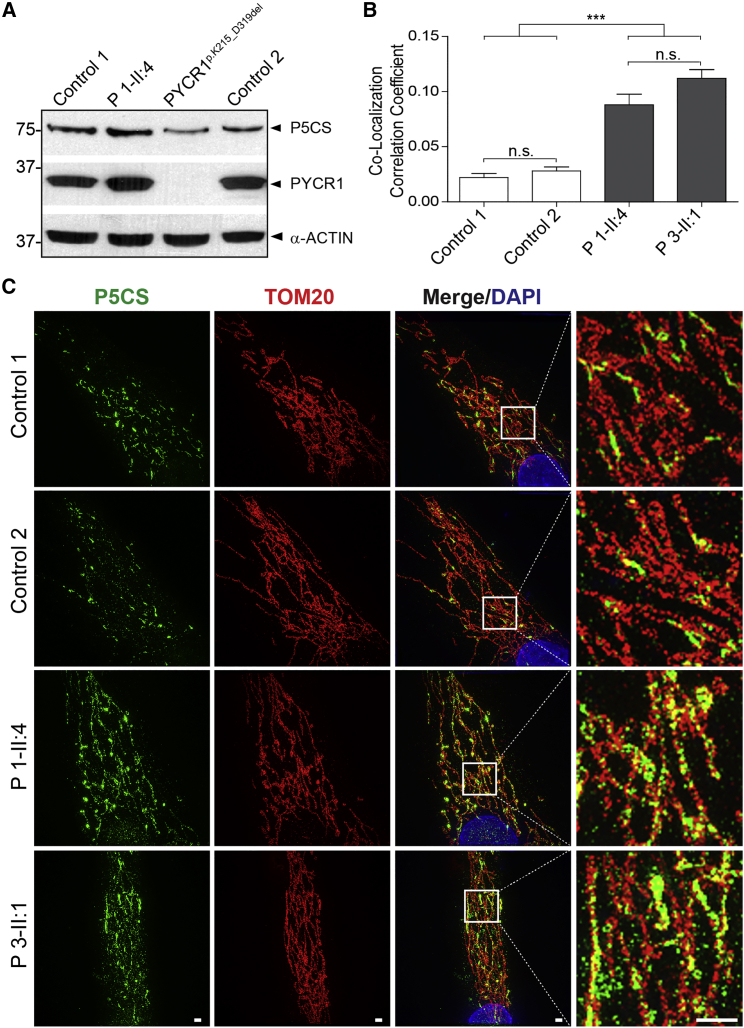
To further investigate the molecular basis of this disease, we documented that formation of the P5CS protein was not significantly reduced in skin fibroblasts bearing the heterozygous p.Arg138Trp substitution (Figures 3A and S2A). The levels of PYCR1 were unaltered or slightly higher. Because the protein was stable, a possible explanation for the pathogenic effect would be an altered targeting of the mutant P5CS protein. Superresolution microscopy revealed that P5CS co-localizes significantly more with the outer mitochondrial membrane marker TOM20 in P5CS mutant cells than in control cells, as shown by the Pearson’s correlation coefficients (Figure 3B). Furthermore, in fibroblasts derived from affected individuals, P5CS showed ribbon-like structures whereas in control cells more discrete puncta were visible (Figure 3C). To selectively assess the subcellular localization of P5CS-p.Arg138Trp, we created cell lines stably producing P5CS-WT and P5CS-p.Arg138Trp using the pLXIN vector system (Clontech) according to manufacturer instructions. Again, a mitochondrial localization was visible, and the mutant protein P5CS-p.Arg138Trp-V5 showed a more even distribution within the mitochondrial network and less accumulation in puncta when compared to V5-tagged wild-type P5CS (Figure S2B). We suspected that the altered localization of the mutant P5CS-p.Arg138Trp enzyme could impact its function and have dominant effects on the wild-type P5CS.
Figure 3.
Normal Levels and Altered Mitochondrial Targeting of the P5CS-p.Arg138Trp Protein
(A) No reduction in P5CS or PYCR1 levels in skin fibroblasts from proband (P) 1-II:4 compared to fibroblasts harboring the homozygous PYCR1 substitution p.Lys215_Asp319del cultivated in DMEM with 4.5 g/l glucose supplemented with 10% FCS, 1% ultraglutamine, and 1% penicillin/streptomycin at 37°C in a 5% CO2 atmosphere.28 For immunoblot, cells were lysed with modified RIPA (50 mM Tris-HCL, 1% NP40, 0.25% Na-deoxycholat, 150 mM NaCl, 1 mM EDTA + Complete proteinase inhibitor cocktail [Roche]) and protein concentrations were determined with the BCA-Kit (Pierce). A total amount of 20 μg protein was separated on a SDS-PAGE gel and proteins were transferred to nitrocellulose membranes. Membranes were blocked for 30 min at RT and primary antibodies (P5CS, Novus; GAPDH, Ambion; PYCR1, PTG) were incubated overnight at 4°C. After washing, the corresponding HRP-conjugated secondary antibodies were incubated for 1 hr at RT. Bands were visualized with ECL reagent (PerkinElmer). All blots were performed at least three times using different cell lysates.
(B) For each sample, five individual cells were imaged and the Pearson’s correlation coefficient calculated with the Coloc2 tools in FIJI.46 The graph shows that the co-localization between TOM20 and P5CS is 3.5- to 5.5-fold higher in cells from affected individuals compared to controls. Error indicates 1 SEM; n.s. indicates not significant; ∗∗∗p < 0.001.
(C) Mitochondrial localization of P5CS (green) complexes containing P5CS-p.Arg138Trp in comparison to the mitochondrial outer membrane marker TOM20 (red) as determined by superresolution microscopy. After fixation and permeabilization, primary antibodies were incubated overnight at 4°C. Secondary antibodies labeled with either 488 or 568 Alexa Fluor dyes (Life technologies) were incubated 1 hr at room temperature. Slides were mounted in VECTASHIELD H-1000 (Vector Laboratories). Superresolution images were acquired with a DeltaVision OMX v4 Blaze (GE Healthcare). The mutated protein forms smaller protein accumulations and is more evenly distributed throughout the mitochondrial network. Scale bars represent 5 μm.
It is known that the prokaryotic ortholog of the N-terminal portion of the bifunctional P5CS enzyme, γ-glutamyl kinase, is a dimer of dimers and a similar structure is also assumed for mammalian P5CS.39,40 In order to know whether substitutions of Arg138 alter the protein interaction profile of P5CS, we performed co-immunoprecipitation with transfected HEK293T cells and found that the substitutions did not interfere with the ability to bind P5CS-WT molecules (Figure 4A). Furthermore, we investigated the endogenous protein complex containing P5CS in fibroblast lysates from affected and control individuals by using native-PAGE followed by immunoblot analysis. In control cells, the total size of the P5CS-containing protein complex was approximately 440 kDa, which is larger than the theoretical 349 kDa for a P5CS homotetramer. Compared to that, the size of the complex containing the P5CS-p.Arg138Trp mutant protein in proband-derived fibroblasts was reproducibly smaller (about 410 kDa) (Figure 4B). The two most obvious explanations for the apparent mass of the P5CS complex would be binding of additional proteins other than P5CS or posttranslational modifications of P5CS itself. The reduced size of the mutant P5CS protein might indicate that one of these mechanisms or the whole 3D conformation is disturbed. Therefore, we asked whether this substitution interferes with the catalytic function of the enzyme. To answer this question, we chose to quantify the flux through the glutamate-proline pathway. We provided 13C515N-labeled glutamic acid to the cells and after 6 and 12 hr we determined the amount of 13C515N-labeled proline by mass spectrometry as previously described by Bicknell et al. (Figure 5A).30 In the controls we found similar amounts of labeled proline at the indicated time points, whereas in the P5CS-deficient cell line and the fibroblast lines bearing the heterozygous p.Arg138Trp substitution a reduced proline accumulation was detectable (Figure 5B).
Figure 4.
Preserved Interaction of P5CS-p.Arg138Trp and Altered Size of Protein Complex
(A) Co-immunoprecipitation of PC-tagged P5CS and P5CS-p.Arg138Trp with HA-tagged P5CS and P5CS-p.Arg138Trp. 3′-tagged human ALDH18A1 wild-type and c.412C>T mutant ORFs, cloned into pCS2+ between the BamHI and XhoI restriction site, were expressed in HEK293 cells after transient transfection with a NEPA21 super Eletroporator (NEPAGENE). Unrelated proteins ASUN-PC and ELMO-PC served as negative controls. Proteins were extracted with lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.1% Nonidet P-40, 0.05% deoxycholate, 1 mM CaCl2) supplemented with protease inhibitor cocktail (Roche). Protein C-tagged proteins were co-immunoprecipitated on Anti-Protein C Affinity Matrix (Roche) during 5 hr at 4°C together with HA-tagged proteins overnight at 4°C. Bound proteins were eluted from the beads via a wash buffer (20 mM Tris [pH 7.5], 0.5 M NaCl, 1 mM CaCl2). Electrophoresis was carried out in reducing loading buffer and analyzed by immunoblotting with indicated antibodies. Immunoblotting of cell lysates with antibodies against PC and HA revealed comparable expression of all tagged proteins.
(B) Native gel electrophoresis of P5CS-containing protein complexes isolated from fibroblasts from control individuals, probands (P) 2-II:1 and 3-II:1, and fibroblasts from an individual with the homozygous P5CS substitution p.Val601Glyfs∗24.31 Cells were lysed with modified RIPA (50 mM Tris-HCl, 1% NP40, 0.25% Na-deoxycholat, 150 mM NaCl, 1 mM EDTA + Complete proteinase inhibitor cocktail [Roche]) and protein concentrations were determined with the BCA-Kit (Pierce). A total amount of 5 μg protein was separated on a native-PAGE gel. Proteins were transferred to a nitrocellulose membrane and incubated with a P5CS antibody (Novus) to visualize the native protein complex. In control cells, a complex of an approximate size of 440 kDa was detected, which appeared smaller in lysates from p.Arg138Trp-expressing fibroblasts. The immunoblots below show identical lysates run in SDS-PAGE under denaturing conditions showing comparable loading by P5CS and GAPDH detection.
Figure 5.
Reduced Proline Accumulation in Fibroblasts from Affected Individuals Harboring the P5CS-p.Arg138Trp Substitution
(A) Overview of the design of the metabolic labeling experiment and the underlying biochemical pathway. Confluent skin fibroblasts derived from healthy controls, the probands (P) 1-II:4 and 3-II:1, and an individual with a homozygous P5CS p.Val601Glyfs∗24 substitution were washed with HBSS (Hanks’ Balanced Salt Solution) and incubated with medium containing 2 mM 13C515N glutamic acid (Silantes) for 6 and 12 hr, respectively. After uptake of the isotopic glutamic acid, the cells (dotted oval) metabolized this substrate and generated 13C515N proline via the indicated pathway. The cells were again washed and collected in 100 μl HBSS. Subsequently, the metabolites were extracted and analyzed via a targeted LC-MS/MS approach.47
(B) 13C515N proline levels relative to Ctrl 1 6 hr values. The resulting ratios are given with a log2 scale. Note clearly reduced efficiency of 13C515N proline accumulation in cells harboring biallelic or monoallelic ALDH18A1 mutations.
In 1968 de Barsy described an affected individual with profound intellectual disability, choreoathetoid movements, clouded corneas, and a progeroid appearance.26 More recently, affected individuals with a very similar phenotype were described to harbor biallelic mutations in ALDH18A1 (ARCL3A) or in PYCR1 (ARCL3B). Only 13 affected individuals from 7 unrelated families carrying biallelic ALDH18A1 mutations have been described so far.31 The present study shows de novo heterozygous mutations in ALDH18A1 affecting the highly conserved amino acid Arg138 of P5CS as the cause of a similar but milder progeroid cutis laxa phenotype, which has important impact for diagnostics and genetic counseling. All probands showed cataract or corneal clouding, thin skin with visible veins and wrinkles, moderate intellectual disability, clenched fingers, and pre- and postnatal growth retardation. Though this phenotype is reminiscent of the milder PYCR1-related cutis laxa phenotype, cataract and/or corneal clouding seem to be distinctive.21 The neurological phenotype of our probands is further characterized by a combination of muscular hypotonia with brisk muscle reflexes, reminiscent of disorders affecting the upper motoneuron like the spastic paraplegias (SPGs). Indeed, heterozygous inherited mutations in different ALDH18A1 exons have recently been shown to be causative for SPG9 (MIM: 601162) with an onset from the first to the fourth decade of life.41 These data further increase the plausibility of a pathogenic effect of heterozygous P5CS variants and expand the disease spectrum caused by ALDH18A1 mutations. Consequently, the phenotype with a pre- or neonatal onset described here lies within the clinical spectrum of disorders caused by biallelic mutations either in PYCR1 or ALDH18A1. We propose to call this disease entity autosomal-dominant cutis laxa with progeroid features (ALDH18A1-ADCL) to distinguish it from ELN-ADCL and FBLN5-ADCL. It should be noted that the parents of the sporadic case initially described by de Barsy et al. were of different nationalities.26 Although this does not exclude a coincidental inheritance of two rare loss-of-function mutations in ALDH18A1 or PYCR1, our data open the possibility that also in this case a de novo ALDH18A1 mutation might have been causative. Furthermore, Martinelli et al. reported on a case carrying the de novo p.Gly93Arg (c.277G>A) substitution in combination with the polymorphism p.Thr299Ile (c.896C>T).33 Interestingly, this proband seems more severely affected because he had cortical atrophy, thin corpus callosum, clonic seizures, and metabolic abnormalities with hyperammonemia and low plasma citrulline levels. Only in a subgroup of recessive cases are alterations of plasma amino acids present.33,42 The absence of such metabolic features does not rule out the diagnosis of ALDH18A1-related cutis laxa, but when present they are a strong indication for P5CS deficiency as in our probands 7-II and 8-II:2.
Most ALDH18A1-related ARCL mutations cluster in the C-terminal region of P5CS between amino acids 601 and 784, which lies within the γ-glutamyl phosphate reductase domain (Figure 2C). Only two mutations residing in the P5CS γ-glutamyl-kinase domain have already been described: a homozygous p.Arg84Gln substitution and, as mentioned above, the heterozygous de novo substitution p.Gly93Arg.33,42 In contrast to biallelic ALDH18A1 mutations, the de novo mutations described here are unlikely to completely destroy P5CS enzymatic function. This is further supported by the strongly reduced proline accumulation in the cell lines harboring p.Arg138Trp in comparison to complete P5CS deficiency. P5CS, which catalyzes the first step of proline biosynthesis, is assumed to be a dimer of dimers.39 Statistically, 6.25% of all formed tetramers can be predicted to still consist of wild-type P5CS molecules if only one allele carries a mutation. Furthermore, it is possible that tetramers containing both wild-type and mutated subunits might still retain some function, especially if the mutated protein is stable and able to interact with the wild-type form, as demonstrated in our study for P5CS-p.Arg138Trp. We believe that this finding is crucial to explain a dominant-negative effect because in the absence of binding, we would end up with 50% of wild-type P5CS complexes, which are obviously sufficient for a healthy development.
P5CS targets to mitochondria through a N-terminal mitochondrial targeting signal (MTS).39 Overexpressed P5CS-p.Arg138Trp also co-localized with mitochondrial markers, but whereas overexpressed P5CS-WT formed large aggregates in vicinity to mitochondria, the mutated protein was more evenly dispersed within the mitochondrial network. An important difference between intrinsic P5CS protein complexes isolated from fibroblasts from control and affected individuals was a size reduction detectable after native-PAGE indicating either loss of an interaction partner, an altered 3D conformation due to the altered charge, or an abnormal modification of the P5CS-p.Arg138Trp-containing complexes. The size of the wild-type complex was approximately 440 kDa. If P5CS is a tetramer, the theoretical size of this complex would be 349 kDa, which would leave room for additional binding partners with a total mass of 90 kDa.39 On the other hand, apart from P5CS itself, eight potential binding partners are mentioned in the IntAct database.43 Of those only ICT1, a 22 kDa subunit of the mitochondrial ribosome, resides in the correct organelle. Further research is needed to determine whether there are relevant P5CS interaction partners. Interestingly, three P5CS lysine residues have been shown to undergo succinylation.44 An altered lysine succinylation could influence the folding state of the P5CS monomers and thereby the migration of the protein complex in the native-PAGE gel. An alternative explanation would be an altered proteolytic cleavage of the P5CS protein. An obvious candidate would be the predicted MTS. In proteins of the mitochondrial intermembrane space like P5CS, the MTS can get cleaved by the inner membrane peptidase (IMP).45 Further experiments will be needed to specify by which mechanism P5CS is targeted and processed.
Taken together, we have shown here that heterozygous P5CS de novo mutations affecting the residue Arg138 lead to subtle changes in the behavior of the P5CS protein complex and its enzymatic function. Our results clearly show that besides mutations that affect components of the ECM, mutations affecting the mitochondrial protein P5CS can cause a form of autosomal-dominant cutis laxa with progeroid features resembling De Barsy syndrome.
Acknowledgments
We are grateful to the affected individuals and their families whose cooperation made this study possible. We thank Beata Lukaszewska-McGreal for excellent technical assistance and metabolite sample preparation. The study was funded by the Deutsche Forschungsgemeinschaft (KO 2891/1-1) to U. Kornak. B.R. is a fellow of the Branco Weiss Foundation, an A∗STAR Investigator, and Young EMBO Investigator. This work was partly funded by the Skin Research Institute of Singapore and a Strategic Positioning Fund on Genetic Orphan Diseases from A∗STAR, Singapore. B.C. is a senior clinical investigator of the Fund for Scientific Research – Flanders. This study was funded in part by a Methusalem grant from the Ghent University (BOF 08/01M01108). This study was funded in part by the Max Planck Society.
Published: August 27, 2015
Footnotes
Supplemental Data include two figures and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.08.001.
Contributor Information
Bruno Reversade, Email: bruno@reversade.com.
Uwe Kornak, Email: uwe.kornak@charite.de.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
Clustal W and Clustal X, http://www.clustal.org/clustal2/
ExAC Browser, http://exac.broadinstitute.org/
GeneTalk, http://www.gene-talk.de/
IntAct Molecular Interaction Database http://www.ebi.ac.uk/intact/
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Morava E., Guillard M., Lefeber D.J., Wevers R.A. Autosomal recessive cutis laxa syndrome revisited. Eur. J. Hum. Genet. 2009;17:1099–1110. doi: 10.1038/ejhg.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Callewaert B., Renard M., Hucthagowder V., Albrecht B., Hausser I., Blair E., Dias C., Albino A., Wachi H., Sato F. New insights into the pathogenesis of autosomal-dominant cutis laxa with report of five ELN mutations. Hum. Mutat. 2011;32:445–455. doi: 10.1002/humu.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Urban Z., Gao J., Pope F.M., Davis E.C. Autosomal dominant cutis laxa with severe lung disease: synthesis and matrix deposition of mutant tropoelastin. J. Invest. Dermatol. 2005;124:1193–1199. doi: 10.1111/j.0022-202X.2005.23758.x. [DOI] [PubMed] [Google Scholar]
- 4.Markova D., Zou Y., Ringpfeil F., Sasaki T., Kostka G., Timpl R., Uitto J., Chu M.L. Genetic heterogeneity of cutis laxa: a heterozygous tandem duplication within the fibulin-5 (FBLN5) gene. Am. J. Hum. Genet. 2003;72:998–1004. doi: 10.1086/373940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardeitchik T., Mohamed M., Fischer B., Lammens M., Lefeber D., Lace B., Parker M., Kim K.J., Lim B.C., Häberle J. Clinical and biochemical features guiding the diagnostics in neurometabolic cutis laxa. Eur. J. Hum. Genet. 2014;22:888–895. doi: 10.1038/ejhg.2013.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hucthagowder V., Sausgruber N., Kim K.H., Angle B., Marmorstein L.Y., Urban Z. Fibulin-4: a novel gene for an autosomal recessive cutis laxa syndrome. Am. J. Hum. Genet. 2006;78:1075–1080. doi: 10.1086/504304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loeys B., Van Maldergem L., Mortier G., Coucke P., Gerniers S., Naeyaert J.M., De Paepe A. Homozygosity for a missense mutation in fibulin-5 (FBLN5) results in a severe form of cutis laxa. Hum. Mol. Genet. 2002;11:2113–2118. doi: 10.1093/hmg/11.18.2113. [DOI] [PubMed] [Google Scholar]
- 8.Urban Z., Hucthagowder V., Schürmann N., Todorovic V., Zilberberg L., Choi J., Sens C., Brown C.W., Clark R.D., Holland K.E. Mutations in LTBP4 cause a syndrome of impaired pulmonary, gastrointestinal, genitourinary, musculoskeletal, and dermal development. Am. J. Hum. Genet. 2009;85:593–605. doi: 10.1016/j.ajhg.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callewaert B., Su C.T., Van Damme T., Vlummens P., Malfait F., Vanakker O., Schulz B., Mac Neal M., Davis E.C., Lee J.G. Comprehensive clinical and molecular analysis of 12 families with type 1 recessive cutis laxa. Hum. Mutat. 2013;34:111–121. doi: 10.1002/humu.22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mégarbané H., Florence J., Sass J.O., Schwonbeck S., Foglio M., de Cid R., Cure S., Saker S., Mégarbané A., Fischer J. An autosomal-recessive form of cutis laxa is due to homozygous elastin mutations, and the phenotype may be modified by a heterozygous fibulin 5 polymorphism. J. Invest. Dermatol. 2009;129:1650–1655. doi: 10.1038/jid.2008.450. [DOI] [PubMed] [Google Scholar]
- 11.Callewaert B.L., Willaert A., Kerstjens-Frederikse W.S., De Backer J., Devriendt K., Albrecht B., Ramos-Arroyo M.A., Doco-Fenzy M., Hennekam R.C., Pyeritz R.E. Arterial tortuosity syndrome: clinical and molecular findings in 12 newly identified families. Hum. Mutat. 2008;29:150–158. doi: 10.1002/humu.20623. [DOI] [PubMed] [Google Scholar]
- 12.Coucke P.J., Willaert A., Wessels M.W., Callewaert B., Zoppi N., De Backer J., Fox J.E., Mancini G.M., Kambouris M., Gardella R. Mutations in the facilitative glucose transporter GLUT10 alter angiogenesis and cause arterial tortuosity syndrome. Nat. Genet. 2006;38:452–457. doi: 10.1038/ng1764. [DOI] [PubMed] [Google Scholar]
- 13.Fischer B., Dimopoulou A., Egerer J., Gardeitchik T., Kidd A., Jost D., Kayserili H., Alanay Y., Tantcheva-Poor I., Mangold E. Further characterization of ATP6V0A2-related autosomal recessive cutis laxa. Hum. Genet. 2012;131:1761–1773. doi: 10.1007/s00439-012-1197-8. [DOI] [PubMed] [Google Scholar]
- 14.Hucthagowder V., Morava E., Kornak U., Lefeber D.J., Fischer B., Dimopoulou A., Aldinger A., Choi J., Davis E.C., Abuelo D.N. Loss-of-function mutations in ATP6V0A2 impair vesicular trafficking, tropoelastin secretion and cell survival. Hum. Mol. Genet. 2009;18:2149–2165. doi: 10.1093/hmg/ddp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kornak U., Reynders E., Dimopoulou A., van Reeuwijk J., Fischer B., Rajab A., Budde B., Nürnberg P., Foulquier F., Lefeber D., ARCL Debré-type Study Group Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2. Nat. Genet. 2008;40:32–34. doi: 10.1038/ng.2007.45. [DOI] [PubMed] [Google Scholar]
- 16.Albrecht B., de Brouwer A.P., Lefeber D.J., Cremer K., Hausser I., Rossen N., Wortmann S.B., Wevers R.A., Kornak U., Morava E. MACS syndrome: A combined collagen and elastin disorder due to abnormal Golgi trafficking. Am. J. Med. Genet. A. 2010;152A:2916–2918. doi: 10.1002/ajmg.a.33712. [DOI] [PubMed] [Google Scholar]
- 17.Basel-Vanagaite L., Sarig O., Hershkovitz D., Fuchs-Telem D., Rapaport D., Gat A., Isman G., Shirazi I., Shohat M., Enk C.D. RIN2 deficiency results in macrocephaly, alopecia, cutis laxa, and scoliosis: MACS syndrome. Am. J. Hum. Genet. 2009;85:254–263. doi: 10.1016/j.ajhg.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egerer J., Emmerich D., Fischer-Zirnsak B., Lee Chan W., Meierhofer D., Tuysuz B., Marschner K., Sauer S., Barr F.A., Mundlos S., Kornak U. GORAB missense mutations disrupt RAB6 and ARF5 binding and golgi targeting. J. Invest. Dermatol. 2015 doi: 10.1038/jid.2015.192. Published online May 22, 2015. [DOI] [PubMed] [Google Scholar]
- 19.Hennies H.C., Kornak U., Zhang H., Egerer J., Zhang X., Seifert W., Kühnisch J., Budde B., Nätebus M., Brancati F. Gerodermia osteodysplastica is caused by mutations in SCYL1BP1, a Rab-6 interacting golgin. Nat. Genet. 2008;40:1410–1412. doi: 10.1038/ng.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajab A., Kornak U., Budde B.S., Hoffmann K., Jaeken J., Nürnberg P., Mundlos S. Geroderma osteodysplasticum hereditaria and wrinkly skin syndrome in 22 patients from Oman. Am. J. Med. Genet. A. 2008;146A:965–976. doi: 10.1002/ajmg.a.32143. [DOI] [PubMed] [Google Scholar]
- 21.Dimopoulou A., Fischer B., Gardeitchik T., Schröter P., Kayserili H., Schlack C., Li Y., Brum J.M., Barisic I., Castori M. Genotype-phenotype spectrum of PYCR1-related autosomal recessive cutis laxa. Mol. Genet. Metab. 2013;110:352–361. doi: 10.1016/j.ymgme.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Kouwenberg D., Gardeitchik T., Wevers R.A., Häberle J., Morava E. Recognizable phenotype with common occurrence of microcephaly, psychomotor retardation, but no spontaneous bone fractures in autosomal recessive cutis laxa type IIB due to PYCR1 mutations. Am. J. Med. Genet. A. 2011;155A:2331–2332. doi: 10.1002/ajmg.a.34154. author reply 2333–2334. [DOI] [PubMed] [Google Scholar]
- 23.Lin D.S., Yeung C.Y., Liu H.L., Ho C.S., Shu C.H., Chuang C.K., Huang Y.W., Wu T.Y., Huang Z.D., Jian Y.R., Lin S.P. A novel mutation in PYCR1 causes an autosomal recessive cutis laxa with premature aging features in a family. Am. J. Med. Genet. A. 2011;155A:1285–1289. doi: 10.1002/ajmg.a.33963. [DOI] [PubMed] [Google Scholar]
- 24.Yildirim Y., Tolun A., Tüysüz B. The phenotype caused by PYCR1 mutations corresponds to geroderma osteodysplasticum rather than autosomal recessive cutis laxa type 2. Am. J. Med. Genet. A. 2011;155A:134–140. doi: 10.1002/ajmg.a.33747. [DOI] [PubMed] [Google Scholar]
- 25.Zampatti S., Castori M., Fischer B., Ferrari P., Garavelli L., Dionisi-Vici C., Agolini E., Wischmeijer A., Morava E., Novelli G. De Barsy Syndrome: a genetically heterogeneous autosomal recessive cutis laxa syndrome related to P5CS and PYCR1 dysfunction. Am. J. Med. Genet. A. 2012;158A:927–931. doi: 10.1002/ajmg.a.35231. [DOI] [PubMed] [Google Scholar]
- 26.de Barsy A.M., Moens E., Dierckx L. Dwarfism, oligophrenia and degeneration of the elastic tissue in skin and cornea. A new syndrome? Helv. Paediatr. Acta. 1968;23:305–313. [PubMed] [Google Scholar]
- 27.Kunze J., Majewski F., Montgomery P., Hockey A., Karkut I., Riebel T. De Barsy syndrome--an autosomal recessive, progeroid syndrome. Eur. J. Pediatr. 1985;144:348–354. doi: 10.1007/BF00441776. [DOI] [PubMed] [Google Scholar]
- 28.Reversade B., Escande-Beillard N., Dimopoulou A., Fischer B., Chng S.C., Li Y., Shboul M., Tham P.Y., Kayserili H., Al-Gazali L. Mutations in PYCR1 cause cutis laxa with progeroid features. Nat. Genet. 2009;41:1016–1021. doi: 10.1038/ng.413. [DOI] [PubMed] [Google Scholar]
- 29.Nakayama T., Al-Maawali A., El-Quessny M., Rajab A., Khalil S., Stoler J.M., Tan W.H., Nasir R., Schmitz-Abe K., Hill R.S. Mutations in PYCR2, encoding pyrroline-5-carboxylate reductase 2, cause microcephaly and hypomyelination. Am. J. Hum. Genet. 2015;96:709–719. doi: 10.1016/j.ajhg.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bicknell L.S., Pitt J., Aftimos S., Ramadas R., Maw M.A., Robertson S.P. A missense mutation in ALDH18A1, encoding Delta1-pyrroline-5-carboxylate synthase (P5CS), causes an autosomal recessive neurocutaneous syndrome. Eur. J. Hum. Genet. 2008;16:1176–1186. doi: 10.1038/ejhg.2008.91. [DOI] [PubMed] [Google Scholar]
- 31.Fischer B., Callewaert B., Schröter P., Coucke P.J., Schlack C., Ott C.E., Morroni M., Homann W., Mundlos S., Morava E. Severe congenital cutis laxa with cardiovascular manifestations due to homozygous deletions in ALDH18A1. Mol. Genet. Metab. 2014;112:310–316. doi: 10.1016/j.ymgme.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Handley M.T., Mégarbané A., Meynert A.M., Brown S., Freyer E., Taylor M.S., Jackson I.J., Aligianis I.A. Loss of ALDH18A1 function is associated with a cellular lipid droplet phenotype suggesting a link between autosomal recessive cutis laxa type 3A and Warburg Micro syndrome. Mol. Genet. Genomic Med. 2014;2:319–325. doi: 10.1002/mgg3.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinelli D., Häberle J., Rubio V., Giunta C., Hausser I., Carrozzo R., Gougeard N., Marco-Marín C., Goffredo B.M., Meschini M.C. Understanding pyrroline-5-carboxylate synthetase deficiency: clinical, molecular, functional, and expression studies, structure-based analysis, and novel therapy with arginine. J. Inherit. Metab. Dis. 2012;35:761–776. doi: 10.1007/s10545-011-9411-8. [DOI] [PubMed] [Google Scholar]
- 34.Skidmore D.L., Chitayat D., Morgan T., Hinek A., Fischer B., Dimopoulou A., Somers G., Halliday W., Blaser S., Diambomba Y. Further expansion of the phenotypic spectrum associated with mutations in ALDH18A1, encoding Δ1-pyrroline-5-carboxylate synthase (P5CS) Am. J. Med. Genet. A. 2011;155A:1848–1856. doi: 10.1002/ajmg.a.34057. [DOI] [PubMed] [Google Scholar]
- 35.Wolthuis D.F., van Asbeck E., Mohamed M., Gardeitchik T., Lim-Melia E.R., Wevers R.A., Morava E. Cutis laxa, fat pads and retinopathy due to ALDH18A1 mutation and review of the literature. Eur. J. Paediatr. Neurol. 2014;18:511–515. doi: 10.1016/j.ejpn.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Mohamed M., Kouwenberg D., Gardeitchik T., Kornak U., Wevers R.A., Morava E. Metabolic cutis laxa syndromes. J. Inherit. Metab. Dis. 2011;34:907–916. doi: 10.1007/s10545-011-9305-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phang J.M., Pandhare J., Liu Y. The metabolism of proline as microenvironmental stress substrate. J. Nutr. 2008;138:2008S–2015S. doi: 10.1093/jn/138.10.2008S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jukkola A., Kauppila S., Risteli L., Vuopala K., Risteli J., Leisti J., Pajunen L. New lethal disease involving type I and III collagen defect resembling geroderma osteodysplastica, De Barsy syndrome, and Ehlers-Danlos syndrome IV. J. Med. Genet. 1998;35:513–518. doi: 10.1136/jmg.35.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pérez-Arellano I., Carmona-Alvarez F., Martínez A.I., Rodríguez-Díaz J., Cervera J. Pyrroline-5-carboxylate synthase and proline biosynthesis: from osmotolerance to rare metabolic disease. Protein Sci. 2010;19:372–382. doi: 10.1002/pro.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marco-Marín C., Gil-Ortiz F., Pérez-Arellano I., Cervera J., Fita I., Rubio V. A novel two-domain architecture within the amino acid kinase enzyme family revealed by the crystal structure of Escherichia coli glutamate 5-kinase. J. Mol. Biol. 2007;367:1431–1446. doi: 10.1016/j.jmb.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 41.Coutelier M., Goizet C., Durr A., Habarou F., Morais S., Dionne-Laporte A., Tao F., Konop J., Stoll M., Charles P. Alteration of ornithine metabolism leads to dominant and recessive hereditary spastic paraplegia. Brain. 2015;138:2191–2205. doi: 10.1093/brain/awv143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baumgartner M.R., Hu C.A., Almashanu S., Steel G., Obie C., Aral B., Rabier D., Kamoun P., Saudubray J.M., Valle D. Hyperammonemia with reduced ornithine, citrulline, arginine and proline: a new inborn error caused by a mutation in the gene encoding delta(1)-pyrroline-5-carboxylate synthase. Hum. Mol. Genet. 2000;9:2853–2858. doi: 10.1093/hmg/9.19.2853. [DOI] [PubMed] [Google Scholar]
- 43.Kerrien S., Aranda B., Breuza L., Bridge A., Broackes-Carter F., Chen C., Duesbury M., Dumousseau M., Feuermann M., Hinz U. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mossmann D., Meisinger C., Vögtle F.N. Processing of mitochondrial presequences. Biochim. Biophys. Acta. 2012;1819:1098–1106. doi: 10.1016/j.bbagrm.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gielisch I., Meierhofer D. Metabolome and proteome profiling of complex I deficiency induced by rotenone. J. Proteome Res. 2015;14:224–235. doi: 10.1021/pr500894v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.