Abstract
Purpose of review
This article will review the genetic evidence implicating ERAP1, which encodes the endoplasmic reticulum-associated amino-peptidase 1, in susceptibility to rheumatic disease.
Recent findings
Genetic variants and haplotypes of ERAP1 are associated with ankylosing spondylitis, psoriasis, and Behçet’s disease in people of varying ancestries. In each of these diseases, disease-associated variants of ERAP1 have been shown to interact with disease-associated class I Human Leukocyte Antigen (HLA) alleles to influence disease risk. Functionally, disease-associated missense variants of ERAP1 concertedly alter ERAP1 enzymatic function, both quantitatively and qualitatively, while other disease-associated variants influence ERAP1 expression. Therefore, ERAP1 haplotypes (or allotypes) should be examined as functional units. Biologically, this amounts to an examination of the gene regulation and function of the protein encoded by each allotype. Genetically, the relationship between disease risk and ERAP1 allotypes should be examined to determine whether allotypes or individual variants produce the most parsimonious risk models.
Summary
Future investigations of ERAP1 should focus on comprehensively characterizing naturally-occurring ERAP1 allotypes, examining the enzymatic function and gene expression of each allotype, and identifying specific allotypes that influence disease susceptibility.
Keywords: major histocompatibility complex, antigen processing, epistasis
Introduction
The gene encoding the endoplasmic reticulum-associated amino-peptidase 1 (ERAP1) protein is highly polymorphic with several common polymorphisms encoding variant amino acids (1). This peptidase trims peptides that have been transported from the intracellular space into the endoplasmic reticulum (ER). Trimming is required for efficient peptide loading onto class I major histocompatibility complex (MHC) molecules, which are displayed on the surface of nearly all cell types. MHC class I-peptide complexes play important roles in immune surveillance and in innate and adaptive immune functions through their interactions with T-cell receptors (TCRs) and several cell activating and inhibitory receptors, including the killer cell immunoglobulin-like receptors (KIRs), the killer cell lectin-like receptors (KLRs), and the leukocyte immunoglobulin-like receptors (LILRs). Similar to TCR-peptide specificity, the inhibitory receptors also exhibit selectivity for peptides bound to MHC class I (2). Several ERAP1 coding variants are associated at genome-wide significance with three rheumatic diseases, ankylosing spondylitis (AS) (3–5), psoriasis (6, 7), and Behçet’s disease (BD) (8), which all have strong associations with MHC class I molecules. Strong interactions of coding ERAP1 variants with the appropriate disease-specific Human Leukocyte Antigen (HLA) class I proteins suggest that ERAP1 trimming of peptides plays a role in susceptibility to these class I HLA-associated diseases (4, 6, 8). Changes to the structure of the ERAP1 protein are likely to influence the nature of peptides bound in the active site and their ability to be trimmed, and therefore could influence the peptidome that is available for class I HLA binding and presentation. Although coding and non-coding ERAP1 SNPs and three or four marker SNP haplotypes have been associated with AS, psoriasis, and BD, the data reported are often insufficient to enable direct comparisons of haplotypes and their disease associations between studies. Recently, coding variation in ERAP1 and alterations in ERAP1 activity have been explored with the idea that ERAP1 amino acid variants may concertedly influence its function (4, 9, 10), and therefore more complete ERAP1 amino acid sequence or allotype information, and even allotype combinations, may be needed to understand ERAP1 function and its role in disease pathogenesis.
In this review, we encapsulate the existing genetic literature implicating ERAP1 variation in rheumatic diseases. The functional consequences of ERAP1 variation are discussed in detail by Tran and colleagues later in this issue [Tran T and Colbert RA, this issue].
Example of ERAP1 allotype assembly from HapMap reference populations
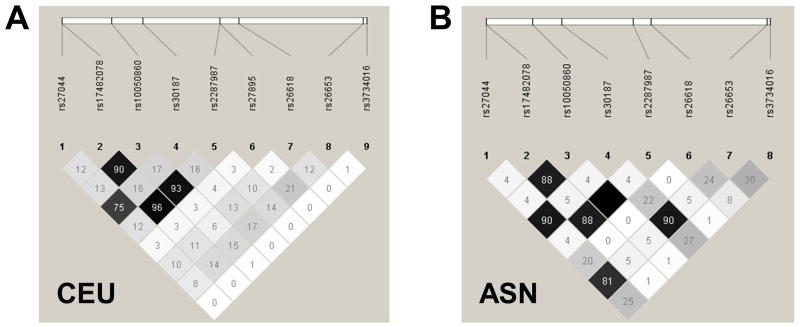
In order to discuss and study ERAP1 haplotypes/allotypes and their role in genetically-complex diseases, it is critical to accurately identify and classify the haplotypes in a standardized way. For disease association analyses, the allotypes should be based on common missense variants in ERAP1, as observed in large, ancestrally-defined reference populations of healthy individuals. As an example, we have examined the 1000 Genomes dataset and identified 10 missense SNPs in ERAP1 that were present at greater than 5% minor allele frequency in at least one super-population (Table 1). Haplotype analysis including the 9 missense variants that were genotyped in the HapMap CEU or ASN (HCB + JPT) individuals reveals 10 haplotypes with a frequency of greater than 1% in one or more of the populations (Table 2). These haplotypes are derived from an ancestral haplotype (Hap1), which bears the alleles found in chimpanzees, orangutans, and macaques (Table 2). An examination of the linkage disequilibrium structure of the common, missense variants in the CEU and ASN populations are shown in Figure 1. In the ensuing discussion of the associations between ERAP1 and rheumatic diseases, we will attempt to unify the discussion using the ERAP1 allotypes defined in the HapMap populations (Table 2).
Table 1.
ERAP1 missense SNPs from the 1000 Genomes Project super-populations*
| SNP ID | Amino acid change (ancestral/non-ancestral) | cDNA change (ancestral/non-ancestral) | EUR Freq. (% ancestral/ % non-ancestral) | AFR Freq. (% ancestral/% non-ancestral) | ASN Freq. (% ancestral/% non-ancestral) |
|---|---|---|---|---|---|
| rs72773968 | T12I | C/T | 86.2/13.8 | 99.5/0.5 | 99.6/0.4 |
| rs3734016 | E56K | G/A | 95.9/4.1 | 91.9/8.1 | 82.8/17.2 |
| rs26653 | P127R | C/G | 71.8/28.2 | 48.2/51.8 | 52.1/47.9 |
| rs26618 | I276M | A/G | 77.3/22.7 | 80.4/19.6 | 71.4/28.6 |
| rs27895 | G346D | G/A | 93.6/6.4 | 76.2/23.8 | 99.9/0.1 |
| rs2287987 | M349V | A/G | 77.5/22.5 | 93.4/6.6 | 94.2/5.8 |
| rs30187 | K528R | A/G | 35.0/65.0 | 40.2/59.8 | 45.3/54.7 |
| rs10050860 | D575N | G/A | 77.1/22.9 | 93.5/6.5 | 94.2/5.8 |
| rs17482078 | R725Q | G/A | 77.6/22.4 | 94.6/5.4 | 94.2/5.8 |
| rs27044 | Q730E | C/G | 28.5/71.5 | 28.9/71.1 | 42.9/57.1 |
Table includes ERAP1 missense SNPs that were present at minor allele frequency > 0.05 in at least 1 of the 3 super-populations of the 1000 Genomes Project data. EUR, European superpopulation; AFR, African superpopulation; ASN, Asian superpopulation.
Table 2.
Haplotypes of common ERAP1 missense variants in HapMap populations*
| Freq. (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA position | 56 | 127 | 276 | 346 | 349 | 528 | 575 | 725 | 730 | CEU | ASN |
| Ancestral AA | E | P | I | G | M | K | D | R | Q | ||
| Non-ancestral AA | K | R | M | D | V | R | N | Q | E | ||
| Hap1 (*013) ancestral | E | P | I | G | M | K | D | R | Q | 12.0 | - |
| Hap2 (*002) | E | R | I | G | M | K | D | R | Q | 13.7 | 43.7 |
| Hap3 | E | R | I | G | M | K | D | R | E | 5.6 | 2.5 |
| Hap4 (*008) | E | R | I | G | M | R | D | R | E | 1.3 | 2.5 |
| Hap5 | E | R | I | D | M | R | D | R | E | 7.5 | - |
| Hap6 | E | P | I | G | M | R | D | R | E | 7.4 | 1.2 |
| Hap7 | K | P | I | G | M | R | D | R | E | 2.5 | 24.4 |
| Hap8 | E | P | M | G | M | R | D | R | E | 21.9 | 20.0 |
| Hap9 | E | P | M | G | M | R | N | R | E | 1.2 | - |
| Hap10 (*001) | E | P | I | G | V | R | N | Q | E | 26.2 | 5.0 |
| Freq. total | 99.3 | 99.3 | |||||||||
Haplotypes were determined in 80 unrelated members of CEU population and 80 unrelated members of the ASN population. Haplotypes with frequencies > 1% in either CEU or ASN population are displayed. Genotypes for rs72773968 (T12I) were not present in the HapMap data. Non-ancestral AAs are shown as grey shaded boxes. AA, amino acid (single letter code); CEU, CEPH, Utah residents with ancestry from northern and western Europe; ASN, east Asian, the combined Japanese in Tokyo (JPT) and Han Chinese in Beijing (CHB).
Figure 1. Linkage disequilibrium (LD) among common missense variants of ERAP1 in CEU and ASN HapMap populations.
The LD plots demonstrate pairwise r2 values from the sets of common, missense variants of ERAP1 derived from 80 unrelated members of the CEU population (A) and 80 unrelated members of the ASN population (B). The minor allele of rs27895 had a frequency of 0.001 in the ASN population, and therefore it was excluded from LD analysis in that population. ASN, east Asian, the combined Japanese in Tokyo (JPT) and Han Chinese in Beijing (CHB) CEU, CEPH, Utah residents with ancestry from northern and western Europe.
Genetic variation of ERAP1 and susceptibility to ankylosing spondylitis
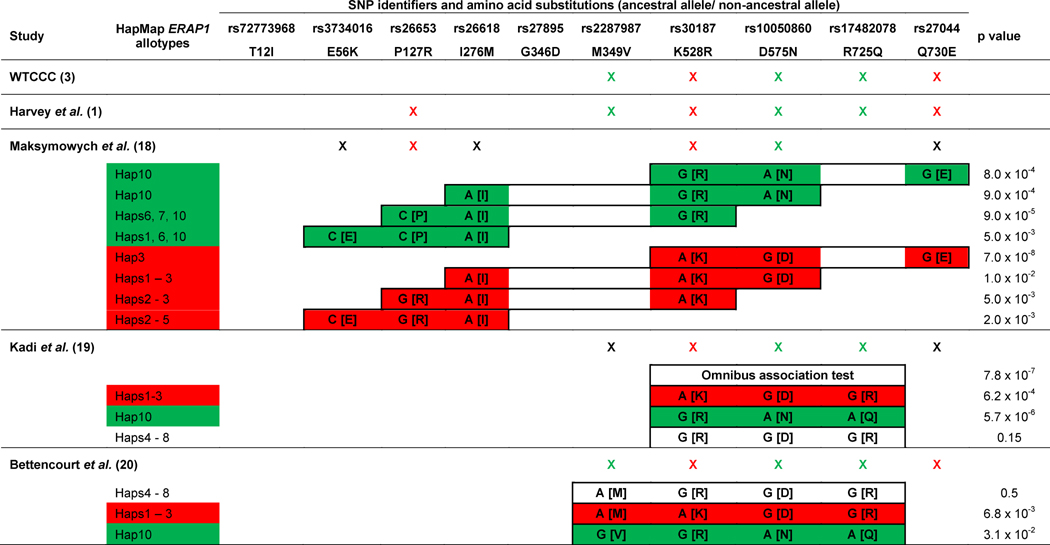
Genetic variants of ERAP1 have clearly been shown to contribute to AS susceptibility, first in U.S. and U.K. cohorts (3), and subsequently in many other populations (4, 11–17). The relationship between AS and ERAP1 (called ARTS1 at the time) was originally identified by the Wellcome Trust Case Control Consortium (WTCCC) and Australo-Anglo-American Spondylitis Consortium genome-wide association study (GWAS) of AS (3). In this study of 1471 AS patients from the U.K. and U.S., 5 non-synonymous SNPs of ERAP1 were found to significantly influence AS risk, with the minor alleles of rs27044 (Q730) and rs30187 (K528) conferring disease risk and of rs10050860 (N575), rs2287987 (V349), and rs17482078 (Q725) protecting against its development (Table 3). The variant most strongly associated with AS in the meta-analysis was K528, however the variant most strongly associated with AS in the U.K. population, Q730, was not examined in the U.S. population or in the meta-analysis. This study did not include haplotypic association testing or conditional analysis of ERAP1 SNPs, and it did not address whether the disease-associated variants represented independent association signals. In a follow-up, fine-mapping study that included 1604 AS cases of European ancestry, Harvey et al. identified six AS-associated ERAP1 missense variants (rs26653 [R127], M349, K528, D575, R725, and Q730; Table 3) (1). Among these, Q730 was the most significantly associated, but regional LD precluded nomination of any single variant as the source of disease risk on the basis of genetics alone. Interestingly, with the exception of R127, all of the AS risk alleles are the ancestral alleles and the derived alleles are associated with disease protection (Table 2). Among the HapMap ERAP1 allotypes (Table 2), the AS-protective V349 and Q725 variants are found exclusively on Hap10, along with the majority of the N575 variants. The K528 risk variant is present in Haps 1–3 and the Q730 risk variant is found in Haps 1 and 2 (Table 2).
Table 3.
Summary of ERAP1 haplotype associations in ankylosing spondylitis and comparison to ERAP1 allotypes from HapMap CEU and ASN populations
|
Individual markers that were tested for association with AS in each study are indicated by an X, where red indicates risk associated with minor allele, green indicates protection associated with minor allele, and black indicates a non-significant association. Haplotypic associations with AS in each study are shown in closed boxes, where red boxes indicate a risk haplotype and green boxes indicate a protective haplotype. Individual markers reported within haplotypes are noted as “cDNA nucleotide [amino acid]”. P values of haplotypic associations are extracted from the respective studies.
The studies that have evaluated ERAP1 haplotypes for association with AS have been limited, both by the markers genotyped and by their sample size. Maksymowich et al. examined 6 ERAP1 variants in three case-control collections that included a total 992 AS patients from Canada (18). They identified 3 ERAP1 variants, P127R, R528K, and D575N, that each significantly influenced AS risk (Table 3). By examining contiguous 3-marker haplotypes of the 6 ERAP1 variants, the authors identified several specific haplotypes that affected AS risk (Table 3). The haplotypes most strongly associated with AS were the haplotype of K528-D575-E730 (KDE), which conferred risk of AS, and the haplotype of P127-I276-R528 (PIR), which protected against AS. Within the HapMap ERAP1 allotypes, the risk KDE haplotype is only found within Hap3, while the PIR protective haplotype is found in Haps 6, 7, and 10. However, if the PIR haplotype tags the same effect as the adjacent, protective 3-marker haplotype (I276-R528-N575 or IRN), then the combined PIRN haplotype is specific to Hap10 (Table 3).
A GWAS by Evans et al. that included over 5000 AS patients refined our understanding of the association of ERAP1 variants with AS (4). The authors examined AS-associated loci for gene-gene interactions and discovered epistasis between HLA-B*27 and ERAP1. In one of the first reports of genetic epistasis in a human disease, they showed that the K528 variant conferred risk of AS specifically in HLA-B*27-positive individuals. Additionally, they used conditional analysis to reveal the presence of two independent effects on AS risk at the ERAP1 locus. These included a primary risk effect, tagged by K528, and a secondary protective effect, tagged by N575. They went on to report that individuals homozygous for R528 and N575 (RN) had a 3- to 4-fold reduction in AS risk. In agreement with previous studies, the RN haplotype predominantly tags Hap10 in the HapMap CEU and ASN populations (Table 2).
Kadi et al. examined the 5 AS-associated ERAP1 variants from the WTCCC AS study in case-control cohorts from France and Belgium that included 436 AS patients (19), but only 3 of these variants were genotyped in both cohorts. The authors replicated the reported associations of AS with the K528 (risk), N575 (protective), and Q725 (protective) variants, with the strongest association between AS and K528. Haplotypic analysis of these 3 variants revealed a significant omnibus association between AS and the 528-575-725 haplotypes (Table 3). An examination of specific haplotypes formed by these residues identified R528-N575-Q725 (RNQ) as an AS protective haplotype and K528-D575-R725 (KDR) as an AS risk haplotype. The protective RNQ haplotype specifically tags Hap10 of the HapMap allotypes, while the risk KDR haplotype is found in Haps 1–3 (Tables 2 and 3).
Similarly, Bettencourt et al. examined the 5 AS-associated variants from the WTCCC study in a collection of 200 HLA-B*27 positive AS patients and 200 HLA-B*27 positive healthy subjects (20). This study replicated the associations of each of these 5 SNPs with AS, identifying the strongest association with the AS risk variant, K528. Haplotype analysis of 4 of these 5 SNPs identified two haplotypes of variants encoding residues 349-528-575-725 that significantly affected AS risk: V349-R528-N575-Q725 (VRNQ) was an AS-protective haplotype and M349-K528-D575-R725 (MKDR) was an AS risk haplotype. Similar to the study from Kadi et al., the MKDR risk haplotype is found within the HapMap Haps 1–3, while the protective VRNQ haplotype is specific to Hap 10.
Taken together, these data indicate that at least two ERAP1 allotypes independently influence susceptibility to AS, one conferring disease risk and the other affording protection against disease. Based on comparisons of the reported AS-associated ERAP1 haplotypes with the ERAP1 allotypes in HapMap individuals (Tables 2 and 3), Hap10 is an AS protective haplotype while one or more of Haps1–3 appear to be AS risk haplotypes. Furthermore, the association of the K528 allele with AS risk was replicated in two studies of East Asians (5, 21). Although K528 is present on Haps1–3 in the CEU population, it is found predominantly on Hap2 in the ASN population, suggesting that Hap2 is the major ERAP1 risk allotype among East Asians.
In a series of two papers, Reeves et al. further investigated combinatorial effects of ERAP1 variants by examining naturally-occurring ERAP1 haplotypes and allotypes from human subjects and cell lines (11, 12). This work suggests that Hap2 (*002 in their study) encodes an ERAP1 molecule that efficiently trims peptides and yields relatively high production of MHC-peptide complexes, while a haplotype that was identical to Hap10 on the basis of its common alleles (*001 in their study) encoded an ERAP1 molecule that trimmed peptides poorly, leading to relatively low production of MHC-peptide complexes (11, 12). These observations are supported by an in vitro study of human ERAP1 that identified enhanced peptide trimming by forms of ERAP1 containing AS-risk variants and reduced peptide trimming by those bearing AS-protective variants (4). This is also consistent with two studies of naturally-occurring ERAP1 polymorphisms and their effects on ERAP1 enzyme activity and the resultant peptidome (22, 23). The authors went on to demonstrate perfect segregation of ERAP1 allotype combinations between cases and controls, leading them to conclude that the effect of the ERAP1 locus on AS risk is due to the net enzymatic effect produced by combinations of ERAP1 allotypes (12).
Although the functional studies of the protein products of ERAP1 allotypes by Reeves et al. were consistent with previous studies, the distribution of ERAP1 variants and allotypes within their collection of 17 AS cases and 19 healthy subjects was surprisingly inconsistent with the existing AS literature. Specifically, the AS risk allele (K528) and risk allotype (Hap2 or *002) were more common among controls than cases, while the AS protective allele (E730) and protective allotype (Hap10 or *001) were more common among cases than controls. The source of this discrepancy is unclear, and this issue is unlikely to be resolved without an analogous, sequencing-based study of the ERAP1 locus in a substantially larger AS case-control collection.
It has also been hypothesized that AS-associated variants or allotypes may influence disease risk by affecting ERAP1 expression. Constantino et al. recently reported that the K528-D575-R725 AS risk haplotype (equivalent to Hap10) was strongly correlated with reduced ERAP1 mRNA levels, more so than was any individual AS-associated variant (24). This raises the possibility that AS-associated ERAP1 allotypes may contain both coding variants that influence AS risk through changes in enzymatic function and noncoding variants that influence AS risk through alterations in gene expression.
Genetic variation of ERAP1 and susceptibility to psoriasis
Genetic variants of ERAP1 have been shown to influence psoriasis susceptibility, originally in populations of European ancestry (6, 25), and subsequently in Han Chinese (7, 26, 27). The original report by Strange et al., a GWAS that included 2178 psoriasis cases, revealed that two ERAP1 variants, rs27524 (noncoding) and rs30187 (K528), were genome-wide significant risk factors for psoriasis (6). Conditional analysis was unable to discriminate between the effects of the two psoriasis-associated variants, suggesting that these variants represent the same association signal. The authors also identified dominant epistasis between HLA-Cw*0602 and the K528 variant of ERAP1. The second study, including 10,588 cases of European ancestry, identified rs27432, an intronic SNP in strong LD with Q730, as the variant most strongly associated with psoriasis (25). Although the associations of K528 and Q730 may implicate one or more of the Haps1–3 as psoriasis risk factors (Table 2), it is unclear whether the effect of ERAP1 on psoriasis will be explained by single variant or allotypic associations.
Several studies have also examined the association of ERAP1 variants with psoriasis in the Han Chinese population. A multi-phase GWAS that included 8312 cases of psoriasis from China identified rs151823, a noncoding SNP ~16 kb upstream of ERAP1, as a psoriasis-protective allele (26). Another association study of psoriasis, which included a combination of exome sequencing and ERAP1 targeted resequencing data from 10,727 cases, identified genome-wide significant protective associations between psoriasis and both the P127 and E730 variants of ERAP1, but surprisingly it failed to replicate the association between psoriasis and K528 (7). Importantly, P127 is in strong LD with E730 in East Asian HapMap samples, but not in those of European ancestry (Figure 1). Finally, Sheng et al. performed a sequencing-based association study of psoriasis that included 15,207 cases from China (27). This revealed an association between psoriasis and rs27043, which was located in intron 16 of ERAP1, but did not establish whether this SNP association was independent from the noncoding SNP, rs151823, or the P127 or E730 variants. The psoriasis risk association with Q730, which specifically tags Hap2 among HapMap ASN haplotypes, but not with the R528 variant that is associated with psoriasis protection in individuals of European ancestry, emphasizes the disease risk associated with Hap2, which is found at high frequency in East Asians, and implies disease protection afforded by Hap10, which is found at high frequency in the HapMap CEU population but at low frequency in East Asians (Table 2).
Genetic variation of ERAP1 and susceptibility to Behçet’s disease
The association between ERAP1 and BD was first identified by Kirino et al. in a large case-control collection of Turkish ancestry that included 1209 cases (8). The authors demonstrated that the N575 and Q725 variants of ERAP1 recessively conferred risk of BD, and similar to AS and psoriasis, the authors also identified genetic epistasis between an ERAP1 variant (Q725) and the BD-associated MHC class I allele, HLA-B*51. If we extrapolate ERAP1 allotypes from these risk variants, Hap10 is the BD risk haplotype. Importantly, the effect of Q725/N575/Hap10 on BD susceptibility is in the opposite direction from their effect on AS and psoriasis risk, conferring risk of BD while protecting against AS and psoriasis. The genetic model under which ERAP1 variants interact with disease-associated MHC class I alleles also differed between these diseases, with a dominant effect in AS and psoriasis and a recessive effect in BD.
A study by Conde-Jaldon et al. examined ERAP1 variants in a collection of 362 cases 460 controls from Spain (28). The study failed to replicate the association between individual variants of ERAP1 and BD. However given its recessive nature, this study was not adequately powered to identify the association.
Conclusion
Genetic variation of ERAP1 influences susceptibility to several class I HLA-associated rheumatic diseases, and in each case, genetic epistasis exists between ERAP1 variants and the disease-associated HLA alleles. In AS, data have consistently demonstrated that the ERAP1 locus has at least two independent effects on disease susceptibility, one affording protection and a second conferring risk. Allotypic extrapolation with HapMap data suggests that these effects may be attributable to allotypes rather than individual genetic variants. In psoriasis, both protective and risk variants have been identified, however no study has established whether these variants represent one or more signals. In BD, a recessive interaction between the ERAP1 locus and HLA-B*51 confers strong disease risk, and interestingly, the same variant/allotype that confers BD risk is protective against AS and likely psoriasis. Going forward, genetic investigations of ERAP1 and its role in the class I HLA-associated rheumatic diseases should focus on identifying naturally-occurring ERAP1 allotypes based on both protein coding and noncoding variants. These naturally-occurring allotypes should be tested for disease association in large case-control collections, and their effect on ERAP1 expression and ERAP1 enzymatic function should be interrogated.
Key Points.
Genetic variants and haplotypes of ERAP1 influence susceptibility to AS, psoriasis and BD, and epistasis between ERAP1 variants and disease-associated class I HLA alleles have been identified in each of these diseases.
Based on comparisons of the existing literature with ERAP1 allotypes assembled from European and East Asian HapMap populations, the most common European allotype (Hap10) affords protection against both AS and likely psoriasis, but it confers risk of BD.
Additionally, Haps1–3 confer risk of AS and psoriasis in European populations, while Hap 2, the most common Asian allotype, is a strong risk factor in East Asians.
Future investigations of ERAP1 in rheumatic diseases should be directed at identifying the naturally-occurring allotypes, testing these allotypes for disease associations, and examining ERAP1 expression levels and the enzymatic function of the ERAP1 protein produced by each allotype.
Acknowledgments
Financial support and sponsorship
This study was supported by the Intramural Research Programs of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (Z01 AR041198) and the National Human Genome Research Institute (Z01 HG200374), National Institutes of Health, Bethesda, U.S.A.
Footnotes
Conflicts of interest
None
References
- 1.Harvey D, Pointon JJ, Evans DM, et al. Investigating the genetic association between ERAP1 and ankylosing spondylitis. Hum Mol Genet. 2009;18(21):4204–12. doi: 10.1093/hmg/ddp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassidy SA, Cheent KS, Khakoo SI. Effects of Peptide on NK cell-mediated MHC I recognition. Front Immunol. 2014;5:133. doi: 10.3389/fimmu.2014.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wellcome Trust Case Control Consortium, Australo-Anglo-American Spondylitis Consortium. Burton PR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39(11):1329–37. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans DM, Spencer CC, Pointon JJ, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43(8):761–7. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International Genetics of Ankylosing Spondylitis Consortium. Cortes A, Hadler J, et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45(7):730–8. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genetic Analysis of Psoriasis Consortium, the Wellcome Trust Case Control Consortium. Strange A, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42(11):985–90. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *7.Tang H, Jin X, Li Y, et al. A large-scale screen for coding variants predisposing to psoriasis. Nat Genet. 2014;46(1):45–50. doi: 10.1038/ng.2827. This study reveals differences in ERAP1-mediated psoriasis risk between European and Asian individuals. It reports that two variants of ERAP1, P127R and E730Q, are protective against psoriasis. These variants are in strong linkage disequilibrium in East Asian HapMap samples, but not in European HapMap samples (Figure 1). This study also found that K528, a common psoriasis risk allele in European populations, was not associated with psoriasis in the East Asian population. [DOI] [PubMed] [Google Scholar]
- **8.Kirino Y, Bertsias G, Ishigatsubo Y, et al. Genome-wide association analysis identifies new susceptibility loci for Behcet’s disease and epistasis between HLA-B*51 and ERAP1. Nat Genet. 2013;45(2):202–7. doi: 10.1038/ng.2520. This is the only paper to identify the association between BD and variants of ERAP1. By demonstrating for the first time that variants of ERAP1 influence BD susceptibility through epistasis with the class I HLA allele, HLA-B*51, this paper reinforces the notion of shared pathophysiologic mechanisms among the class I HLA-associated diseases, BD, AS and psoriasis. Interestingly, the variants that confer risk of BD paradoxically confer protection against AS and likely psoriasis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *9.Reeves E, Edwards CJ, Elliott T, James E. Naturally occurring ERAP1 haplotypes encode functionally distinct alleles with fine substrate specificity. J Immunol. 2013;191(1):35–43. doi: 10.4049/jimmunol.1300598. This paper began to explore the hypothesis that AS-associated ERAP1 variants act together to concertedly influence its function, which in turn influences AS risk. This set the table for subsequent studies seeking to explore naturally-occuring ERAP1 haplotypes/allotypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **10.Reeves E, Colebatch-Bourn A, Elliott T, et al. Functionally distinct ERAP1 allotype combinations distinguish individuals with Ankylosing Spondylitis. Proc Natl Acad Sci U S A. 2014;111(49):17594–9. doi: 10.1073/pnas.1408882111. This paper explores the effects of naturally-occurring ERAP1 allotypes and allotype combinations on ERAP1 function, reporting that the allotype combinations found in AS patients are functionally different than those found in healthy subjects. Importantly, this study’s use of rare and novel genetic variants in the assembly of the proposed ERAP1 allotypic nomenclature may preclude the application of this nomenclature outside of their study population. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pimentel-Santos FM, Ligeiro D, Matos M, et al. Association of IL23R and ERAP1 genes with ankylosing spondylitis in a Portuguese population. Clin Exp Rheumatol. 2009;27(5):800–6. [PubMed] [Google Scholar]
- 12.Davidson SI, Wu X, Liu Y, et al. Association of ERAP1, but not IL23R, with ankylosing spondylitis in a Han Chinese population. Arthritis Rheum. 2009;60(11):3263–8. doi: 10.1002/art.24933. [DOI] [PubMed] [Google Scholar]
- 13.Australo-Anglo-American Spondyloarthritis Consortium. Reveille JD, Sims AM, et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat Genet. 2010;42(2):123–7. doi: 10.1038/ng.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pazar B, Safrany E, Gergely P, et al. Association of ARTS1 gene polymorphisms with ankylosing spondylitis in the Hungarian population: the rs27044 variant is associated with HLA-B*2705 subtype in Hungarian patients with ankylosing spondylitis. J Rheumatol. 2010;37(2):379–84. doi: 10.3899/jrheum.090806. [DOI] [PubMed] [Google Scholar]
- 15.Li C, Lin Z, Xie Y, et al. ERAP1 is associated with ankylosing spondylitis in Han Chinese. J Rheumatol. 2011;38(2):317–21. doi: 10.3899/jrheum.100013. [DOI] [PubMed] [Google Scholar]
- 16.Szczypiorska M, Sanchez A, Bartolome N, et al. ERAP1 polymorphisms and haplotypes are associated with ankylosing spondylitis susceptibility and functional severity in a Spanish population. Rheumatology (Oxford) 2011;50(11):1969–75. doi: 10.1093/rheumatology/ker229. [DOI] [PubMed] [Google Scholar]
- 17.Davidson SI, Liu Y, Danoy PA, et al. Association of STAT3 and TNFRSF1A with ankylosing spondylitis in Han Chinese. Ann Rheum Dis. 2011;70(2):289–92. doi: 10.1136/ard.2010.133322. [DOI] [PubMed] [Google Scholar]
- 18.Maksymowych WP, Inman RD, Gladman DD, et al. Association of a specific ERAP1/ARTS1 haplotype with disease susceptibility in ankylosing spondylitis. Arthritis Rheum. 2009;60(5):1317–23. doi: 10.1002/art.24467. [DOI] [PubMed] [Google Scholar]
- 19.Kadi A, Izac B, Said-Nahal R, et al. Investigating the genetic association between ERAP1 and spondyloarthritis. Ann Rheum Dis. 2013;72(4):608–13. doi: 10.1136/annrheumdis-2012-201783. [DOI] [PubMed] [Google Scholar]
- 20.Bettencourt BF, Rocha FL, Alves H, et al. Protective effect of an ERAP1 haplotype in ankylosing spondylitis: investigating non-MHC genes in HLA-B27-positive individuals. Rheumatology (Oxford) 2013;52(12):2168–76. doi: 10.1093/rheumatology/ket269. [DOI] [PubMed] [Google Scholar]
- 21.Lin Z, Bei JX, Shen M, et al. A genome-wide association study in Han Chinese identifies new susceptibility loci for ankylosing spondylitis. Nat Genet. 2012;44(1):73–7. doi: 10.1038/ng.1005. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Medel N, Sanz-Bravo A, Van Nguyen D, et al. Functional interaction of the ankylosing spondylitis-associated endoplasmic reticulum aminopeptidase 1 polymorphism and HLA-B27 in vivo. Molecular & cellular proteomics : MCP. 2012;11(11):1416–29. doi: 10.1074/mcp.M112.019588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanz-Bravo A, Campos J, Mazariegos MS, Lopez de Castro JA. Dominant Role of the ERAP1 Polymorphism R528K in Shaping the HLA-B27 Peptidome Through Differential Processing Determined by Multiple Peptide Residues. Arthritis Rheumatol. 2015;67(3):692–701. doi: 10.1002/art.38980. [DOI] [PubMed] [Google Scholar]
- **24.Costantino F, Talpin A, Evnouchidou I, et al. ERAP1 gene expression is influenced by non-synonymous polymorphisms associated with predisposition to spondyloarthritis. Arthritis Rheumatol. 2015 doi: 10.1002/art.39072. This is the first study to demonstrate that ERAP1 expression correlates with AS-associated missense variants. This finding brings to light the possibility that in addition to their effect on ERAP1 structure and function, AS-associated allotypes may also influence disease susceptibility through alterations in ERAP1 expression. [DOI] [PubMed] [Google Scholar]
- 25.Tsoi LC, Spain SL, Knight J, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44(12):1341–8. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun LD, Cheng H, Wang ZX, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet. 2010;42(11):1005–9. doi: 10.1038/ng.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *27.Sheng Y, Jin X, Xu J, et al. Sequencing-based approach identified three new susceptibility loci for psoriasis. Nat Commun. 2014;5:4331. doi: 10.1038/ncomms5331. By performing a large-scale, sequencing-based association test of psoriasis susceptibility in a very large Han Chinese case-control population, this study identified a non-coding variant of ERAP1, rs27043, as the SNP most strongly associated with psoriasis. It remains to be seen whether this variant itself may influence disease risk, perhaps through its effect on ERAP1 expression, or whether it is a surrogate for the causative variant or allotype. [DOI] [PubMed] [Google Scholar]
- *28.Conde-Jaldon M, Montes-Cano MA, Garcia-Lozano JR, et al. Epistatic interaction of ERAP1 and HLA-B in Behcet disease: a replication study in the Spanish population. PLoS One. 2014;9(7):e102100. doi: 10.1371/journal.pone.0102100. This study attempted to replicate the association between BD and the ERAP1 locus, but the association did not achieve statistical significance. Given that this association acts in a recessive manner, this study was inadequately powered to identify this association. [DOI] [PMC free article] [PubMed] [Google Scholar]