Abstract
Ornithine transcarbamylase (OTC) deficiency is an X-linked trait that accounts for nearly half of all inherited disorders of the urea cycle. OTC is one of the enzymes common to both the urea cycle and the bacterial arginine biosynthesis pathway; however, the role of OTC has changed over evolution. For animals with a urea cycle, defects in OTC can trigger hyperammonemic episodes that can lead to brain damage and death. This is the fifth mutation update for human OTC with previous updates reported in 1993, 1995, 2002, and 2006. In the 2006 update, 341 mutations were reported. This current update contains 417 disease-causing mutations, and also is the first report of this series to incorporate information about natural variation of the OTC gene in the general population through examination of publically available genomic data and examination of phenotype/genotype correlations from patients participating in the Urea Cycle Disorders Consortium Longitudinal Study and the first to evaluate the suitability of systematic computational approaches to predict severity of disease associated with different types of OTC mutations.
Keywords: Ornithine transcarbamylase, Mutation, Ornithine transcarbamylase deficiency, Urea cycle, Hyperammonemia
INTRODUCTION
Ornithine transcarbamylase (OTC, MIM# 311250, RefSeq NP_000522) is a mitochondrial urea cycle enzyme that catalyzes the reaction between carbamyl phosphate and ornithine to form citrulline and phosphate. OTC is essential for the conversion of neurotoxic ammonia into non-toxic urea in mammals and other ureotelic animals (Fig. 1, (Brusilow and Horwich, 2001). In microbes and plants, OTC catalyzes the sixth step of arginine biosynthesis (Glansdorff and Xu, 2006) while the function of OTC in animals that excrete nitrogenous waste as either uric acid or ammonia remains to be elucidated.
Fig. 1.
Schematic of a hepatocyte highlighting the various enzymes, transporters, and metabolites of the urea cycle.
Shown in yellow is a mitochondrion containing the proximal enzymes N-acetylglutamate synthase (NAGS), carbamylphosphate synthetase 1 (CPS1), ornithine transcarbamylase (OTC), and the ornithine transporter (ORNT) and citrin transporters. Citrulline is transported out of the mitochondrion, and converted by argininosuccinate synthetase (ASS), argininosuccinate lyase (ASL), and arginase I (ARG1) to produce urea and regenerate ornithine that is reutilized by OTC.
The human OTC gene is located on the X-chromosome within band Xp21.1 (Lindgren et al., 1984). Ten exons and nine introns spanning 73 kb comprise the human OTC gene with an open reading frame of 1062 nucleotides (Horwich et al., 1984; Hata et al., 1986). The precursor protein contains 354 amino acids and has a calculated molecular weight of 39.9 kDa. Upon import into the mitochondria, a 32 amino acid N-terminal leader sequence is removed in two steps (Horwich et al., 1986). The mature OTC protein contains 322 amino acids and has a calculated molecular weight of 36.1 kDa. The functional OTC is a homotrimer; it has three-fold symmetry and three active sites, located at the interface between the protein monomers (Shi et al., 1998). OTC is expressed in the liver, the only organ that expresses all urea cycle enzymes, and in the intestinal mucosa. In the liver, ammonia is converted into urea, while in the intestinal mucosa, where N-acetylglutamate synthase, carbamylphosphate synthetase and OTC are also found, conversion stops at citrulline, a precursor of arginine and an intermediate in NO-signaling (Brusilow and Horwich, 2001).
OTC deficiency (OTCD) is the most common inherited defect of ureagenesis because the OTC gene is located on the X-chromosome. OTCD accounts for about half of all urea cycle defects (Brusilow and Horwich, 2001; Seminara et al., 2010). The estimated prevalence of OTCD is one in 14,000 (Brusilow and Maestri, 1996), but more recent estimates based on a review of national medical records and comparisons of newborn screening data with the number of patients with urea cycle disorders indicate a prevalence of one in 62,000–77,000 (Dionisi-Vici et al., 2002; Keskinen et al., 2008; Balasubramaniam et al., 2010; Summar et al., 2013). Most of the patients with OTCD are hemizygous males; approximately 20% of female carriers of OTC mutations also present symptoms of OTCD (Maestri et al., 1996; Maestri et al., 1998). The onset of OTCD symptoms is extremely variable. Heterozygous females and males with partial defects in the OTC can present later in life and well into adulthood, while hemizygous males with complete OTCD present with acute hyperammonemia within the first week of life (Hudak et al., 1985; McCullough et al., 2000). Neonatal presentation usually correlates with the absence of liver OTC activity (Tuchman et al., 1998) and null alleles (McCullough et al., 2000). Approximately 50% of patients with partial OTCD present later in life or even in adulthood (Finkelstein et al., 1990; Tuchman and Holzknecht, 1991). The clinical symptoms of OTCD result from the toxic effects of ammonia on the brain and can include recurrent vomiting, a clinical picture resembling Reye syndrome (Glasgow and Middleton, 2001), neurobehavioral changes or seizures. In addition to elevated plasma ammonia, biochemical symptoms of OTCD include elevated plasma glutamine, low or absent plasma citrulline and elevated urinary orotic acid, which distinguishes OTCD from other proximal urea cycle disorders.
This is the fifth mutation update for the human OTC gene (Tuchman and Plante, 1995; Tuchman et al., 1998; Tuchman et al., 2002; Yamaguchi et al., 2006). In addition to 417 disease-causing mutations, this report contains information about natural variation in the human OTC gene and the severity of disease associated with different types of OTC mutations.
MUTATIONS AND POLYMORPHISMS IN THE OTC GENE
A total of 417 disease-causing mutations in the OTC gene, including 29 mutations reported here for the first time, are listed in Table S1. Twenty-three of the newly reported mutations were identified by the longitudinal study of urea cycle disorders (Batshaw et al., 2014). Previously undetected chromosomal defects in intronic and regulatory regions of the OTC gene can now be diagnosed due to advances in sequencing technologies. Fifty-two patients with OTCD due to deletions, duplications or complex rearrangements involving OTC gene have also been identified (Table S2). Of the identified disease-causing mutations reported for OTC, the majority are amino acid replacements, followed by RNA splicing defects, premature protein terminations and deletions (Table 1). Many sequence changes within the OTC coding sequence result in more than one missense mutations of the same codon and some, but not all, of these mutations occur within codons that overlap with CpG dinucleotides on both strands (Fig. 2).
Table 1.
Types of mutations that cause OTCD.
| Mutation type | Number of mutations |
|---|---|
| Missense | 264 |
| Nonsense | 39 |
| Frame shift | 40 |
| In frame indels | 11 |
| Splice site errors | 60 |
| Extendinga | 2 |
| Regulatory | 1 |
| Disease presentation | |
| Neonatal | 163 |
| Late | 87 |
| Female | 144 |
| No information | 23 |
| Total | 417 |
| New mutations identified through UCDCb | 23 |
Extending mutations are sequence changes within the OTC translation termination codon that extend the OTC open reading frame and protein.
UCDC, Urea Cycle Disorders Consortium.
Fig. 2.
Missense and nonsense mutations in OTC and their overlap with CpG dinucleotides. The number of different missense and nonsense mutations that cause OTCD was plotted against their codon number. Blue symbols, missense mutations. Orange symbols, nonsense mutations. Gray bars, codons that overlap with CpG dinucleotides on sense and antisense strands.
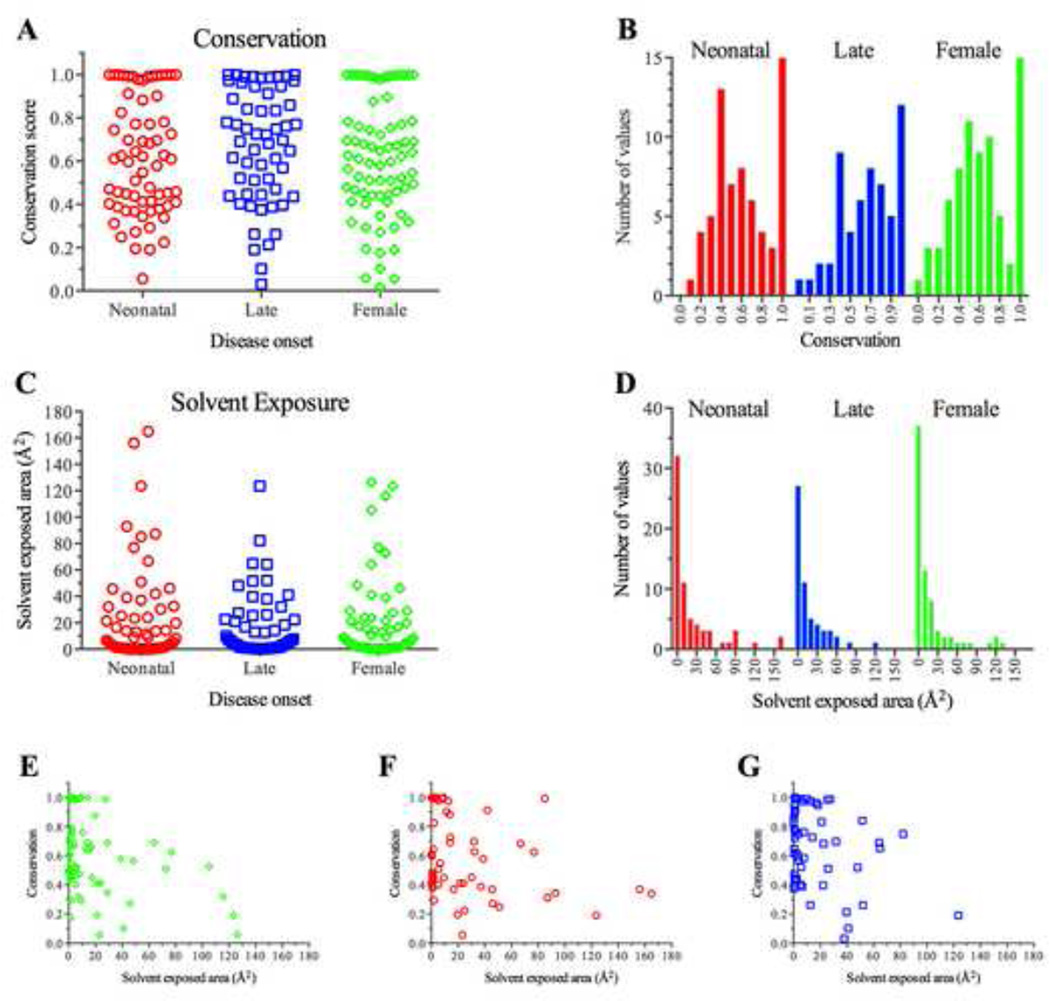
A complete loss of OTC function due to large deletions, frameshift and nonsense mutations as well as missense mutations that abolish either enzymatic activity or folding of OTC is known to cause neonatal onset disease in hemizygous males and symptoms of OTCD in some heterozygous females (Tuchman, 1993; Tuchman et al., 1997; Tuchman et al., 2002; Yamaguchi et al., 2006). Late onset disease in hemizygous males is caused by missense mutations that retain some OTC activity but have destabilized protein or lower enzymatic activity or decreased substrate affinity. A few female carriers of hypomorphic OTC alleles can also present with symptoms of OTCD (Pinner et al., 2010). Disease presentation and severity in OTCD patients with missense mutations were rationalized by assessing the position and function of the affected amino acid in the OTC enzyme (Tuchman et al., 1997; Tuchman et al., 2002). However, these studies did not attempt to find predictors of the severity of OTCD based on the biochemical properties of mutant OTC. As patients undergoing whole exome sequencing become more common, uncovering OTC variants is only likely to increase, making it important to have an understanding of the relationship between specific mutations and disease. Buried amino acids have more stringent steric restrictions in a folded protein, and examination of the degree of amino acid conservation, and solvent accessibility of mutation sites identified in male and symptomatic OTCD patients revealed that the largest numbers of missense mutations affect the most conserved and least solvent exposed residues (Fig. 3A and C). In both symptomatic females and male patients (Fig. 3B), the second most common missense mutations affect moderately conserved residues that also have little solvent accessible surface (Fig. 3E – G). These moderately conserved residues tend to be buried in the protein interior (Fig. 3F and G). Most mutant OTC proteins found in patients with OTCD are predicted to be less stable that the wild type (Fig. 4A and B) but the distributions of calculated differences in free energies between mutant and wild type OTC differed between the two methods for calculation used here (Fig. 4C and D). Most symptomatic females had missense mutations that were highly destabilizing to the mutant protein while there was a paucity of such mutations in males with late onset OTCD (Fig. 4D).
Fig. 3.
Conservation and solvent accessible area of amino acids affected by missense mutations in patients with OTCD.
A: Conservation scores of amino acids affected by missense mutations found in females (green, n = 73) and males with either neonatal (red, n = 66) or late onset (blue, n = 57) OTCD. B: Frequency distribution of conservation scores of the amino acids affected by missense mutations that cause OTCD. C: Solvent accessible areas of amino acids affected by missense mutations found in females and males with either neonatal or late onset OTCD. D: Frequency distribution of the solvent accessible areas of amino acids affected by missense mutations that cause OTCD. E–G: Scatter plots of conservation scores vs. solvent accessible areas of amino acids affected by mutations that cause OTCD in female patients (E) and male patients with either neonatal (F) or late (G) onset OTCD.
Fig. 4.
Destabilization of the OTC protein by amino acid replacements found in patients with OTCD.
A: Calculated difference between stability of mutant and wild type OTC using support vector machines. B: Calculated difference between wild type and mutant OTC using force fields. C: Frequency distribution of ΔΔG values calculated using support vector machines. D: Frequency distribution of ΔΔG values calculated using force fields. Red, males with neonatal onset OTCD (n = 87). Blue, males with late onset OTCD (n = 70). Green, females with OTCD (n = 95)
In addition to disease-causing mutations, 44 rare sequence variants (MAF < 1%) have been found in the OTC gene (Table S3). In previous reports, 12 of these SNPs were considered to be polymorphisms (non-disease causing) because they have been found through screens of healthy individuals ((Hata et al., 1988; Matsuura et al., 1993; Tuchman and Plante, 1995; Plante and Tuchman, 1998; Climent and Rubio, 2002a, b; Tanaka et al., 2005). The remaining 32 rare SNPs were found through querying the 1000 Genomes project and NHLBI Exome Variant Server (ESP6500SI-V2); 18 rare SNPs have been found in heterozygous females and are predicted to affect either splicing of the OTC mRNA or function of the OTC enzyme (Kumar et al., 2009; Adzhubei et al., 2010; Schwarz et al., 2010) suggesting that female carriers of these alleles might be predisposed to OTCD. Eighteen common polymorphisms (MAF ≥ 1%) have been found in the regulatory region, within exons and in the introns in close proximity of intron/exon boundaries (Table 2). Allele frequencies of the majority of common polymorphisms that were found through population screens by individual researchers (Tuchman and Plante, 1995; Plante and Tuchman, 1998; Azevedo et al., 2002a; Azevedo et al., 2002b; Climent and Rubio, 2002a; Azevedo et al., 2003) are similar to those reported in the dbSNP, 1000 Genome project and NHLBI Exome Variant Server. Two variants, rs73196229 (c.299-8A>T) and rs1800326 (c.387-7G>A), however, appear to differ in populations of healthy individuals (1000 Genomes projects, ESP6500SI-V2 and Azevedo et al(Azevedo et al., 2002a). and among patients suspected of OTCD (Tuchman and Plante, 1995; Plante and Tuchman, 1998; Climent and Rubio, 2002a). It is possible that these two variants may affect splicing of the OTC mRNA and result in symptoms of OTCD.
Table 2.
Common polymorphisms (MAF > 1%) in the upstream regulatory region, exons and introns of the human OTC gene.
| No. | Region | Nucleotide/amino acid change/SNP ID |
Allele frequency | Reference/submitter |
|---|---|---|---|---|
| 1. | Upstream region | c.–520_–518delCT rs200016637 |
CTTCTT: 36.88% (610/1659); CTT: 63.12% (1049/1659) | 1000 genomesa |
| 2. | Upstream region | c.–514_–511delTCTA rs201351170 |
TCTATCTA: 36.09% (600/1659); TCTA: 63.91% (1059/1659) | 1000 genomes |
| 3. | Upstream region | c.–512insTTCT rs57752938 |
TTCTTTCT: 28%, 38%, 29%; TTCT: 72%, 62%, 71%b | Azevedo et al., 2003 |
| 4. | Upstream region | c.–441G>A rs5917572 |
A: 23%, 38%, 24%; G: 77%, 62%, 76%b A: 31.28% (519/1659); G: 68.72% (1140/1659) A: 30.30% (547/1805); G: 68.42 (1235/1805); O: 1.27% (23/1805) |
Azevedo et al., 2003 1000 genomes dbSNP build 138c |
| 5. | Upstream region | c.–365G>A rs5963030 |
A: 15%, 16%, 5%; G: 85%, 84%, 95%b A: 12.18 (201/1659); G: 87.82% (1458/1659) |
Azevedo et al., 2003 1000 genomes |
| 6. | Upstream region | c.–359A>G rs5917573 |
G: 23%, 31%, 27%; A: 77%, 69%, 73%b G: 31.28% (519/1659); A: 68.72% (1140/1659) G: 31.22 (1142/3646); A: 68.68% (2504/3646) |
Azevedo et al., 2003 1000 genomes dbSNP build 138 |
| 7. | Upstream region | c.–294G>A rs139948134 |
A: 1.33% (22/1654); G:98.67% (1632/1654) | 1000 genomes |
| 8. | Exon 2 | c.137A>G p.Arg46Lys rs1800321d |
G: 32%; A: 68% (71 alleles assayed)e G: 28%; A: 72% (123 alleles)f; G: 46%; A: 54% (46 alleles)g G: 19.70% (327/1659); A: 80.30% (1332/1659) G: 25.24% (2189/8671); A: 72.07% (6249/8671); O h: 2.69% (233/8671) G: 29.03% (3066/10560); A: 70.97% (7494/10560) |
Plante and Tuchman, 1998 Azevedo et al., 2002a 1000 genomes dbSNP build 138 NHLBI-WESi |
| 9. | Intron 3 | c.299–39_299–40insT rs148081863 rs143626513 |
GT: 34%; G: 66% (86 alleles screened) GT: 3.80% (141/3701); G: 96.20% (3560/3701) GT: 11.61% (192/1654); G: 88.39% (1462/1654) GT: 16.37% (1664/10167); G: 83.67% (8503/10167) |
Climent and Rubio, 2002a 1000 genomes dbSNP build 138 NHLBI-WESi |
| 10. | Intron 3 | c.299–17delT rs200509203 |
T: 0%; TT: 100% (87 alleles screened) T: 1.33% (22/1659); TT: 98.67% (1632/1659) |
Climent and Rubio, 2002a 1000 genomes |
| 11. | Intron 3 | c.299–8A>T rs73196229 |
A: 50%; T: 50% (76 alleles)e A: 34%; T: 66% (93 alleles)e A: 11.60% (192/1659); T: 88.4% (1467/1659) A: 11.76% (195/1658); T: 88.24% (1463/1658) A: 15.65% (1652/10558); T: 84.35% (8906/10558) |
Tuchman and Plante, 1995 Climent and Rubio, 2002a 1000 genomes dbSNP build 138 NHLBI-WES |
| 12. | Intron 4 | c.387–7G>Aj rs1800326 |
G: 29%; A: 71% (45 alleles screened)e A: 0%; G: 100% (164 alleles) |
Plante and Tuchman, 1998 Azevedo et al., 2002a |
| 13. | Exon 5 | c.429T>C p.Tyr143Tyr rs145777402 |
C: 0%; T: 100% (51 alleles)e C: 2%; T: 98% (59 alleles)e C: 4.22% (45/10561); T: 95.78% (10516/10561) |
Climent and Rubio, 2002a Tuchman and Plante, 1995 NHLBI-WES |
| 14. | Intron 5 | c.541–63G>A rs2235125 |
G: 62%, 71%, 30%; A: 38%, 29%, 70%b G: 47.90% (794/1659); A: 52.10% (865/1659) G: 40.37% (1694/4196); A: 54.12% (2271/4196); O: 5.48% (230/4196); Nh: 0.02% (1/4196) |
Azevedo et al., 2003 1000 genomes dbSNP build 138 |
| 15. | Intron 7 | c.718–14T>C rs55722856 |
C: 5.13% (85/1657); T: 94.87% (1572/1657) C: 4.90% (82/1659); T: 95.10% (1577/1659) C: 6.92% (731/10561); T: 93.08% (9830/10561) |
dbSNP build 138 1000 genomes NHLBI-WES |
| 16. | Exon 8 | c.809A>G p.Gln270Argk rs1800328 |
G: 12.5%; A: 87.5% (56 alleles)e G: 4%; A: 96% (169 alleles) G: 2% (34/1659); A: 98% (1625/1659) G: 3.05% (191/6272); A: 96.95% (6081/6272) G: 3.32% (351/10561); A: 96.68% (10210/10561) |
Plante and Tuchman, 1998 Azevedo et al., 2002a 1000 genomes dbSNP build 138 NHLBI-WES |
| 17. | Intron 8 | c.867+35G>T rs62622415 |
G: 38%; T: 62% (26 alleles) G: 1.30% (22/1659); T: 98.70% (1637/1659) G: 1.33% (22/1654); T: 98.67% (1632/1654) G: 2.71% (286/10534); T: 97.28% (10248/10534) |
Azevedo et al., 2002a 1000 genomes dbSNP build 138 NHLBI-WES |
| 18. | Intron 9 | c.1006–70C>T rs12557315 |
T: 21%, 26%, 0%; C: 79%, 74%, 100%b T: 21.50% (356/1659); C: 78.5% (1303/1659) T: 17.40% (731/4201); C: 79.53% (3341/4201); O: 3.07% (129/4201) T: 16.17% (582/3600); C: 83.83% (3018/3600) |
Azevedo et al., 2003 1000 genomes dbSNP build 138 NHLBI-WES |
Variant frequency reported in the 1000 Genomes Project. The 1000 Genomes Project seeks to provide a comprehensive resource on human genetic variation.
Screening was done in healthy males from Portugal (n = 85), Czech Republic (n = 100–142) and Mozambique (n = 40). Not all individuals were screened for all SNPs in the Czech samples.
Variant frequency reported in the dbSNP build 138 track of the UCSC Genome Browser. The dbSNP database is curated by the National Center for Biotechnical Informatics containing short genetic variations. Build 138 of the database was used for this table.
Reference sequence used is RefSeq NM_000531.3. In RefSeq NM_000531.3 codon 46 encodes a Lys. We more commonly see Arg at this position.
Samples were from subjects suspected of OTC deficiency.
Screening was done in Portuguese healthy males.
Screening was done in healthy males from Mozambique.
Nucleotides other than A, T, G and C. N represents any nucleotide; O would normally represent a null allele in a hemizygote, but may also include sequencing errors.
Variant frequency reported in the NHLBI Exome Variant Server. The NHLBI-WES contains sequence information from over 200,000 individuals.
Annotated as splice site variant in dbSNP 138.
This is a commonly seen polymorphism; however, SIFT and Polyphen2 scores for this are deleterious (0.00) and probably damaging (0.977), respectively.
CLINICAL, DIAGNOSTIC, AND BIOLOGICAL RELEVANCE
Mutations that completely abolish either OTC mRNA production and processing or OTC enzymatic activity manifest with acute neonatal onset hyperammonemia. Individuals with sequence variants that allow residual OTC activity can present with hyperammonemia at any point in life, while others remain asymptomatic. In patients with late onset OTCD, acute hyperammonemia can be triggered by a high protein meals (Ben-Ari et al., 2010; Thurlow et al., 2010; Cavicchi et al., 2014), fasting (Marcus et al., 2008), infections (McGuire et al., 2013), invasive medical procedures (Chiong et al., 2007; Hu et al., 2007; Bezinover et al., 2010), chemotherapy (Lipskind et al., 2011; Cavicchi et al., 2014), or other environmental insults that result in increased protein catabolism and ammonia production (Seminara et al., 2010). Missense mutations that cause partial OTCD reduce OTC enzymatic activity or stability while mutations in the vicinity of consensus splice sites can potentially affect mRNA processing and result in decreased abundance of the OTC enzyme. Large phenotypic heterogeneity is observed among patients with hypomorphic OTC alleles, even among members of the same family. This is likely due to environmental factors and genetic modifiers, which are poorly understood at present. A better understanding of the correlation between genotype and phenotype of OTCD is needed to interpret the effects of OTC sequence variants discovered through sequencing of whole exomes and genomes and could be achieved by studying natural history of OTCD (Batshaw et al., 2014).
GENOTYPE AND PHENOTYPE OF PATIENTS IN THE LONGITUDINAL STUDY OF OTCD
We wanted to determine whether the severity of OTCD, measured by disease onset, biochemical markers and intellectual outcomes, could be correlated with changes in the OTC protein that can be calculated and/or predicted from the known three dimensional structure of the OTC trimer (Ding and Dokholyan, 2006; Yin et al., 2007a, b; Adzhubei et al., 2010). A total of 398 participants with OTCD and their asymptomatic relatives have been enrolled in the longitudinal study of urea cycle disorders for the study of natural history of OTCD. Gender, disease onset and liver transplant status have been recorded for each participant. The OTC genotypes were determined for 293 study participants (197 females, 80 males and 16 deceased patients without gender information). Table S4 lists the participants’ gender, onset of their disease and whether they received a liver transplant. In addition to the study participants listed in Table S4, six asymptomatic males and 88 asymptomatic females as well as six males and 12 female study subjects with unknown disease onset have been enrolled in the longitudinal study of OTCD. Participants in the study are examined regularly; biomarkers of liver function are measured at each visit (Batshaw et al., 2014); neuropsychological evaluations of study participants are performed when they are six months, four, eight, 15 and 18 years old and once in adulthood (Weisbren et al., 2014). Of the 293 participants with a known genotype, 18 participants have large deletions of the whole or parts of OTC gene while remaining 275 have either point mutations or small insertions and deletions. The majority of participants, 66 male and 140 female, have missense mutations, 26 participants (3 male and 23 female) have nonsense mutations, 11 participants (2 male and 9 female) have frameshift mutations and 14 (5 male and 9 female) have mutations that affect splicing. Three female participants have mutations within their stop codon that result in extending of the OTC protein and one male patient has a 9 bp in frame insertion that results in the insertion of three amino acids in the OTC protein. Many point mutations occur repeatedly and they are in codons that either contain or overlap with CpG dinucleotides (Fig. 5).
Fig. 5.
Recurrent mutations found in participants enrolled in the longitudinal study of OTCD. Number of study subjects with each missense and nonsense mutation and their overlap between affected codons and CpG dinucleotides on sense and antisense strands of the OTC coding region were plotted for each codon of the OTC open reading frame. Blue symbols, positions of missense and nonsense mutations in the OTC coding sequence. Gray bars, codons that overlap with CpG dinucleotides on sense and antisense strands.
Several measures of mutation severity were used to assess whether certain types of mutations are associated with neonatal or late onset OTCD. A majority of both male and female study subjects had OTCD due to missense mutations (Table 3). The second largest group of study participants had defects that resulted in the complete loss of functional OTC enzyme. One male study subject with late onset OTCD had a mutation in a consensus splice site that would have been expected to result in a neonatal presentation, which could not be determined whether the late onset was due to partial splicing of the RNA or due to somatic mosaicism (Table 3). The severity of missense mutations was assessed using conservation and solvent accessible area of the replaced amino acid, calculated destabilization of mutant proteins and their SIFT and PolyPhen2 scores (Figs. S1–S3). Most missense mutations found in males with neonatal OTCD and females with late onset OTCD were found to be replacements of highly conserved amino acids, while males with late onset disease and asymptomatic males and females had replacements of less conserved residues (Fig. S1A and B). Most mutated residues were either completely or nearly completely buried in the protein interior (Fig. S1C). Mutant OTC proteins in all study subjects were predicted to be less stable than the wild type by the machine learning method (Fig. S2A), whereas force field calculations predicted that 41 mutant proteins found in asymptomatic females and 30 mutant proteins found in females with late onset OTCD would be more stable than the wild type (Fig. S2B). Additionally, five mutant OTC found in males with neonatal onset OTCD, four mutant proteins found in males with late onset OTCD and one mutant found in an asymptomatic male were predicted to be more stable than the wild type (Fig. S2B). Tools such as SIFT and PolyPhen2 that have been used in genomics studies to predict effects of sequence variants on protein function rely on protein conservation and force field calculations based on existing three-dimensional structure (Cheng et al., 2006; Ding and Dokholyan, 2006; Yin et al., 2007a, b). Tradeoffs where increased protein stability reduces enzyme activity have been experimentally explored in T4 lysozyme by Shoichet and colleagues (Shoichet et al., 1995). This leads to experimentally testable hypotheses regarding changes in enzyme stability for these mutations. Most missense mutations found in study subjects were correctly predicted to affect function of the OTC by both SIFT and PolyPhen2; however, amino acid replacements R26Q, D41G, A135E, E239D and A152V were not predicted to affect OTC function by either tool (Fig. S3). Computational tools to evaluate the effect that sequence variants have on protein function are still not reliable, but improvements in this area are necessary as the ability to detect new OTC variants will increase with the adoption of genomics approaches.
Table 3.
Disease onset and the type of defect in the OTC gene in patients enrolled in the longitudinal study of OTCD.
| Disease onset | Gender | OTC defect | |||
|---|---|---|---|---|---|
| Loss of function |
Amino acid replacement |
Variant in introns |
|||
| Neonatal | Male | 32 | 7 | 22 | 3 |
| Female | 5 | 2 | 3 | 0 | |
| Late | Male | 36 | 1 | 33 | 2 |
| Female | 92 | 29 | 62 | 1 | |
| Asymptomatic | Male | 6 | 0 | 6 | 0 |
| Female | 88 | 17 | 70 | 1 | |
Neuropsychological Outcomes in Patients with OTCD
In addition to disease onset, the disease severity in participants of the longitudinal study of the OTCD was evaluated using results of neuropsychological tests, number of hyperammonemic episodes and biomarkers of liver dysfunction. Most female study subjects have average or above average IQ (Fig. 6A). In male participants, earlier disease onset is associated with poorer neurocognitive outcomes (Fig. 6B). These data are consistent with the recent report of neurocognitive outcomes in patients with urea cycle disorders, including OTCD (Seminara et al., 2010; Weisbren et al., 2014).
Fig. 6.
Neurocognitive outcomes in female and male participants of the longitudinal study of OTCD.
A: IQ scores of the neonatal onset females (dark purple), late onset females (light purple), asymptomatic females (lavender) study subjects. B: IQ scores of the neonatal onset males (navy), late onet males (light blue), asymptomatic males (cyan) study subjects. Error bars represent median and interquartile ranges. Normal IQ ranges are shown in grey.
Hyperammonemic Episodes in Patients with OTCD
Because declining neurocognitive outcomes correlated with the number of hyperammonemic episodes (Weisbren et al., 2014), we examined whether the type of mutation in study subjects correlated with the number of hyperammonemic episodes they experience. The majority of study subjects, who were grouped by gender and disease onset, did not report illness or hyperammonemic episodes during their participation in the longitudinal study (Fig. 7A – C). Mutations found in study subjects were classified as loss of function (LOF), which includes small and large deletions of the parts or entire OTC gene, nonsense mutations and sequence changes within consensus splice sites, amino acid replacements and variants in OTC introns, which presumably decrease efficiency but do not abolish splicing of the OTC mRNA. There was no correlation between the number of hyperammonemic episodes and either the type of mutation or the length of participation in the study among males with neonatal OTCD who did not receive a liver transplant (Fig. 7D). Examples from this group include two study subjects with LOF mutations, each of whom only had one hyperammonemic episode, while a study subject with what is considered a hypomorphic OTC allele had the most episodes (Fig. 7D). The number of hyperammonemic episodes in male study subjects with late onset OTCD due to amino acid replacements correlated with the length of their participation in the longitudinal study (Fig. 7E). One male participant with mutation in the consensus splice site, which is considered to be a LOF mutation, and two study subjects with mutations in OTC introns experienced no hyperammonemic episodes (Fig. 7E). The patterns of correlation between the number of hyperammonemic episodes and the length of participation in the study differed in male study subjects with neonatal and late onset OTCD; this could be because males with neonatal OTCD, which is considered to be more severe than late onset disease, leave the study before they have a chance to experience many hyperammonemic episodes. Female study subjects with late onset OTCD due to amino acid replacements and LOF mutations also tended to have more hyperammonemic episodes that correlated with the length of participation (Fig. 7F).
Fig. 7.
Hyperammonemic and illness episodes experienced by the participants in the longitudinal study of the OTCD.
Distribution of reported illness (gray) and hyperammonemic (cyan) episodes by the male study subjects with either neonatal onset (A) or late onset (B) OTCD and female study subjects with late onset OTCD (C). Frequency of hyperammonemic episodes reported by male study subjects with either neonatal onset (D) or late onset (E) OTCD and female study subjects with late onset OTCD (F) due to missense (blue), loss of function (orange) or intronic (magenta) mutations.
Liver Damage in Patients with OTCD
Some patients with OTCD experience liver damage evidenced by elevated plasma alanine aminotransferase (ALT) and prolonged coagulation time resulting in increased international normalized ratio (INR) (Burlina et al., 2006; Mustafa and Clarke, 2006; Ihara et al., 2013; Gallagher et al., 2014). Among female study subjects who did not receive liver transplant, two with neonatal onset OTCD, 25 with late onset OTCD and 12 asymptomatic participants had mildly elevated plasma ALT when they joined the study (Fig. 8A). Five female participants with late onset disease had moderately elevated ALT and one asymptomatic study subject had severe elevation of plasma ALT when they joined the study (Fig. 8A). Mildly elevated plasma ALT was also present in three male study subjects with neonatal onset OTCD and six male participants with late onset OTCD when they joined the study (Fig. 8B). Mildly elevated INR was present in four female subjects with late onset OTCD and one asymptomatic female while one female with late onset OTCD and one asymptomatic female had moderately elevated INR when they joined the study (Fig. 8C). Two male study subjects with neonatal onset disease had mildly elevated INR and one male with late onset OTCD had moderately elevated INR when they joined the study (Fig. 8D). None of the patients described here had liver transplants. When both baseline ALT and INR were measured, study subjects who had elevated plasma ALT when they joined the study did not have elevated INR and vice versa. In females with symptoms of OTCD, mildly to moderately elevated baseline INR and plasma ALT was present in 3.3% (1 out of 30) and 36.7% (11 out of 30) of participants with LOF mutations, respectively, and 6.8% (4 out of 59) and 40.7% (24 out of 59) of study subjects with missense mutations, respectively (Fig. S4A and B). In asymptomatic female study subjects, elevated plasma ALT at baseline was present in 18.8% (3 out of 16) and 16.9% (11 out of 65) of participants with LOF mutations and amino acid replacements, respectively, while 4.7% (2 out of 43) of participants with amino acid replacements had elevated INR at baseline (Fig. S4C and D). Mildly to moderately elevated INR and mildly elevated plasma ALT at baseline were found in 12.0% (3 out of 25) and 30.9% (13 out of 42) male study subjects with OTCD due to amino acid replacements (Fig. S4E and F). Asymptomatic male study subjects had normal INR and plasma ALT when they joined the study. Subjects with severe (LOF) or mild (amino acid replacements) OTCD showed similar elevations in INR and plasma ALT, suggesting that factors other than the type of OTC allele determine whether OTCD will be accompanied with liver dysfunction. Additionally, male and female study subjects had similar incidences of OTCD with mildly elevated plasma ALT (30.9% and 33.9%, respectively), suggesting that both genders are equally susceptible to liver dysfunction associated with OTCD. If liver damage is cumulative, and females with OTCD live longer, they may be at greater risk for detectable liver damage as they age.
Fig. 8.
Biomarkers of liver damage in participants of the longitudinal study of OTCD.
Baseline values of plasma ALT in female (A) and male (B) study subjects; baseline INR values in female (C) and male (D) study subjects. Plasma ALT activities of 35–105 U/L and 40–120 U/L were considered mildly elevated in females and males, respectively. Plasma ALT activities of 105–175 U/L and 120–200 U/L were considered moderately elevated in females and males, respectively. Plasma ALT activities above 175 and 200 U/l were considered severely elevated in females and males, respectively. INR values between 1.2 and 1.5 were considered mildly elevated, while values between 1.5 and 2.5 were considered moderately elevated. Gray areas indicate normal ranges of plasma ALT and INR values. Neonatal onset females (dark purple), late onset females (light purple), asymptomatic females (lavender), neonatal onset males (navy), late onset males (light blue), asymptomatic males (cyan)
A number of participants in the longitudinal study of OTCD had normal liver function when they joined the study but subsequently developed symptoms of hepatic dysfunction. Most study subjects with symptoms of liver dysfunction had mildly elevated plasma ALT (Table S5). One male study subject with neonatal onset OTCD, five female participants with late onset OTCD and three asymptomatic female participants had moderately elevated plasma ALT at least once during their participation in the study (Table S5). Four study subjects, a male with neonatal onset OTCD and three females with late onset OTCD, had severely elevated plasma ALT at least once during their participation in the study (Table S5). A majority of study subjects who experienced elevated INR al least once during participation in the study had moderately elevated INR, while one female participant with late onset OTCD and three asymptomatic females had severely elevated INR (Table S6). A number of study subjects, one male and two females with neonatal onset OTCD, five males and eight females with late onset disease and two asymptomatic females, had both elevated INR and plasma ALT during their participation in the study. Some study subjects with most elevated markers of liver dysfunction had hyperammonemic episodes while others did not. A male study subject with neonatal onset OTCD who had severely elevated ALT and moderately elevated INR experienced two hyperammonemic episodes during two years of participation in the study. Of the three female study subjects with late onset OTCD and severely elevated plasma ALT, one had moderately elevated INR and experienced one hyperammonemic episode in four years of participation in the study, one had normal INR and one hyperammonemic episode in five years and one had normal INR and no hyperammonemic episodes during five years of her participation in the study. Of the four female study subjects with severely elevated INR, one had no hyperammonemic episodes during five years of her participation in the study and other three are asymptomatic carriers of OTC mutations. This suggests a complex relationship between health of the liver and its ability to maintain normal ammonia levels in the blood.
MOLECULAR DIAGNOSTIC STRATEGIES
OTCD is presently most often diagnosed by sequencing 10 OTC exons and their intron/exon boundaries due to the non-invasive nature of collecting leukocytes for DNA isolation and improvement and decreasing costs of sequencing technology. Advances in comparative genome and oligonucleotide hybridization techniques led to improvements and increases in diagnosing OTCD due to large deletions, duplications or rearrangements in the OTC gene (Arranz et al., 2007; Jakubiczka et al., 2007; Deardorff et al., 2008; Wong et al., 2008; Shchelochkov et al., 2009; Balasubramaniam et al., 2010; Quintero-Rivera et al., 2010). With these methods, OTCD can be confirmed at molecular level in 80%–90% of patients with clinical symptoms of the disease (Yamaguchi et al., 2006; Shchelochkov et al., 2009). As the use of next-generation sequencing becomes standard practice, we expect additional disease-causing mutations will be found in previously unexamined OTC gene regulatory regions as well as in the OTC introns enabling confirmatory molecular diagnoses for most, if not all, patients with clinical symptoms of OTCD. Another consequence of the wide use of whole exome and genome sequencing will be identification of OTC sequence variants in patients who may or may not have clinical symptoms of OTCD.
FUTURE DIRECTIONS
As sequencing of whole exomes and genomes becomes common clinical practice, OTC variants will be found in patients who do not and may never present with symptoms of OTCD. Interpretation of this clinically important incidental information is relevant because mutations in the OTC gene put patients at risk of developing hyperammonemia, which can have devastating consequences if left untreated, and are associated with nonverbal learning disability (Gyato et al., 2004; Kim et al., 2014), appear to be associated with liver dysfunction (Burlina et al., 2006; Mustafa and Clarke, 2006; Ihara et al., 2013; Gallagher et al., 2014), and have implications for family planning. Bioinformatic tools can be used to evaluate whether sequence variants in introns either disrupt splicing through creating new splice sites or decrease splicing efficiency of OTC mRNA. Predicting effects of sequence variants in regulatory regions of the OTC gene requires detailed understanding of the regulation of expression of the OTC gene. Enzymatic activity assays have been performed from livers obtained after transplantation, from deceased patients, and biopsy tissue; however, these typically are from liver homogenates with only a few examples where OTC was purified (Snodgrass, 1968; Pierson et al., 1977; Kalousek et al., 1978). Biochemical characterization of human OTC has been relatively limited as numerous factors can confound interpretation. Levels of normal OTC fluctuate with dietary protein intake, illness, and other environmental changes (Klein et al., 2008; McGuire et al., 2013; McGuire et al., 2014). We have previously characterized several OTC mutations based on the enzyme kinetics of purified recombinant human protein expressed in E. coli (Morizono et al., 1997a; Morizono et al., 1997b). Limitations of this approach were poor purification efficiencies for some mutant proteins. An affinity column with an immobilized bisubstrate analog of carbamylphosphate and ornithine, N-(phosphonoacetyl)-L-ornithine (PALO) was used. For mutations that affect substrate binding, the PALO affinity column approach was not effective. Simply using affinity tags does not discriminate between soluble trimers and small OTC aggregates that normally occur even when expressing wild type OTC. Expression, purification and characterization of mutant OTC from bacteria are very labor intensive. Enzyme activities have been measured from crude mammalian cell culture lysates in which mutant or wild type OTC was expressed (Nishiyori et al., 1997; Kogo et al., 1998; Augustin et al., 2000; Kim et al., 2006). These approaches can sometimes provide additional information such as OTC mutations that affect mitochondrial targeting and processing (Mavinakere et al., 2001) but are inappropriate for obtaining detailed enzyme kinetics. High throughput functional assays in vitro or in cultured cells will still be needed to confirm and validate predicted effects of detected variants. Advances in genome editing will provide the ability to engineer mutations and examine their effects in vivo using animal models. The current computational approaches are imperfect, but help identify regions in the protein and mutations that may benefit from further functional characterization. Mutations in which the in silico predictions differ from experimental measurements can provide the basis for generating new hypotheses about the structure and function relationships within OTC, and also help with refining prediction algorithms.
Another challenge associated with finding OTC sequence variants, found using standard or next generation sequencing methods, is prediction of severity of disease in people with those sequence changes. We have documented that most participants with OTCD enrolled in the longitudinal study did not experience episodes of elevated plasma ammonia whereas some had dozens of hyperammonemic episodes. Additionally, there was no correlation between type of mutation and severity of disease measured by either number of hyperammonemic episodes or liver dysfunction in patients with late onset OTCD. This suggests that complex relationship between genetic and environmental factors affects disease outcome and better understanding of these factors can improve our ability to avoid devastating consequences of hyperammonemia. Genome wide association studies (GWAS) could be used to identify genetic modifiers of OTCD but the small number of patients available for such studies will require development of new computational and statistical methods to analyze the data. International collaborations among consortia that study natural histories of OTCD and other urea cycle disorders (Seminara et al., 2010; Batshaw et al., 2014; Summar et al., 2014) will help overcome the limited number of subjects available for such GWAS. These studies of the natural history of OTCD are also likely to identify environmental factors that contribute to the variability of OTCD presentation. Encouraging the development of international patient registries and standardization of reporting will be essential next steps in refining our understanding of factors that contribute to the severity of rare diseases such as OTCD.
Supplementary Material
ACKNOWLEDGEMENT
The Urea Cycle Disorders Consortium (U54HD061221) is a part of the National Institutes of Health (NIH) Rare Disease Clinical Research Network (RDCRN), supported through collaboration between the Office of Rare Diseases Research (ORDR), the National Center for Advancing Translational Science (NCATS) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). The Urea Cycle Disorders Consortium is also supported by the O’Malley Foundation, the Rotenberg Family Fund, the Dietmar-Hopp Foundation, and the Kettering Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTARY DATA
Fig. S1. Conservation and solvent accessible area of amino acids affected by missense mutations in patients enrolled in the longitudinal study of OTCD.
Fig. S2. Destabilization of the OTC protein by amino acid replacements found in patients enrolled in the longitudinal study of OTCD.
Fig. S3. Predicted effects of missense mutations on the function of OTC protein in the participants enrolled in the longitudinal study of OTCD.
Fig. S4. Biomarkers of liver damage in participants of the longitudinal study of OTCD.
Table S1. Mutations in the OTC gene associated with OTC deficiency.
Table S2. Large duplications, deletions and rearrangements associated with OTC deficiency.
Table S3. Rare polymorphisms (MAF < 1%) in the OTC gene.
Table S4. Disease onset, gender and liver transplant status of OTC patients with neonatal and late onset OTCD enrolled in the longitudinal study of urea cycle disorders.
Table S5. Patients with OTCD who did not receive liver transplant and had symptoms of liver dysfunction at any point during longitudinal study of urea cycle defects.
Table S6. Patients with OTCD who did not receive liver transplant and had elevated plasma INR at any point during longitudinal study of urea cycle defects.
REFERENCES
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arranz JA, Madrigal I, Riudor E, Armengol L, Mila M. Complete deletion of ornithine transcarbamylase gene confirmed by CGH array of X chromosome. J. Inherit. Metab. Dis. 2007;30:813. doi: 10.1007/s10545-007-0578-y. [DOI] [PubMed] [Google Scholar]
- Augustin L, Mavinakere M, Morizono H, Tuchman M. Expression of wild-type and mutant human ornithine transcarbamylase genes in Chinese hamster ovary cells and lack of dominant negative effect of R141Q and R40H mutants. Pediatr. Res. 2000;48:842–846. doi: 10.1203/00006450-200012000-00023. [DOI] [PubMed] [Google Scholar]
- Azevedo L, Calafell F, Vilarinho L, Amorim A. Haplotype analysis and phylogeny of ornithine transcarbamylase polymorphisms. Ann. Hum. Genet. 2002a;66:379–385. doi: 10.1046/j.1469-1809.2002.00129.x. [DOI] [PubMed] [Google Scholar]
- Azevedo L, Stolnaja L, Tietzeova E, Hrebicek M, Hruba E, Vilarinho L, Amorim A, Dvorakova L. New polymorphic sites within ornithine transcarbamylase gene: population genetics studies and implications for diagnosis. Mol Genet Metab. 2003;78:152–157. doi: 10.1016/s1096-7192(03)00019-2. [DOI] [PubMed] [Google Scholar]
- Azevedo L, Vilarinho L, Teles EL, Amorim A. Ornithine transcarbamylase deficiency: a novel splice site mutation in a family with meiotic recombination and a new useful SNP for diagnosis. Mol. Genet. Metab. 2002b;76:68–70. doi: 10.1016/s1096-7192(02)00013-6. [DOI] [PubMed] [Google Scholar]
- Balasubramaniam S, Rudduck C, Bennetts B, Peters G, Wilcken B, Ellaway C. Contiguous gene deletion syndrome in a female with ornithine transcarbamylase deficiency. Mol. Genet. Metab. 2010;99:34–41. doi: 10.1016/j.ymgme.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Batshaw ML, Tuchman M, Summar M, Seminara J. A longitudinal study of urea cycle disorders. Mol. Genet. Metab. 2014;113:127–130. doi: 10.1016/j.ymgme.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Z, Dalal A, Morry A, Pitlik S, Zinger P, Cohen J, Fattal I, Galili-Mosberg R, Tessler D, Baruch RG, Nuoffer JM, Largiader CR, Mandel H. Adult-onset ornithine transcarbamylase (OTC) deficiency unmasked by the Atkins’ diet. J. Hepatol. 2010;52:292–295. doi: 10.1016/j.jhep.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Bezinover D, Douthitt L, McQuillan PM, Khan A, Dalal P, Stene J, Uemura T, Kadry Z, Janicki PK. Fatal hyperammonemia after renal transplant due to late-onset urea cycle deficiency: a case report. Transplant Proc. 2010;42:1982–1985. doi: 10.1016/j.transproceed.2010.03.142. [DOI] [PubMed] [Google Scholar]
- Brusilow SW, Horwich AL. Urea Cycle Enzymes. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic & Molecular Bases of Inherited Disease. McGraw-Hill: 2001. pp. 1909–1963. [Google Scholar]
- Brusilow SW, Maestri NE. Urea cycle disorders: diagnosis, pathophysiology, and therapy. Adv. Pediatr. 1996;43:127–170. [PubMed] [Google Scholar]
- Burlina AB, Peduto A, Di Palma A, Bellizzi A, Sperli D, Morrone A, Burlina AP. An unusual clinical and biochemical presentation of ornithine transcarbamylase deficiency in a male patient. J. Inherit. Metab. Dis. 2006;29:179–181. doi: 10.1007/s10545-006-0193-3. [DOI] [PubMed] [Google Scholar]
- Cavicchi C, Donati M, Parini R, Rigoldi M, Bernardi M, Orfei F, Gentiloni Silveri N, Colasante A, Funghini S, Catarzi S, Pasquini E, la Marca G, Mooney S, Guerrini R, Morrone A. Sudden unexpected fatal encephalopathy in adults with OTC gene mutations-Clues for early diagnosis and timely treatment. Orphanet J. Rare. Dis. 2014;9:105. doi: 10.1186/s13023-014-0105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Randall A, Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62:1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- Chiong MA, Bennetts BH, Strasser SI, Wilcken B. Fatal late-onset ornithine transcarbamylase deficiency after coronary artery bypass surgery. Med. J. Aust. 2007;186:418–419. doi: 10.5694/j.1326-5377.2007.tb00976.x. [DOI] [PubMed] [Google Scholar]
- Climent C, Rubio V. H intragenic polymorphisms and haplotype analysis in the ornithine transcarbamylase (OTC) gene and their relevance for tracking the inheritance of OTC deficiency. Hum. Mutat. 2002a;20:407–408. doi: 10.1002/humu.9076. [DOI] [PubMed] [Google Scholar]
- Climent C, Rubio V. Identification of seven novel missense mutations, two splice-site mutations, two microdeletions and a polymorphic amino acid substitution in the gene for ornithine transcarbamylase (OTC) in patients with OTC deficiency. Hum. Mutat. 2002b;19:185–186. doi: 10.1002/humu.9011. [DOI] [PubMed] [Google Scholar]
- Deardorff MA, Gaddipati H, Kaplan P, Sanchez-Lara PA, Sondheimer N, Spinner NB, Hakonarson H, Ficicioglu C, Ganesh J, Markello T, Loechelt B, Zand DJ, Yudkoff M, Lichter-Konecki U. Complex management of a patient with a contiguous Xp11.4 gene deletion involving ornithine transcarbamylase: a role for detailed molecular analysis in complex presentations of classical diseases. Mol. Genet. Metab. 2008;94:498–502. doi: 10.1016/j.ymgme.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Dokholyan NV. Emergence of protein fold families through rational design. PLoS Comput. Biol. 2006;2:e85. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisi-Vici C, Rizzo C, Burlina AB, Caruso U, Sabetta G, Uziel G, Abeni D. Inborn errors of metabolism in the Italian pediatric population: a national retrospective survey. J. Pediatr. 2002;140:321–327. doi: 10.1067/mpd.2002.122394. [DOI] [PubMed] [Google Scholar]
- Finkelstein JE, Hauser ER, Leonard CO, Brusilow SW. Late-onset ornithine transcarbamylase deficiency in male patients. J. Pediatr. 1990;117:897–902. doi: 10.1016/s0022-3476(05)80129-5. [DOI] [PubMed] [Google Scholar]
- Gallagher RC, Lam C, Wong D, Cederbaum S, Sokol RJ. Significant hepatic involvement in patients with ornithine transcarbamylase deficiency. J. Pediatr. 2014;164:720–725. doi: 10.1016/j.jpeds.2013.12.024. e726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glansdorff N, Xu Y. Microbial arginine biosynthesis: pathway, regulation and industrial production. In: Wendisch VF, editor. Amino Acid Biosynthesis – Pathways, Regulation and Metabolic Engineering. Berlin: Springer; 2006. pp. 219–257. [Google Scholar]
- Glasgow JF, Middleton B. Reye syndrome--insights on causation and prognosis. Arch. Dis. Child. 2001;85:351–353. doi: 10.1136/adc.85.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyato K, Wray J, Huang ZJ, Yudkoff M, Batshaw ML. Metabolic and neuropsychological phenotype in women heterozygous for ornithine transcarbamylase deficiency. Ann. Neurol. 2004;55:80–86. doi: 10.1002/ana.10794. [DOI] [PubMed] [Google Scholar]
- Hata A, Tsuzuki T, Shimada K, Takiguchi M, Mori M, Matsuda I. Isolation and characterization of the human ornithine transcarbamylase gene: structure of the 5’-end region. J. Biochem. 1986;100:717–725. doi: 10.1093/oxfordjournals.jbchem.a121764. [DOI] [PubMed] [Google Scholar]
- Hata A, Tsuzuki T, Shimada K, Takiguchi M, Mori M, Matsuda I. Structure of the human ornithine transcarbamylase gene. J. Biochem. 1988;103:302–308. doi: 10.1093/oxfordjournals.jbchem.a122265. [DOI] [PubMed] [Google Scholar]
- Horwich AL, Fenton WA, Williams KR, Kalousek F, Kraus JP, Doolittle RF, Konigsberg W, Rosenberg LE. Structure and expression of a complementary DNA for the nuclear coded precursor of human mitochondrial ornithine transcarbamylase. Science. 1984;224:1068–1074. doi: 10.1126/science.6372096. [DOI] [PubMed] [Google Scholar]
- Horwich AL, Kalousek F, Fenton WA, Pollock RA, Rosenberg LE. Targeting of pre-ornithine transcarbamylase to mitochondria: definition of critical regions and residues in the leader peptide. Cell. 1986;44:451–459. doi: 10.1016/0092-8674(86)90466-6. [DOI] [PubMed] [Google Scholar]
- Hu WT, Kantarci OH, Merritt JL, 2nd, McGrann P, Dyck PJ, Lucchinetti CF, Tippmann-Peikert M. Ornithine transcarbamylase deficiency presenting as encephalopathy during adulthood following bariatric surgery. Arch. Neurol. 2007;64:126–128. doi: 10.1001/archneur.64.1.126. [DOI] [PubMed] [Google Scholar]
- Hudak ML, Jones MD, Jr, Brusilow SW. Differentiation of transient hyperammonemia of the newborn and urea cycle enzyme defects by clinical presentation. J. Pediatr. 1985;107:712–719. doi: 10.1016/s0022-3476(85)80398-x. [DOI] [PubMed] [Google Scholar]
- Ihara K, Yoshino M, Hoshina T, Harada N, Kojima-Ishii K, Makimura M, Hasegawa Y, Watanabe Y, Yamaguchi S, Hara T. Coagulopathy in patients with late-onset ornithine transcarbamylase deficiency in remission state: a previously unrecognized complication. Pediatrics. 2013;131:e327–e330. doi: 10.1542/peds.2012-0030. [DOI] [PubMed] [Google Scholar]
- Jakubiczka S, Bettecken T, Mohnike K, Schneppenheim R, Stumm M, Tonnies H, Volleth M, Wieacker P. Symptoms of OTC deficiency but not DMD in a female carrier of an Xp21.1 deletion including the genes for dystrophin and OTC. Eur. J. Pediatr. 2007;166:743–745. doi: 10.1007/s00431-006-0303-0. [DOI] [PubMed] [Google Scholar]
- Kalousek F, Francois B, Rosenberg LE. Isolation and characterization of ornithine transcarbamylase from normal human liver. J. Biol. Chem. 1978;253:3939–3944. [PubMed] [Google Scholar]
- Keskinen P, Siitonen A, Salo M. Hereditary urea cycle diseases in Finland. Acta Paediatr. 2008;97:1412–1419. doi: 10.1111/j.1651-2227.2008.00923.x. [DOI] [PubMed] [Google Scholar]
- Kim GH, Choi JH, Lee HH, Park S, Kim SS, Yoo HW. Identification of novel mutations in the human ornithine transcarbamylase (OTC) gene of Korean patients with OTC deficiency and transient expression of the mutant proteins in vitro. Hum. Mutat. 2006;27:1159. doi: 10.1002/humu.9465. [DOI] [PubMed] [Google Scholar]
- Kim SH, Lee JS, Lim BC, Kim KJ, Hwang YS, Park JD, Cheon JE, Kim IO, Kim BN, Chae JH. A female carrier of ornithine carbamoyltransferase deficiency masquerading as attention deficit-hyperactivity disorder. Brain Dev. 2014;36:734–737. doi: 10.1016/j.braindev.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Klein OD, Kostiner DR, Weisiger K, Moffatt E, Lindeman N, Goodman S, Tuchman M, Packman S. Acute fatal presentation of ornithine transcarbamylase deficiency in a previously healthy male. Hepatol. Int. 2008;2:390–394. doi: 10.1007/s12072-008-9078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo T, Satoh Y, Kanazawa M, Yamamoto S, Takayanagi M, Ohtake A, Mori M, Niimi H. Expression analysis of two mutant human ornithine transcarbamylases in COS-7 cells. J. Hum. Genet. 1998;43:54–58. doi: 10.1007/s100380050037. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lindgren V, de Martinville B, Horwich AL, Rosenberg LE, Francke U. Human ornithine transcarbamylase locus mapped to band Xp21.1 near the Duchenne muscular dystrophy locus. Science. 1984;226:698–700. doi: 10.1126/science.6494904. [DOI] [PubMed] [Google Scholar]
- Lipskind S, Loanzon S, Simi E, Ouyang DW. Hyperammonemic coma in an ornithine transcarbamylase mutation carrier following antepartum corticosteroids. J. Perinatol. 2011;31:682–684. doi: 10.1038/jp.2011.23. [DOI] [PubMed] [Google Scholar]
- Maestri NE, Brusilow SW, Clissold DB, Bassett SS. Long-term treatment of girls with ornithine transcarbamylase deficiency. N. Engl. J. Med. 1996;335:855–859. doi: 10.1056/NEJM199609193351204. [DOI] [PubMed] [Google Scholar]
- Maestri NE, Lord C, Glynn M, Bale A, Brusilow SW. The phenotype of ostensibly healthy women who are carriers for ornithine transcarbamylase deficiency. Medicine (Baltimore) 1998;77:389–397. [PubMed] [Google Scholar]
- Marcus N, Scheuerman O, Hoffer V, Zilbershot-Fink E, Reiter J, Garty BZ. Stupor in an adolescent following Yom Kippur fast, due to late-onset ornithine transcarbamylase deficiency. Isr. Med. Assoc. J. 2008;10:395–396. [PubMed] [Google Scholar]
- Matsuura T, Hoshide R, Setoyama C, Shimada K, Hase Y, Yanagawa T, Kajita M, Matsuda I. Four novel gene mutations in five Japanese male patients with neonatal or late onset OTC deficiency: application of PCR-single-strand conformation polymorphisms for all exons and adjacent introns [corrected] Hum. Genet. 1993;92:49–56. doi: 10.1007/BF00216144. [DOI] [PubMed] [Google Scholar]
- Mavinakere M, Morizono H, Shi D, Allewell NM, Tuchman M. The clinically variable R40H mutant ornithine carbamoyltransferase shows cytosolic degradation of the precursor protein in CHO cells. J. Inherit. Metab. Dis. 2001;24:614–622. doi: 10.1023/a:1012726207870. [DOI] [PubMed] [Google Scholar]
- McCullough BA, Yudkoff M, Batshaw ML, Wilson JM, Raper SE, Tuchman M. Genotype spectrum of ornithine transcarbamylase deficiency: correlation with the clinical and biochemical phenotype. Am. J. Med. Genet. 2000;93:313–319. doi: 10.1002/1096-8628(20000814)93:4<313::aid-ajmg11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- McGuire PJ, Lee HS, Summar ML. Infectious precipitants of acute hyperammonemia are associated with indicators of increased morbidity in patients with urea cycle disorders. J. Pediatr. 2013;163:1705–1710. doi: 10.1016/j.jpeds.2013.08.029. e1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire PJ, Tarasenko TN, Wang T, Levy E, Zerfas PM, Moran T, Lee HS, Bequette BJ, Diaz GA. Acute metabolic decompensation due to influenza in a mouse model of ornithine transcarbamylase deficiency. Dis. Model. Mech. 2014;7:205–213. doi: 10.1242/dmm.013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizono H, Listrom CD, Rajagopal BS, Aoyagi M, McCann MT, Allewell NM, Tuchman M. ‘Late onset’ ornithine transcarbamylase deficiency: function of three purified recombinant mutant enzymes. Hum. Mol. Genet. 1997a;6:963–968. doi: 10.1093/hmg/6.6.963. [DOI] [PubMed] [Google Scholar]
- Morizono H, Tuchman M, Rajagopal BS, McCann MT, Listrom CD, Yuan X, Venugopal D, Barany G, Allewell NM. Expression, purification and kinetic characterization of wild-type human ornithine transcarbamylase and a recurrent mutant that produces ‘late onset’ hyperammonaemia. Biochem. J. 1997b;322(Pt 2):625–631. doi: 10.1042/bj3220625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa A, Clarke JT. Ornithine transcarbamoylase deficiency presenting with acute liver failure. J. Inherit. Metab. Dis. 2006;29:586. doi: 10.1007/s10545-006-0303-2. [DOI] [PubMed] [Google Scholar]
- Nishiyori A, Yoshino M, Kato H, Matsuura T, Hoshide R, Matsuda I, Kuno T, Miyazaki S, Hirose S, Kuromaru R, Mori M. The R40H mutation in a late onset type of human ornithine transcarbamylase deficiency in male patients. Hum. Genet. 1997;99:171–176. doi: 10.1007/s004390050333. [DOI] [PubMed] [Google Scholar]
- Pierson DL, Cox SL, Gilbert BE. Human ornithine transcarbamylase. Purification and characterization of the enzyme from normal liver and the liver of a Reye’s syndrome patient. J. Biol. Chem. 1977;252:6464–6469. [PubMed] [Google Scholar]
- Pinner JR, Freckmann ML, Kirk EP, Yoshino M. Female heterozygotes for the hypomorphic R40H mutation can have ornithine transcarbamylase deficiency and present in early adolescence: a case report and review of the literature. J. Med. Case Rep. 2010;4:361. doi: 10.1186/1752-1947-4-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante RJ, Tuchman M. Polymorphisms in the human ornithine transcarbamylase gene useful for allele tracking. Mutations in brief no. 193. Online. Hum. Mutat. 1998;12:289–290. [PubMed] [Google Scholar]
- Quintero-Rivera F, Deignan JL, Peredo J, Grody WW, Crandall B, Sims M, Cederbaum SD. An exon 1 deletion in OTC identified using chromosomal microarray analysis in a mother and her two affected deceased newborns: implications for the prenatal diagnosis of ornithine transcarbamylase deficiency. Mol. Genet. Metab. 2010;101:413–416. doi: 10.1016/j.ymgme.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Seminara J, Tuchman M, Krivitzky L, Krischer J, Lee HS, Lemons C, Baumgartner M, Cederbaum S, Diaz GA, Feigenbaum A, Gallagher RC, Harding CO, Kerr DS, Lanpher B, Lee B, Lichter-Konecki U, McCandless SE, Merritt JL, Oster-Granite ML, Seashore MR, Stricker T, Summar M, Waisbren S, Yudkoff M, Batshaw ML. Establishing a consortium for the study of rare diseases: The Urea Cycle Disorders Consortium. Mol. Genet. Metab. 2010;100(Suppl. 1):S97–S105. doi: 10.1016/j.ymgme.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchelochkov OA, Li FY, Geraghty MT, Gallagher RC, Van Hove JL, Lichter-Konecki U, Fernhoff PM, Copeland S, Reimschisel T, Cederbaum S, Lee B, Chinault AC, Wong LJ. High-frequency detection of deletions and variable rearrangements at the ornithine transcarbamylase (OTC) locus by oligonucleotide array CGH. Mol. Genet. Metab. 2009;96:97–105. doi: 10.1016/j.ymgme.2008.11.167. [DOI] [PubMed] [Google Scholar]
- Shi D, Morizono H, Ha Y, Aoyagi M, Tuchman M, Allewell NM. 1.85-A resolution crystal structure of human ornithine transcarbamoylase complexed with N-phosphonacetyl-L-ornithine. Catalytic mechanism and correlation with inherited deficiency. J. Biol. Chem. 1998;273:34247–34254. doi: 10.1074/jbc.273.51.34247. [DOI] [PubMed] [Google Scholar]
- Shoichet BK, Baase WA, Kuroki R, Matthews BW. A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. USA. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snodgrass PJ. The effects of pH on the kinetics of human liver Ornithine--carbamyl phosphate transferase. Biochemistry. 1968;7:3047–3051. doi: 10.1021/bi00849a004. [DOI] [PubMed] [Google Scholar]
- Summar ML, Endo F, Kolker S. On the Creation, Utility and Sustaining of Rare Diseases Research Networks: Lessons learned from the Urea Cycle Disorders Consortium, the Japanese Urea Cycle Disorders Consortium and the European Registry and Network for Intoxication Type Metabolic Diseases. Mol. Genet. Metab. 2014;113:105–108. doi: 10.1016/j.ymgme.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summar ML, Koelker S, Freedenberg D, Le Mons C, Haberle J, Lee HS, Kirmse B. The incidence of urea cycle disorders. Mol. Genet. Metab. 2013;110:179–180. doi: 10.1016/j.ymgme.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Wada T, Maruyama M, Tanaka A, Takikawa H, Komatsu Y. Hyperammonemia-induced encephalopathy due to ornithine transcarbamylase deficiency in an adult woman: identification of novel missense mutations. J. Gastroenterol. 2005;40:106–107. doi: 10.1007/s00535-004-1502-y. [DOI] [PubMed] [Google Scholar]
- Thurlow VR, Asafu-Adjaye M, Agalou S, Rahman Y. Fatal ammonia toxicity in an adult due to an undiagnosed urea cycle defect: under-recognition of ornithine transcarbamylase deficiency. Ann. Clin. Biochem. 2010;47:279–281. doi: 10.1258/acb.2010.009250. [DOI] [PubMed] [Google Scholar]
- Tuchman M. Mutations and polymorphisms in the human ornithine transcarbamylase gene. Hum. Mutat. 1993;2:174–178. doi: 10.1002/humu.1380020304. [DOI] [PubMed] [Google Scholar]
- Tuchman M, Holzknecht RA. Heterogeneity of patients with late onset ornithine transcarbamylase deficiency. Clin. Invest. Med. 1991;14:320–324. [PubMed] [Google Scholar]
- Tuchman M, Jaleel N, Morizono H, Sheehy L, Lynch MG. Mutations and polymorphisms in the human ornithine transcarbamylase gene. Hum. Mutat. 2002;19:93–107. doi: 10.1002/humu.10035. [DOI] [PubMed] [Google Scholar]
- Tuchman M, Morizono H, Rajagopal BS, Plante RJ, Allewell NM. Identification of ‘private’ mutations in patients with ornithine transcarbamylase deficiency. J. Inherit. Metab. Dis. 1997;20:525–527. doi: 10.1023/a:1005301513465. [DOI] [PubMed] [Google Scholar]
- Tuchman M, Morizono H, Rajagopal BS, Plante RJ, Allewell NM. The biochemical and molecular spectrum of ornithine transcarbamylase deficiency. J. Inherit. Metab. Dis. 1998;21:40–58. doi: 10.1023/a:1005353407220. [DOI] [PubMed] [Google Scholar]
- Tuchman M, Plante RJ. Mutations and polymorphisms in the human ornithine transcarbamylase gene: mutation update addendum. Hum. Mutat. 1995;5:293–295. doi: 10.1002/humu.1380050404. [DOI] [PubMed] [Google Scholar]
- Weisbren SE, McCarter R, He J, Krivitzky L, Caudle S, Wilkening G, Burgard P, Sanz J, Nguyen MD, Batshaw M. Neuropsychological outcomes in the longitudinal study of urea cycle disorders. Mol. Genet. Metab. 2014 in press. [Google Scholar]
- Wong LJ, Dimmock D, Geraghty MT, Quan R, Lichter-Konecki U, Wang J, Brundage EK, Scaglia F, Chinault AC. Utility of oligonucleotide array-based comparative genomic hybridization for detection of target gene deletions. Clin. Chem. 2008;54:1141–1148. doi: 10.1373/clinchem.2008.103721. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Brailey LL, Morizono H, Bale AE, Tuchman M. Mutations and polymorphisms in the human ornithine transcarbamylase (OTC) gene. Hum. Mutat. 2006;27:626–632. doi: 10.1002/humu.20339. [DOI] [PubMed] [Google Scholar]
- Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nat. Methods. 2007a;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- Yin S, Ding F, Dokholyan NV. Modeling backbone flexibility improves protein stability estimation. Structure. 2007b;15:1567–1576. doi: 10.1016/j.str.2007.09.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.