Abstract
Biopatterning has been increasingly used for well-defined cellular microenvironment, patterned surface topology, and guided biological cues; however, it meets additional challenges on biocompatibility, temperature and chemical sensitivity and limited reagent volume. In this paper, we target at combining the desired features from the non-contact inkjet printing and the dot-matrix impact printing to establish a versatile multiplexed micropatterning platform, referred to as Microfluidic Impact Printer (MI-Printer), for emerging biomedical applications. Using this platform, we can achieve the distinct features of no cross-contamination, minute volume manipulation with minimal dead volume, high-throughput and biocompatible printing process, multiplexed patterning with automatic alignment, printing availability for complex medium (cell suspension or colloidal solutions), interchangeable/disposable microfluidic cartridge design with out-of-cleanroom microfabrication, simple printing system assembly and configuration, all highly desirable towards biological applications. Specifically, the printing resolution of the MI-printer platform has been experimentally characterized and theoretically analyzed. Printed droplets with 80µm in diameter have been repeatedly obtained. Furthermore, two unique features of MI-printer platform, multiplexed printing and self-alignment printing, have been successfully experimentally demonstrated (less than 10µm misalignment). In addition, combinatorial patterning and biological patterning, which utilizes the multiplexed and self-alignment printing nature of the MI-printer, have been devised to demonstrate the applicability of this robust printing technique for emerging biomedical applications.
Introduction
Creating well-defined micro-nanoscopic patterns of biomaterials (e.g., cells, proteins, nucleic acids, and polysaccharides) can be of particular interest for a variety of academic and industrial applications, including composite material investigation, electronic and optic system development, combinatorial chemistry, cell biology, tissue engineering, and medical sciences. [1–9] Recently, biological micropatterning has been increasingly explored by biologists, bioengineers, and medical researchers for well-defined cellular microenvironment, patterned surface topology, and guided biological cues. [10–15] For instance, micro/nano-patterned intracellular and extracellular protein arrays have been widely used for the investigation of signaling pathway, ligand interactions and cellular responses. [16–18] Well-aligned single-cell arrays have been utilized to analyze individual cellular responses, cytoskeletal structures, and ligand-receptor interactions. [19–21] Moreover, printed combinatorial biomolecular libraries (e.g., peptides and oligonucleotides) have been extended to multiplexed high-throughput screening, including cancerous biomarker detection, drug discovery, and genomic identification. [4, 22–25] Unlike the conventional micro-nanopatterning (e.g., for microelectronics), biopatterning methods encounter additional challenges, such as biocompatibility, temperature and chemical sensitivity as well as limited reagent amount.
Specifically, a number of micro-nanopatterning techniques have been developed over the past decades with an emphasis on biological and medical uses, which can be divided into the following categories: photolithography, screen printing, and inkjet printing. Photolithography employs high-intensity UV light source to selectively photo-activate biomaterials through high-precision photomasks with its resolution down to a sub-micrometer range. Derived from photolithography and 3D printing, the light-enabled printing technique has been developed to rapidly prototype biodegradable cellular matrixes for medical implants, such as artificial bones and organs, in a stereo fashion. [26] However, wet chemical processing and UV exposure step can potentially cause biomolecular degradation (protein denaturation and aggregation) and cellular damage. Furthermore, photolithography typically requires a high-maintenance cleanroom environment and expensive processing equipment (e.g., spincoaters and mask aligners), which may not be available to many biological and biomedical research laboratories. [13,27–30] Screening printing transfers wet biological samples to the desired locations through a selectively blocked stencil on the substrate. Limited positioning precision and fabrication complexity of the stencil can be the major drawbacks of this type of techniques. [18, 31] In comparison with the other patterning techniques, the inkjet-based printing offers several apparent advantages, for instance, the non-contact nature eliminates the potential cross-contamination from the source to the substrate, which can be highly advantageous to biological applications. Moreover, ink-jet printing has been fully automated with ultrahigh throughput, benefiting from its huge commercial success, which allows the design-to-printing cycle to be completed in a computer aided design tool. [32, 33] The modern inkjet printing technology employs either thermal expansion, piezoelectric actuator, or a combination to drive the printing fluid. [33–35] For example, inkjet printing systems have been devised to create 3D artificial organs through layer-by-layer deposition of multiple cell types and matrix materials together. [36, 37] The goal is to establish branched vasculature tissues surrounded by supporting matrixes at a sub-millimeter resolution. [6, 36–38] Moreover, biomolecules, such as proteins and DNA, can be co-located on various solid supports through inkjet printing for high-throughput screening, multiplexed bio-sensing and immunoassaying. [39–41] However, the standardized ink cartridge of the inkjet printing requires at least milliliter volume of samples to initiate the printing process, which is unsuitable for research with scarce biological samples. [34,35] In addition, the cross contamination and limited multiplexibility of the ink cartridge can be additional concerns for applying inkjet printing to biological research. On the other hand, the thermal expansion approach is incompatible with most of the biological reagents, while the piezoelectric actuator can be difficult and costly to integrate with a customized cartridge.
As a preceding technology to the mainstream inkjet printing, impact printing (also known as dot-matrix printing) utilizes an array of electromagnetically actuated pins to strike ink-soaked color ribbons onto the printing substrate. Once the contact is made between the ribbon and the substrate under the striking pressure applied by the pin, a minute color dot will display on the substrate. Although it has been almost abandoned industrially, for the noisy operation, low printing speed, limited color options and low graphic resolution [42, 43], several intriguing features make it potentially attractive to custom bio-printing applications. Specifically, it includes a highly adaptive and efficient electromechanical design (i.e., striking pins with several millimeter travel distance), a multiplexed pin matrix control (e.g., offering up to 24 independently actuatable elements with simple control circuitry, compared with 3- or 4-independent cartridges in the inkjets), and low-cost interchangeable ribbon rolls (e.g., the separable actuator and ink units, unlike the integrated piezoelectric drive design), in addition to its simple operations without any chemical or thermal treatment. [44]
In this paper, we target at combining the desired features from the non-contact inkjet printing and the dot-matrix impact printing to establish a versatile multiplexed micropatterning platform, referred to as Microfluidic Impact Printer (MI-Printer), for emerging microfluidic and biological applications. [45] In particular, the MI-printer is comprised of a conventional impact matrix head, an interchangeable microfluidic cartridge set with multiplexed ink channels, and a programmable traveling stage holding the printing substrates. Using the MI-printer platform, we can achieve the following distinct features, which are highly desirable for printing or patterning biological objects. First of all, the modular design of the MI-printer ensures no direct contact among the printer head, the liquid inks, and the targeted substrate, which avoid the cross-contamination among different chemical/biological reagents. Secondly, the highly customized cartridge set allows adapting to a wide range of aqueous and non-aqueous fluids with an extremely small loading volume (as little as 0.6µL) and an optimized dead volume (less than 0.05µL), which can be highly attractive and economic to the academic researchers. It can also be utilized to print complex fluids, for instance, cell suspensions and colloidal solutions. Moreover, the microfluidic cartridge is built on a low-cost polymer (i.e., polydimethylsiloxane or PDMS) with an easily interchangeable and disposable design. The fabrication of the cartridge only requires a single-layer lithography using laser micromachining and plasma bonding [46, 47]. In addition, the dot-matrix printer head can host up to 24 independently programmable pins, which can be extended to control and printing of 24 different inks simultaneously. Importantly, in the MI-printer, both printer head and travel stage as well as their computer control module can be adopted from the existing dot-matrix printers with minor alternations on the controlling circuitry, which makes the entire system easy to assemble and configure. Table 1 compares the MI-printing system with the existing biopatterning techniques. In brief, the MI-printing platform offers a versatile, high-throughput, low-cost micropatterning solution, which is highly desired for microfluidic and biofluidic research, by simply reconfiguring an obsolete printing technology. Fig. 1 illustrates a prototype of the MI-printer system with the printer head adopted from a dot-matrix printer.
TABLE 1.
Comparison among different biopatterning processes including MI-printing system
| Patterning techniques |
Advantages | Disadvantages |
|---|---|---|
| Photolithography |
|
|
| Screening printing |
|
|
| Inkjet printing |
|
|
| Microfluidic impact printing |
|
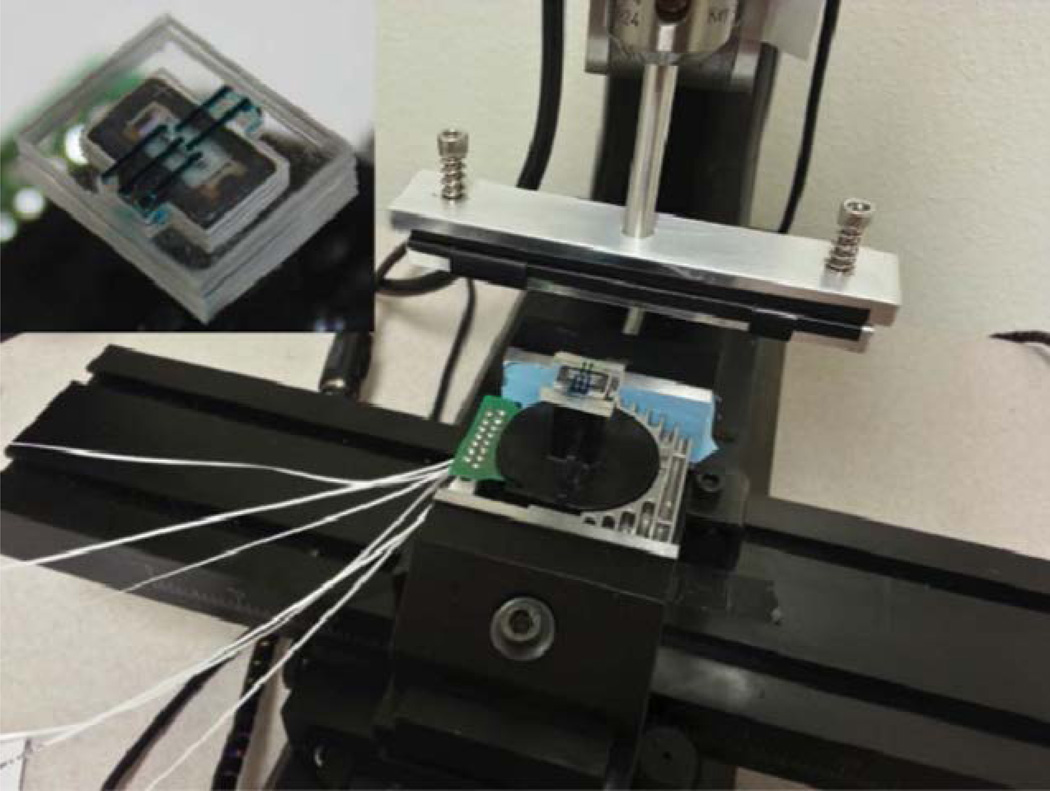
Fig. 1.
Prototype of MI-printer system (inset: prototype of microfluidic cartridge).
Experimental Methods
MI-printer system
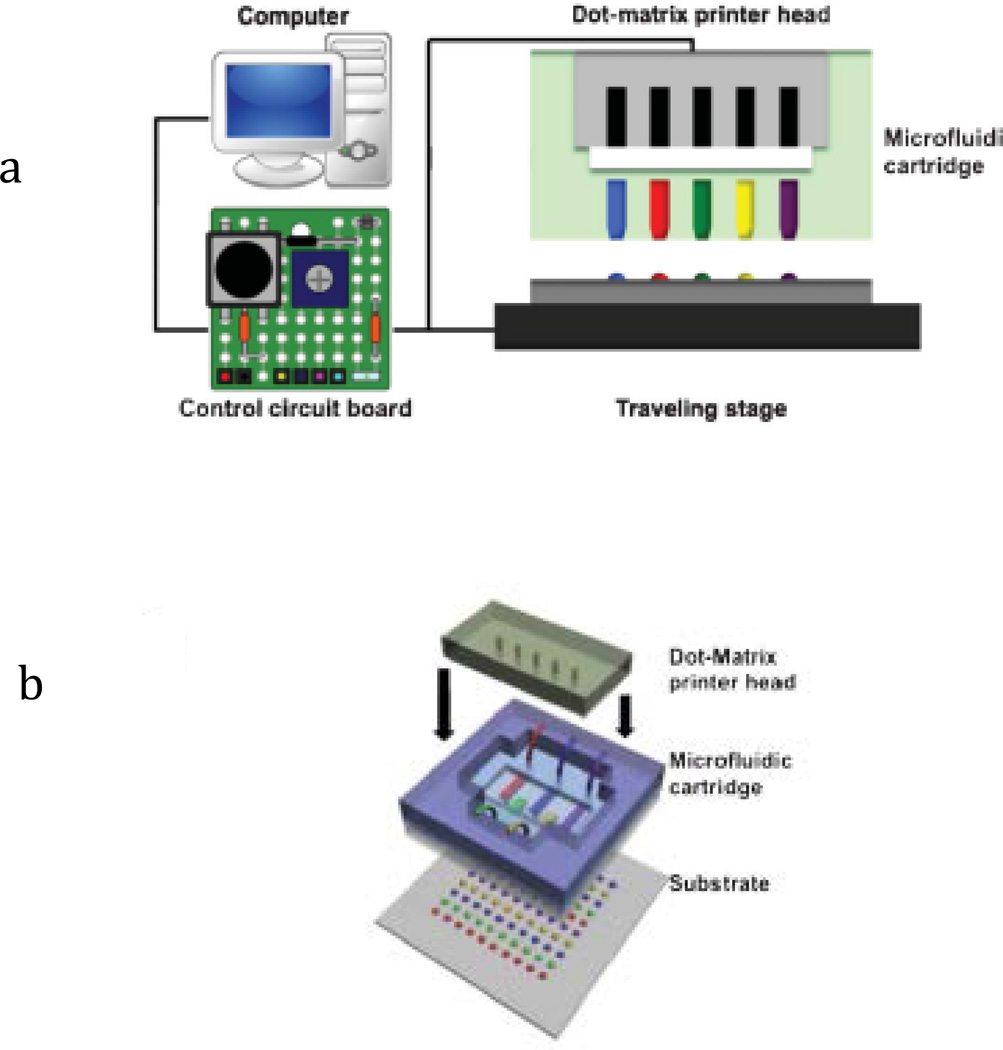
As aforementioned, the MI-printer consists of a dot-matrix printer head, a traveling stage, and a microfluidic cartridge set. As illustrated in Fig. 2a, a microfluidic cartridge set is fixed on the dot-matrix printer head (Panasonic KX-P1150) by a simple fitting structure, and the whole printer head is mechanically attached to a multi-axis traveling stage (Sherline, CNC 2000) for rapid motion and accurate positioning. In order to establish the fully automatic printing process, the interfacial circuits and firmware (Linux EMC2) have been modified to control the printer head and motion stage simultaneously. In brief, we use EMC2 program to control planar motion (in both x- and y-axes) of the traveling stage, while the other two axial controls (with 4 parallel data lines) are employed to control the driving circuitry of up to 8 electromagnetic pins.
Fig. 2.
(a) Illustration of MI-printer system with computer control and interface circuit; (b) Illustration of the microfluidic cartridge.
Microfluidic cartridge
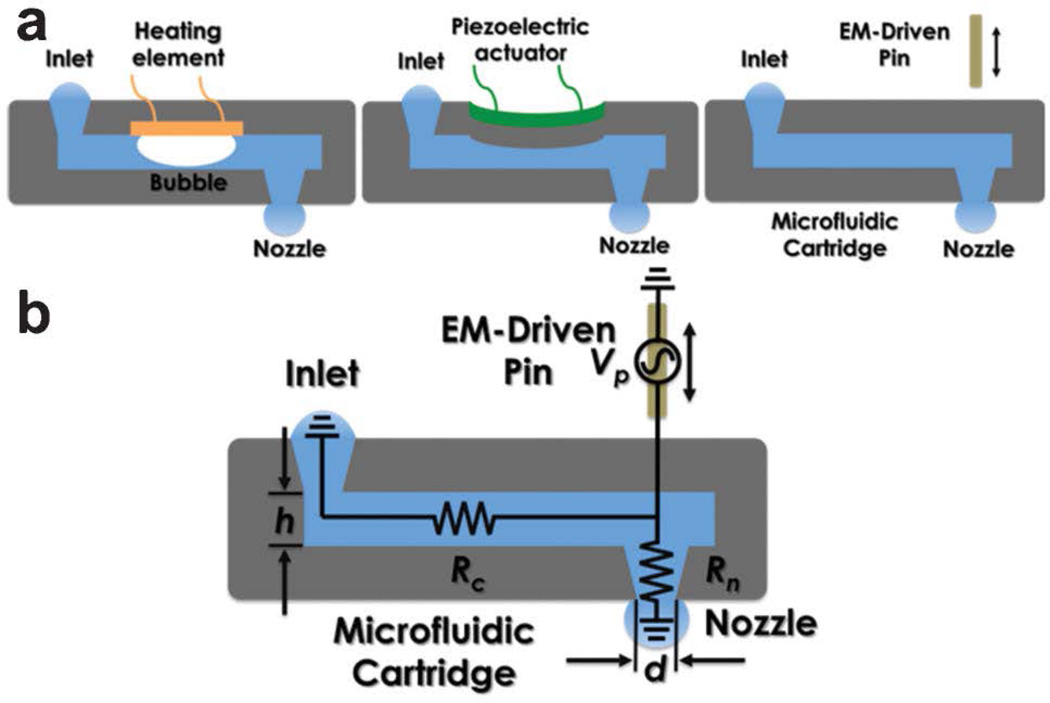
As a key component of the non-contact printing, the microfluidic cartridge includes multiplexed channels with matched printing nozzles to the pin matrix. As shown in Fig. 2b, it is structured by stacking five layers of PDMS from the top to the bottom: an alignment/fitting structure to the printer head, a spacer, a deformable membrane which is contact with the pins during the printing process, a microchannel layer, and finally, the nozzle arrays. The alignment/fitting layer allows the microfluidic cartridge is easily fitted and aligned to the dot-matrix printer head while the space determines the overall deformation caused by the striking pins. As the mechanically induced deformation increases, the hydrostatic pressure elevates accordingly inside the microchannel, and thus, induces flow motion towards the nozzle. As the geometry of the orifices tapers, the flow accelerates along the nozzle and shoots out as a droplet at the nozzle opening, which will be deposited onto the target substrate. Fig. 3a compares the different operation principles among inkjet and MI printing. As can be seen, the thermal inkjet printing uses micro-bubble expansion caused by the electrical heating element to drive discrete droplets out of the nozzle opening, while the piezoelectric-actuated printer utilizes the channel deformation caused by the piezoelectric element to generate droplets from the nozzle. Whereas, the proposed MI-printing process, which inherits the dot matrix printing mechanism, is enabled by the pin-induced membrane deformation.
Fig. 3.
(a) Comparison among various printing principles including inkjet and MIprinting; (b) Illustration of the principle of the MI-printing process.
Laser micromachining technique has been used as the primary means to fabricate the multilayer PDMS-based microfluidic cartridge. Specifically, the fabrication process starts at layout design of each structural layer in the graphic design software (i.e., CorelDRAW). Following the structural design, PDMS pre-polymer is prepared at a mixing ratio of the base to curing agents at 10:1 weight ratio (SYLGARD 184, Dow Corning), which can be either casted on petri dishes or spun on glass slides based on the thickness requirement for each layer. The PDMS film is then thermally cured at 80°C for 2 hours. Subsequently, a computer-controlled CO2-laser engraver with focusing optics (Universal Laser Systems, VersaLaser 2.30) is used to photo-etch the microfluidic network design on the PDMS layer, followed by the standard oxygen plasma treatment (at 90W for 30 seconds, Diener electronic). Bonding between different PDMS structural layers is subsequently carried out for cartridge assembly. [48]
Printing characterization
Planar PDMS substrates are used as the calibration surface for characterization of the printed aqueous droplets (e.g., the sizes and shapes), given the low surface energy preferred in analyzing the droplet geometry and preventing adjacent cross-contamination. The aqueous-based solution is typically subject to evaporation under the MI printing, due to the large surface-to-volume ratio of the droplets. Therefore, dimethyl sulfoxide (DMSO) has been introduced to the printing solutions for the reduced evaporation. [49] Three color dyes (bromophenol blue, methyl green, and phenol red) are respectively added to the aqueous solutions for easy observation. The aqueous mixtures are at the mixing ratio of 5%:47.5%:47.5% wt, Color Dye: DMSO: deionized water for the calibration test and measurement. For each printing parameter, an array of identical droplets (with more than 36 elements) is deposited onto the PDMS surface, among which six droplets are arbitrarily selected and measured. The diameter of the droplets and misalignment of the printed array are measured under a standard optical microscope. For characterization of printing speed, the MI-printer unit is driven by tunable frequency driving waveforms, which are generated by a standard function generator (33220A, Agilent).
Sample preparation
In this report, we have demonstrated the MI printing can be applied to various biological fluids, including protein solutions, cell suspensions, and agarose gel matrix. Specifically, Protein solutions with fluorescent labels are prepared by diluting the original protein-fluorophore conjugates (BSA-FITC, BSA-TR, and streptavidin-MB) in DMSO solution (50%: 50% wt, DMSO: DI water) to a concentration of 0.1mg/ml for the following bio-printing experiments. Printed cell suspension (U937 human monocyte cell line, 1×106/ml for original cell density) is prepared by mixing 0.2ml of the original cell suspension with 0.2ml DMSO solution. Agarose gel sample is prepared by mixing 0.1g of agarose powder in 10ml DI water at 42°C. Prior to sample loading, microfluidic cartridges are sterilized in a 70% ethanol solution for 20 minutes. Subsequently, the samples (fluorescent proteins, cells and agarose gel) are injected successively into the sterilized microfluidic cartridges by a 1ml syringe and printed onto hydrophobic PDMS substrates by the impact printing head. The printing results are recorded by a high-resolution boom stereo microscope (EMZ5-V15, Omano) and a fluorescent microscope (Axiovert 200M, Carl Zeiss).
Result and discussion
Printing process
MI printing process utilizes the kinetic energy from a moving pin, driven by an electromagnetic actuator, to strike on an elastic membrane above the microfluidic channel with an aligned nozzle. Upon the stroke, the deformed membrane displaces the fluid inside both the horizontal microchannel and the vertical nozzle, from which an ejection of a small droplet volume is resulted. Fig. 3b illustrates a simplified fluidic circuit model to describe the printing process with key controlling parameters. Essentially, the fluid dynamic inside the microchannel can be modeled as a displaced flow (of volume Vp) caused by the pin movement-induced membrane deformation. The displaced flow moves towards either the nozzle openings or the adjacent connecting channel, of which the flow resistances are Rn and Rc, respectively. Subsequently, the extra volume fluid entering the nozzle opening (Vn) accelerates along the tapered geometry and is ejected into a discrete droplet. According to this description, Eq. 1 expresses such a simple relationship between the printing droplet volume and the geometric parameters of the microfluidic channel and the nozzle opening:
| Eq. 1 |
where S is the tip area of the pin, ΔH is pin-induced membrane deflection, l, w, and h represent the channel length, width, and height, respectively, whereas d and t indicate the nozzle diameter and length.
Moreover, we have further derived the printed droplet shape on the surface by considering the surface energy of the substrate and surface tension of the fluid. Eq. 2 links the diameter of the printed droplet (D) to the ejected volume (Vn) and the liquid contact angle (θ) and N indicates the number of the ejections:
| Eq. 2 |
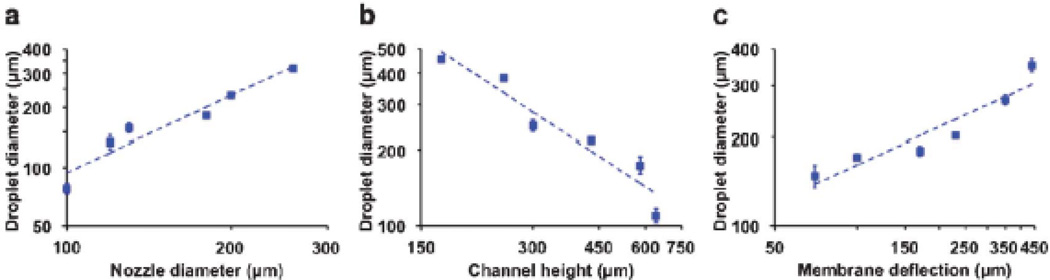
where f(θ) is a function of liquid contact angle and g (L, S, t, w) stands for a geometrical factor derived from the dimensions of the microfluidic cartridge (the detailed derivation can be found in ESI). Based on the importance and controllability in the theoretical model of the MI printing, three design parameters have been selected for further characterization of the printing process, i.e., the nozzle diameter (d), the channel height (h), and the pin-induced membrane deflection (Vp). The relationship between each parameter and the printed droplet size has been experimentally measured and theoretically analyzed.
Parameter characterization
Nozzle diameter
As predicted by Eq. 1, the nozzle dimension plays a dominant role (4th power) in determining the printed droplet volume. The diameters of printed droplets have been experimentally evaluated by varying the nozzle size, arranging from 100 to 280µm, while the other design parameters of the microfluidic cartridge keep unaltered. Fig. 4a summarizes the measurement results of the printed droplet with theoretical analysis. As expected, the printed droplet diameter exhibits a 4/3rd power dependence on the nozzle size, which is in a general agreement with the theoretical predictions, given the droplet diameter following a cubic root of the droplet volume in Eq. 2. Importantly, under our cartridge design, we intend to incorporate a higher resistance in the nozzle head than that of the microchannel (e.g., 15 times higher), leading to a reduced volume to be ejected (similar to a current divider in the circuitry). On the other hand, the further reducing the nozzle size could result in drastic increase in the nozzle resistance, a 4th power dependence according to Poiseuville’s law, and eventually could prevent the ink solution to be ejected due to the surface tension [50]. In addition, the upper limit of the nozzle boundary is fixed by the width of microchannels.
Fig. 4.
Characterization of printing resolution with key design parameters: (a) nozzle diameter, (b) channel height, (c) pin-induced membrane deflection.
Channel height
Moreover, as the second most influential design parameter, the channel height can be inversely proportional to the printed resolution as predicted by the model. Measurements on the printed droplets are varied with the channel heights, ranging from 170 to 640µm. Fig. 4b illustrates such an inversely linear relationship in both experimental measurements and theoretically analyses. Practically, the channel height can be controlled through the thickness of the microfluidic layer for ink loading. Higher profile ink channels with lower flow resistance (and thereby a lower Rc/Rn ratio) would be preferred to generate finer dots in the MI printing. In the channel of 640µm in height, aqueous droplets of around 100µm in diameter can be repetitively printed.
Pin-induced deflection: As shown in the model, the pin-induced membrane deflection causes the overall fluidic displacement in the microchannel (Vp), which is directly proportional to the printed droplet volume (Vn). Fig. 4c compares the printing resolutions from various deflections on the PDMS membranes, arranging from 70 to 440µm. The trend of the experimental measurements clearly indicates that the printed droplet size follows the theoretical predictions, in which the droplet diameter is related to the 1/3rd power of the membrane deflection. However, the maximal membrane deflection is limited by the fixed travel distance (up to 800µm) of the electromagnetically driven pins.
Printing speed and precision characterization
Under the current automated configuration, the MI-printing speed is set at 100 ejections per minute, which can be facilitated by altering the frequency of the driving waveform to a higher value (up to 200 Hz). The positioning precision of the MIprinter system is primarily provided by the mechanical tolerance of the computer-controlled traveling stage, which offers high alignment precision (less than 40 mm misalignment at a motion speed of 300 mm min−1, which is consistent with the commercially available printing platforms) without any additional calibration. [51]
Bio-Patterning Demonstrations
Multiplexed Printing
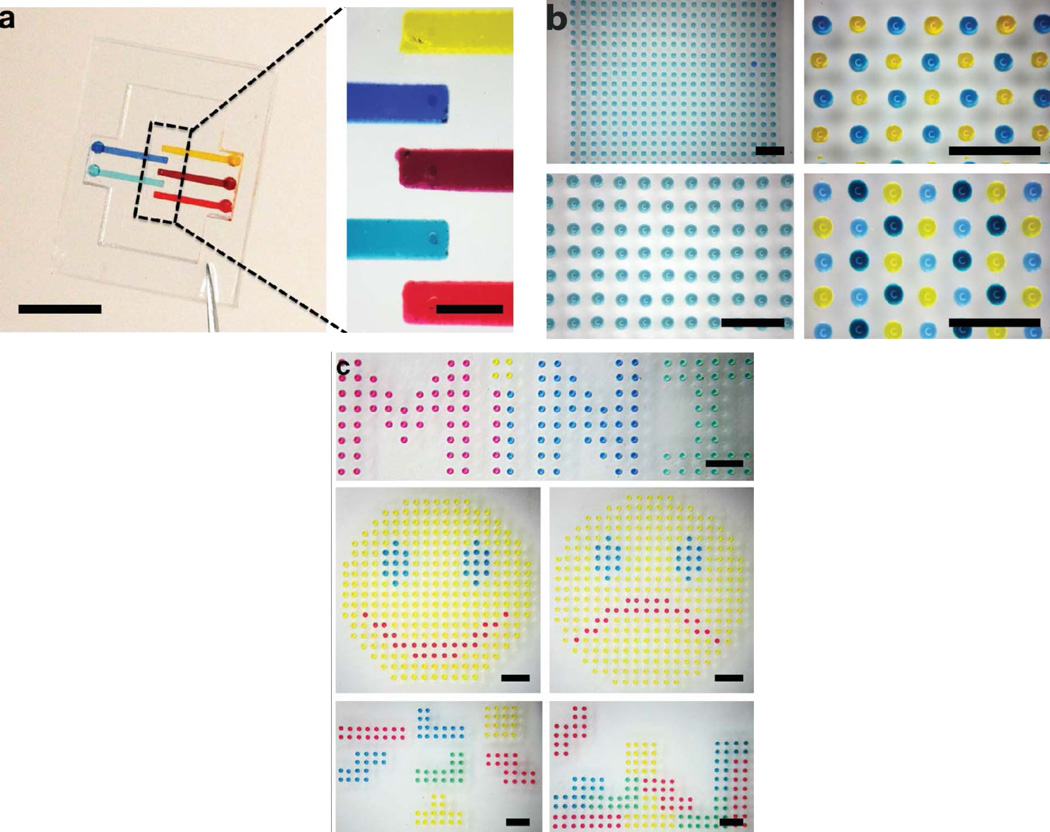
The trend of contemporary biological and medical research has highlighted the significance of high-throughput patterning of multiplexed biological and/or molecular objects for the quantitative investigation on cellular behaviors and responses within specific microenvironments at a single- or sub-cellular scale. [52–54] The MI-printer provides a facile means to establish such a complex multiplexed array patterns. In particular, we have devised a multi-channel microfluidic cartridge loaded with 5 different colored solutions, enabling simultaneous printing of five different reagents or biological objects, as illustrated in Fig. 5a. Each microchannel contains a volume of 0.6µL, which can be self-primed due to the hydrophilic nature inside. The nozzle is located at the end of the microchannel, which permits most loaded agents can be printed/ejected with an extremely low dead volume. As characterized in Fig. 4, the ejected droplet size is dependent on the microchannel and nozzle dimensions, for a droplet of 250pL, the current micro-cartridge design can produce more than 2000 droplets with each loading. As indicated before, this minute load volume is one of the key advantages of the MI printing setup, which only requires a minimal volume to perform the printing function. This feature can be highly desirable for small-scale research and development and biological/biochemical prototyping where the reagents are typically limited and expensive. For large-scale droplet generation and extended printing applications, we can easily expand our loading reservoirs to store extra volume of inks.
Fig. 5.
(a) Five-channel microfluidic cartridge device (scale bar: 5 mm for left, 1 mm for right); (b) Monocolor and multicolor droplet array printed on planar substrate (scale bar: 1 mm); (c) Multiplexed droplet-based micropatterns (MiNI logo, smiley and sad faces, and Tetris blocks, scale bar: 1 mm).
As the proof-of-concept experiment, multiple colored aqueous solutions are used as the inks for the calibration tests. Fig. 5b shows that various monocolor and multicolor droplet arrays have been printed on the planar PDMS substrates accordingly. Besides the multiplexed droplet microarray generation, multiplexed printing of the MI-printer can be also used to create complex droplet-based micropatterns for potential applications of tissue engineering and regenerative medicine. [5, 6, 36–38] Fig. 5c shows several multiplexed micropatterned images (including our lab logo, smiley and sad faces, and Tetris blocks) generated by the MI-printer.
Combinatorial Printing with Self-Alignment
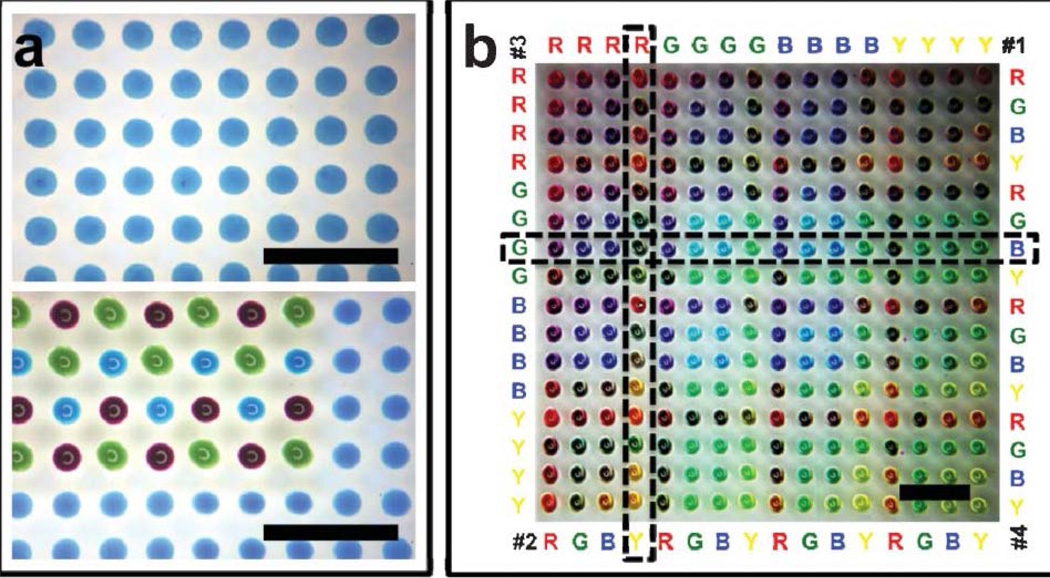
Unlike the conventional printing process, quantitative biological studies usually require multiple additive steps of biological reagents to achieve the analytical aims. Thus, it is crucial to ensure each chemical cue can be precisely deposited on the desired spot. This can be achieved by using a manual microscopic alignment approach, which is lengthy and tedious with inherent inaccuracy of operation. [32] In our MI-printing method, we have implemented a wetting contrast-enabled self-alignment strategy, in addition to computer-programmed precise movements of the automated traveling stage. In the self-alignment process, we first deposit hydrophilic aqueous droplet as the positioning anchor, followed by the consecutive aqueous printings. As a common matrix for cell culture, agarose gel is intrinsically hydrophilic and can be self-primed. [55] When deposited onto the chemically inert PDMS substrate, the agarose droplet retain its spherical shape. During the following printing steps, the ejected aqueous droplets, once in contact with the substrate, will autonomously move under the high wetting contrast towards the agrarose patterns. Fig. 6c illustrates formation of a multicolor droplet array on a pre-defined aqueous array using the self-alignment approach. As can be seen, comparing the droplet printing results, the printing alignment (less than 10µm misalignment) has been evidentially approved.
Fig. 6.
(a) Multicolor printing on pre-defined agarose array using a self alignment approach (scale bar: 1 mm); (b) four-color combinatorial printing on the planar PDMS substrate (scale bar: 1 mm).
Enabled by the multiplexed printing using wettability alignment, the MI-printing can be extended to build a large array of combinatorial mixtures of cellular and molecular substances. As illustrated in Fig. 6b, a four-color combinatorial array (of Red, Green, Blue and Yellow) has been created on the planar PDMS substrate. Exhaustive combinations (of 256 = 4×4×4×4) among four colored solutions have been represented in the printed 16×16 array. In order to generate a four-color exhaustive combinatorial array, four printing cycles with distinguished orders are needed. The first printing cycle is successively delivering quadruple sequences of each color (e.g. RRRR GGGG BBBB YYYY) in the parallel direction while the second printing cycle is delivering quadruple groups of four-color sequence (e.g. RGBY RGBY RGBY RGBY) in the parallel direction. These two printing cycles are repeated in the vertical direction for the third and fourth printing cycles. (Fig. 6b) In such a fashion, a combinatorial library with exhaustive combinations of 256 (= 4×4×4×4) elements from the four colored solutions can be represented in the printed 16×16 array. Different color droplets have been successively deposited for each micro-mixture at the identical location. As can be seen, the color hue of each mixture dot is determined by the number of different color droplets printed. For example, the combination of RYGB represents one droplets of red dye, one droplet of yellow dye, one droplet of green dye and one droplet of yellow dye are sequentially deposited at the same spot.
Biomolecular and Cellular Printing
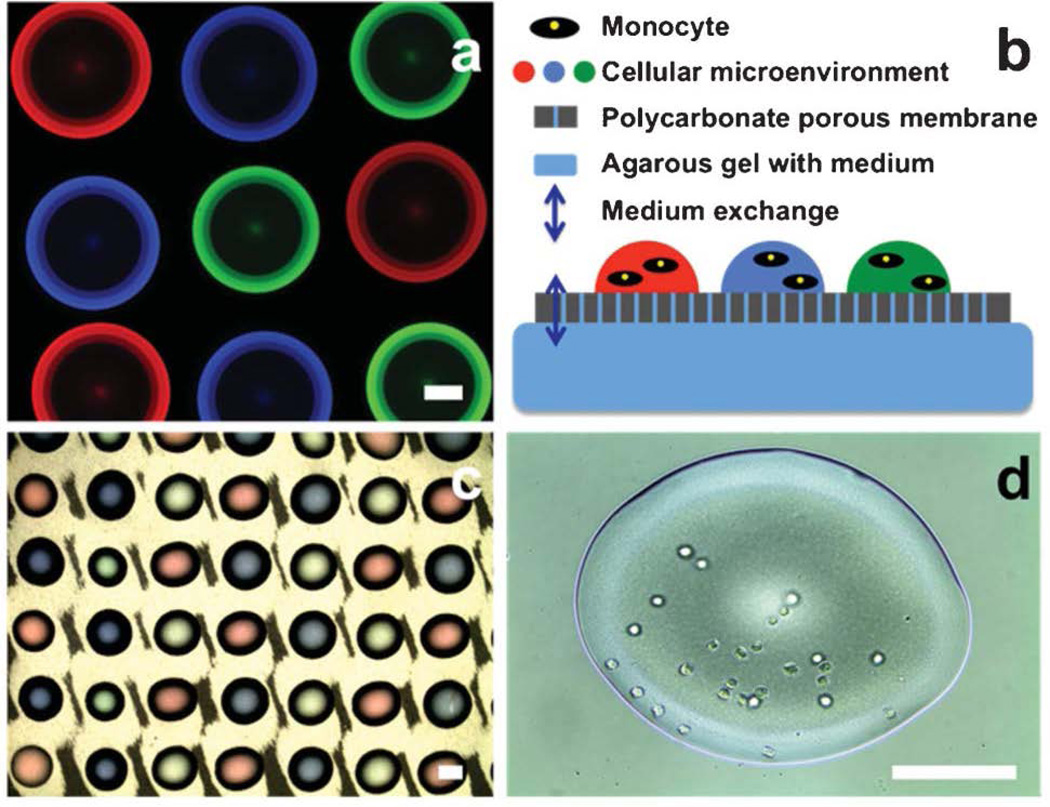
Following the proof-of-concept printing of multiplexed and combinatorial micropatterns using colored aqueous fluids, we extend the MI-printing platform to the biochemical and biological samples. Here, we will use fluorescent protein solutions and cell suspension as the printing inks. In particular, three fluorescent proteins (BSA-FITC, BSA-TR, and streptavidin-MB) are dissolved in a pre-mixed water-DMSO solution (note: DMSO is added to reduce the water evaporation [49]). These ink solutions are loaded into the microfluidic cartridge, respectively. Using the programmable printing stage, a multiplexed protein microarray can be established, as shown in Fig. 7a. Because of the low energy of PDMS substrate and non-contact nature of the printing process, no cross-contamination has been observed among adjacent printing droplets. Facile formation of micropatterned biomolecule arrays can be of extended use for embedding biological cues in cellular microenvironment [57] and defining molecular probes for high-throughput screening [58].
Fig. 7.
(a) Multiplexed fluorescent protein droplet array (scale bar: 0.1 mm); (b) Scheme of cellular printing for long-term cell culture and manipulation; (c) Cellular printing on the polycarbonate porous membrane (scale bar: 0.1 mm); (d) U937 monocytes in the printed droplet (scale bar: 0.1 mm).
Furthermore, we can employ the MI-printing to build a living cell array in a high-throughput manner. Prior to the cellular printing, a sterilization step using 70% ethanol for 20-minute ultrasonication is applied to the microfluidic cartridge [59], followed by sample injection (color dye solutions, monocyte suspensions and agarose gel) into each channels of the sterilized microfluidic cartridge. A biomimic four-color combinatorial array is first created by previous combinatorial patterning protocol. Each colored combination indicates one distinguished complex biological microenvironment. Subsequently, identical droplets of monocyte suspension are printed on each combination spot through unique automatic alignment process, followed by transferring the patterned polycarbonate membrane onto the agarose gel with cell culture medium to form a transwell-like droplet-based cellular array for long-term cell culture and manipulation. [60,61] Fig. 7b illustrates the proposed scheme of the transwell-like droplet-based cellular array with potential use for pharmaceutical cell assays.58,60,62 The nutrition and ionic molecules in the droplets and the agarose gel can be exchanged through the nanoporous polycarbonate membrane (200 nm in diameter of nanopores). Fig.7c and 7d illustrates the cellular printing results on the polycarbonate membrane created by the MI-printer. The high-magnification photo in Fig. 7d shows monocytes presented in the biomimic microenvironment. The number of the monocytes in an individual droplet can be controlled by the printed droplet volume, which can be precisely controlled by the key geometric parameters of the MI-printer, as shown in the section of parameter characterization. Moreover, the MI-printing technique shows no appreciable adverse effect on cell viability during the patterning process due to the biocompatible material (PDMS) and non-contact process of the printing platform. Thus, these cellular printing results clearly demonstrate the potential of the MI-printer platform as a convenient and powerful tool for the investigation in cellular responses to the complex (and combinatorial) biological responses, which can be of potential benefit to medicinal chemistry, drug screening, analysis of cellular processes and intercellular communication, and regeneration of tissues and organs. [21, 36, 58] It is worth noting that DMSO at high concentration (greater than 10 vol%) may potentially cause protein denaturation and cytotoxic effects. [63, 64] As an alternative, various biocompatible hydrogels, such as Matrigel, polyethylene glycol (PEG) hydrogel, and agarose gel, can be adopted as the non-evaporative reagent to the microfluidic impact printing platform to prevent dehydration with minimal bio- or cytotoxicity.
Conclusion
In this paper, we have developed a versatile multiplexed micropatterning platform, referred to as microfluidic impact printer (MI-printer) that combines ink-jet printing and dot-matrix printing for advanced biological patterning applications. Compare to conventional biopatterning process, the MI-printer platform achieves several distinct features, including (a) no cross-contamination enabled by the non-contact nature of the MI printing technique; (b) minute volume manipulation with minimal dead volume achieved by the microfluidic cartridge design; (c) multiplexed printing with self-alignment nature (less than 10µm misalignment); (d) biocompatible printing without any thermal or chemical treatments; (e) patterning availability for complex medium (cell suspensions or colloidal solution); (f) interchangeable/disposable microfluidic cartridge design with simple microfabrication process; (g) high throughput printing enabled by the pin numbers of the dot-matrix printer head (up to 24 pins); (h) easy assembly and configuration of the MI printer using existing dot-matrix printer head, traveling stage, computer and minor-alternated controlling circuits. In summary, the novel MI-printer system offers a simple, low-cost and powerful solution for high-throughput, large-scale, out-of-cleanroom biological patterning, which can be widely employed in drug discovery, biosensing, regenerative medicine, and combinatorial chemistry.
Acknowledgement
This work was supported by the NSF CAREER Award (ECCS-0846502). Authors gratefully acknowledge the assistance from Shaun Garland for the fluorescent imaging, Arnold Chen for the sample holder fabrication, Qing Zhou and Kyungjin Son for the cell culture of U937 monocytes.
References cited
- 1.Aizenberg J, Muller DA, Grazul JL, Hamann DR. Science. 2003;299:1205–1208. doi: 10.1126/science.1079204. [DOI] [PubMed] [Google Scholar]
- 2.Kim C. Science. 2000;288:831–833. doi: 10.1126/science.288.5467.831. [DOI] [PubMed] [Google Scholar]
- 3.Perry H, Gopinath A, Kaplan DL, Dal Negro L, Omenetto FG. Adv. Mater. 2008;20:3070–3072. [Google Scholar]
- 4.LeProust E, Pellois JP, Yu P, Zhang H, Gao X. J. Comb. Chem. 2000;2:349–354. doi: 10.1021/cc000009x. [DOI] [PubMed] [Google Scholar]
- 5.Fukuda J, Khademhosseini A, Yeh J, Eng G, Cheng J, Farokhzad OC, Langer R. Biomaterials. 2006;27:1479–1486. doi: 10.1016/j.biomaterials.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Sarkar S, Lee GY, Wong JY, Desai TA. Biomaterials. 2006;27:4775–4782. doi: 10.1016/j.biomaterials.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 7.Falconnet D, Csucs G, Grandin HM, Textor M. Biomaterials. 2006;27:3044–3063. doi: 10.1016/j.biomaterials.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 8.Xia Y, Whitesides GW. Annu. Rev. Mater. Sci. 1998;28:153–184. [Google Scholar]
- 9.Shi J, Ahmed D, Mao X, Lin SC, Lawit A, Huang TJ. Lab Chip. 2009;9:2890–2895. doi: 10.1039/b910595f. [DOI] [PubMed] [Google Scholar]
- 10.Atsuta K, Noji H, Takeuchi S. Lab Chip. 2004;4:333–336. doi: 10.1039/b400774c. [DOI] [PubMed] [Google Scholar]
- 11.Abhyankar VV, Beebe DJ. Anal. Chem. 2007;79:4066–4073. doi: 10.1021/ac062371p. [DOI] [PubMed] [Google Scholar]
- 12.Csucs G, Michel R, Lussi JW, Textor M, Danuser G. Biomaterials. 2003;24:1713–1720. doi: 10.1016/s0142-9612(02)00568-9. [DOI] [PubMed] [Google Scholar]
- 13.Revzin A, Tompkins RG, Toner M. Langmuir. 2003;19:9855–9862. [Google Scholar]
- 14.Takayama S, Ostuni E, Qian X, McDonald JC, Jiang X, LeDuc P, Wu M-H, Ingber DE, Whitesides GW. Adv. Mater. 2001;13:570–574. [Google Scholar]
- 15.Schwarzenbacher M, Kaltenbrunner M, Brameshuber M, Hesch C, Paster W, Weghuber J, Heise B, Sonnleitner A, Stockinger H, Schutz GJ. Nat. Methods. 2008;5:1053–1060. doi: 10.1038/nmeth.1268. [DOI] [PubMed] [Google Scholar]
- 16.Hwang YS, Chung BG, Ortmann D, Hattori N, Moeller HC, Khademhosseini A. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16978–16983. doi: 10.1073/pnas.0905550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falconnet D, Koenig A, Assi F, Textor M. Adv. Funct. Mater. 2004;14:749–756. [Google Scholar]
- 18.Zhao S, Chen A, Revzin A, Pan T. Lab Chip. 2011;11:224–230. doi: 10.1039/c0lc00275e. [DOI] [PubMed] [Google Scholar]
- 19.Azioune A, Storch M, Bornens M, Thery M, Piel M. Lab Chip. 2009;9:1640–1642. doi: 10.1039/b821581m. [DOI] [PubMed] [Google Scholar]
- 20.Di Carlo D, Wu LY, Lee LP. Lab Chip. 2006;6:1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 21.Raghavan S, Chen CS. Adv. Mater. 2004;16:1303–1313. [Google Scholar]
- 22.Falsey JR, Renil M, Park S, Li S, Lam KS. Bioconjugate Chem. 2001;12:346–353. doi: 10.1021/bc000141q. [DOI] [PubMed] [Google Scholar]
- 23.Khandurina J, Guttman A. Curr. Opin. Chem. Biol. 2002;6:359–366. doi: 10.1016/s1367-5931(02)00323-x. [DOI] [PubMed] [Google Scholar]
- 24.Gosalia DN, Diamond SL. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8721–8726. doi: 10.1073/pnas.1530261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu X, Landry JP, Sun Y-S, Gregg JP, Lam KS, Guo X. Appl. Opt. 2007;46:1890–1895. doi: 10.1364/ao.46.001890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liska R, Schuster M, Inführ R, Turecek C, Fritscher C, Seidl B, Schmidt V, Kuna L, Haase A, Varga F, Lichtenegger H, Stampfl J. J. Coat. Technol. Res. 2007;4:505–510. [Google Scholar]
- 27.Sorribas H, Padeste C, Tiefenauer L. Biomaterials. 2002;23:893–900. doi: 10.1016/s0142-9612(01)00199-5. [DOI] [PubMed] [Google Scholar]
- 28.Lee JY, Shah SS, Zimmer CC, Liu G-Y, Revzin A. Langmuir. 2008;24:2232–2239. doi: 10.1021/la702883d. [DOI] [PubMed] [Google Scholar]
- 29.Yan J, Pedrosa VA, Simonian AL, Revzin A. ACS Appl. Mater. Interfaces. 2010;2:748–755. doi: 10.1021/am9007819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goubko CA, Cao X. Mater. Sci. Eng., C. 2009;29:1855–1868. [Google Scholar]
- 31.Zhao S, Cong H, Pan T. Lab Chip. 2009;9:1128–1132. doi: 10.1039/b817925e. [DOI] [PubMed] [Google Scholar]
- 32.Jacot-Descombes L, Gullo MR, Cadarso VJ, Brugger J. J. Micromech. Microeng. 2012;22:074012. [Google Scholar]
- 33.Sirringhaus H. Science. 2000;290:2123–2126. doi: 10.1126/science.290.5499.2123. [DOI] [PubMed] [Google Scholar]
- 34.Setti L, Fraleoni-Morgera A, Ballarin B, Filippini A, Frascaro D, Piana C. Biosens. Bioelectron. 2005;20:2019–2026. doi: 10.1016/j.bios.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 35.Hossain SMZ, Luckham RE, Smith AM, Lebert JM, Davies LM, Pelton RH, Filipe CDM, Brennan JD. Anal. Chem. 2009;81:5474–5483. doi: 10.1021/ac900660p. [DOI] [PubMed] [Google Scholar]
- 36.Mironov V, Visconti RP, Kasyanov V, Forgacs G, Drake CJ, Markwald RR. Biomaterials. 2009;30:2164–2174. doi: 10.1016/j.biomaterials.2008.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mironov V, Kasyanov V, Markwald RR. Curr. Opin. Biotechnol. 2011;22:667–673. doi: 10.1016/j.copbio.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Guillotin B, Guillemot F. Trends Biotechnol. 2011;29:183–190. doi: 10.1016/j.tibtech.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Goldmann T, Gonzalez JS. J. Biochem. Biophys. Methods. 2000;42:105–110. doi: 10.1016/s0165-022x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 40.Delaney JT, Smith PJ, Schubert US. Soft Matter. 2009;5:4866. [Google Scholar]
- 41.Xu T, Jin J, Gregory C, Hickman JJ, Boland T. Biomaterials. 2005;26:93–99. doi: 10.1016/j.biomaterials.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 42.Handriks F. IBM J. Res. Develop. 1983;27 [Google Scholar]
- 43.Kamphoefner FJ. IEEE Trans. Electron Devices. 1972;19:584–593. [Google Scholar]
- 44.Boglietti A, Chiampi M, Tartaglia M, Cattaneo S, Contessa M, Garramone A. IEEE Trans. Magn. 1989;25:3587–3589. [Google Scholar]
- 45.Pan T, Wang W. Ann. Biomed. Eng. 2010;39:600–620. doi: 10.1007/s10439-010-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klank H, Kutter JP, Geschke O. Lab Chip. 2002;2:242–246. doi: 10.1039/b206409j. [DOI] [PubMed] [Google Scholar]
- 47.Duffy DC, McDonald JC, Schueller OJA, Whitesides GW. Anal. Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 48.Ding Y, Hong L, Nie B, Lam KS, Pan T. Lab Chip. 2011;11:1464–1469. doi: 10.1039/c0lc00710b. [DOI] [PubMed] [Google Scholar]
- 49.Park K, Kim N, Morisette DT, Aluru NR, Bashir R. J. Microelectromech. Syst. 2012;21:702–711. [Google Scholar]
- 50.Man PF, Mastrangelo CH, Burns MA, Burke DT. Micro Electro Mechanical Systems, 1998. MEMS 98; Proceedings., The Eleventh Annual International Workshop on; 1998. pp. 45–50. [Google Scholar]
- 51.Wang W, Zhao S, Pan T. Lab Chip. 2009;9:1133–1137. doi: 10.1039/b816287e. [DOI] [PubMed] [Google Scholar]
- 52.Xu J, Lynch M, Nettikadan S, Mosher C, Vegasandra S, Henderson E. Sens. Actuators, B. 2006;113:1034–1041. [Google Scholar]
- 53.Sekula S, Fuchs J, Weg-Remers S, Nagel P, Schuppler S, Fragala J, Theilacker N, Franzreb M, Wingren C, Ellmark P, Borrebaeck CA, Mirkin CA, Fuchs H, Lenhert S. Small. 2008;4:1785–1793. doi: 10.1002/smll.200800949. [DOI] [PubMed] [Google Scholar]
- 54.Zheng Z, Daniel WL, Giam LR, Huo F, Senesi AJ, Zheng G, Mirkin CA. Angew. Chem., Int. Ed. 2009;48:7626–7629. doi: 10.1002/anie.200902649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serwer P. Electrophoresis. 1983;4:375–382. [Google Scholar]
- 56.Blossey R. Nat. Mater. 2003;2:301–306. doi: 10.1038/nmat856. [DOI] [PubMed] [Google Scholar]
- 57.Folch A, Toner M. Annu. Rev. Biomed. Eng. 2000;02:227–256. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 58.Lam KS, Renil M. Curr. Opin. Chem. Biol. 2002;6:353–358. doi: 10.1016/s1367-5931(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Bindra DS, Barrau M-B, Wilson GS. Biosens. Bioelectron. 1991;6:653–661. doi: 10.1016/0956-5663(91)87018-7. [DOI] [PubMed] [Google Scholar]
- 60.Salmon SE, Liu-Stevens RH, Zhao Y, Lebl M, Krchnak V, Wertman K, Sepetov N, Lam KS. Mol. Diversity. 1996;2:57–63. doi: 10.1007/BF01718701. [DOI] [PubMed] [Google Scholar]
- 61.Mann BK, Gobin AS, Tsai AT, Schmedlen RH, West JL. Biomaterials. 2001;22:3045–3051. doi: 10.1016/s0142-9612(01)00051-5. [DOI] [PubMed] [Google Scholar]
- 62.Townsend JB, Shaheen F, Liu R, Lam KS. J. Comb. Chem. 2010;12:700–712. doi: 10.1021/cc100083f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tjernberg A, Markova N, Griffiths WJ, Hallen D. J. Biomol. Screening. 2005;11:131–137. doi: 10.1177/1087057105284218. [DOI] [PubMed] [Google Scholar]
- 64.Violante GD, Zerrouk N, Richard I, Provot G, Chaumeil JC, Arnaud P. Biol. Pharm. Bull. 2002;25:1600–1603. doi: 10.1248/bpb.25.1600. [DOI] [PubMed] [Google Scholar]
- 65.Kleinman HK, Martin GR. Semin. Cancer Biol. 2005;15:378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 66.Burdick JA, Anseth KS. Biomaterials. 2002;23:4315–4323. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]