Abstract
Human plasma contains proteins that reflect overall health and represents a rich source of proteins for identifying and understanding disease pathophysiology. However, few studies have investigated changes in plasma phosphoproteins. In addition, little is known about the normal variations in these phosphoproteins, especially with respect to specific sites of modification. To address these questions, we evaluated variability in plasma protein phosphorylation in healthy individuals using multiple reaction monitoring (MRM) and SWATH MS2 data-independent acquisition. First, we developed a discovery workflow for phosphopeptide enrichment from plasma and identified targets for MRM assays. Next, we analyzed plasma from healthy donors using an analytical workflow consisting of MRM and SWATH MS2 that targeted phosphopeptides from 58 and 68 phosphoproteins, respectively. These two methods produced similar results showing low variability in 13 phosphosites from 10 phosphoproteins (CVinter <30%) and high interpersonal variation of 16 phosphosites from 14 phosphoproteins (CVinter >30%). Moreover, these phosphopeptides originate from phosphoproteins involved in cellular processes governing homeostasis, immune response, cell-extracellular matrix interactions, lipid and sugar metabolism, and cell signaling. This limited assessment of technical and biological variability in phosphopeptides generated from plasma phosphoproteins among healthy volunteers constitutes a reference for future studies that target protein phosphorylation as biomarkers.
Keywords: Biomarkers, Cancer, Mass Spectrometry, Phosphorylation
1 Introduction
Protein phosphorylation is a dynamic and reversible posttranslational modification known to regulate cellular signaling pathways that control multiple biological processes. Significant changes in protein phosphorylation occur in many disease states, including cancer [1]. Some cancer-related phosphoproteins enter the blood-stream where they could potentially serve as biomarkers for disease detection or in monitoring the efficacy of therapeutic strategies [2, 3]. Similarly phosphorylation status of native plasma proteins may reflect the disease status of the organism during systemic responses like inflammation [4]. Biological fluids, including blood, are the preferred and most accessible samples for disease diagnosis and monitoring as they reflect processes in distant parts of the body and are relatively easily available. Plasma phosphoproteome has been a largely overlooked source of potential biomarkers with only limited studies performed [5-8]. Our recent work has shown potential for use of plasma phosphopeptides as biomarkers of breast cancer differentiating subtypes of this disease [3]. However, despite growing numbers of plasma protein phosphorylation sites discovered, nothing is known about biological variability in these sites among healthy human population, let alone cancer patients. Moreover, most plasma and serum collection protocols are not designed to stabilize phosphoproteins against phosphatase activity and therefore we sought to implement plasma collection protocol incorporating the addition of phosphatase inhibitors early after blood draw.
Mass spectrometry (MS)-based proteomics combined with selective phosphopeptide enrichment strategies is a powerful technique for large scale comprehensive characterization of the phosphoproteome [3]. Additionally, quantitative MS approaches, namely multiple reaction monitoring (MRM), are increasingly used for biomarker verification and validation in clinical samples due to their reproducibility and sensitivity [9]. However, MRM in its most quantitative version requires synthetic stable isotope-labeled peptides for normalization and endogenous peptide verification. MRM assays also require triple quadrupole instrumentation with significant upfront development efforts and are further constrained by the number of target peptides (typically less than <150) that can be measured in a single analysis. Another recently developed targeting MS approach alternative to MRM, SWATH-MS2, is based on a data-independent acquisition (DIA) mass spectrometric method [10]. In this approach, data is typically acquired on a fast high resolution QqTOF instrument by repeated cycling through sequential isolation m/z windows over the entire chromatographic separation. In contrast to MRM, SWATH M2S acquisitions record the fragment ion spectra of all analytes detectable in a sample, which are then used for identification and quantification using a targeted data extraction method. These data can be mined post-acquisition and the method does not require extensive development. Overall, SWATH MS2 has strengths of shotgun proteomics to detect large numbers of analytes and also produce accurate quantification comparable with MRM reproducibility and sensitivity as recently shown for N-linked glycoproteins in plasma [11] and other PTMs of biomarkers [12].
In this study, we developed an optimized and robust discovery workflow to first identify phosphopeptides in plasma, and second, an analytical workflow that used much smaller plasma volumes to assess variations among a target set of phosphopeptides among healthy individuals using MRM-MS assays. Subsequently, an independent set of SWATH-MS2 acquisitions was obtained to assess the sensitivity and quantitative capabilities of this newer method in comparison with the gold standard MRM-MS protocol. The two methods produced similar, but complementary, results with overlap of over 40 phosphopeptides which showed reproducible quantitation by both MRM and SWATH MS2. Biological variability of phosphopeptides from numerous proteins that have known roles in cellular processes and signaling pathways altered in cancer were also assessed. These independent analyses showed that while there was a relatively high level of biological variation among these targeted phosphopeptides in healthy individuals, a significant number of phosphopeptides showed low biological interpersonal CVs (<30%). To our knowledge, this is the first effort to quantify relative differences in plasma protein phosphorylation by MRM-MS and SWATH-MS2 in healthy human subjects.
2 Materials and methods
2.1 Plasma preparation and immunodepletion
Human blood samples from healthy volunteers (18-50 years; 4 males, 6 females) were collected at the University of California San Francisco according to the CPTAC blood collection protocol (http://wikisites.mcgill.ca/djgroup/images/9/9f/CPTAC_plasma_protocol_20080110.pdf) after informed written consent was obtained. The protocol was approved by the UCSF Human Research Protection Program Committee on Human Research (IRB #10-03275) and the Buck Institute BUA B1022.
Within 30 min of collection, the blood was centrifuged twice at 4 °C (1,500 × g and 2,000 × g for 15 min each) and phosphatase and protease inhibitors (PhosSTOP and Complete Mini EDTA-free protease inhibitor cocktail tablets, Roche) were added to the plasma. The aliquots of plasma were stored at −80 °C until further processing [3]. The fourteen most abundant plasma proteins were immunodepleted according to the manufacturer's instructions using a Multiple Affinity Removal System (MARS) Human-14 column (10 × 100 mm) from Agilent Technologies on a Waters 1525 HPLC system. The depleted proteins constituted about 92% of the total plasma protein and the protein concentration of the depleted fraction was between 4-5 mg/ml. The protein flow-through fractions were collected and re-adjusted to the original volume of 200 μL per injection using 5K MWCO centrifugal concentrators (Agilent Technologies). Henceforth, an immunodepleted plasma volume refers to the original plasma volume (plasma equivalents, PE).
2.2 Trypsin digestion of plasma proteins
The MARS Hu-14 depleted plasma (1 mL PE) were denatured with 6 M urea, reduced with 20 mM DTT (30 min at 37 °C), alkylated with 50 mM iodoacetamide (30 min at RT), and digested overnight at 37°C with 1:50 enzyme:substrate ratio (wt/wt) of sequencing grade trypsin (Promega, Madison, WI) as previously described [13]. Following digestion, the samples were acidified with formic acid, and desalted using HLB Oasis SPE cartridges (Waters, Milford, MA), and concentrated by vacuum centrifugation. Peptides were stored at −80°C until use.
2.3 Stable isotope-labeled heavy phosphopeptides as internal standards
Pure, stable isotope-labeled synthetic peptides, labeled at the C-terminus with 13C6, 15N4-Arg (R) of phosphorylated peptides were synthesized by Thermo as AQUA >97% purity peptides (10 nmol): AAIpSGENAGLVR from ITIH1, ISApSAEELR from APOA4, LPTDpSELAPR from SEPP1.
2.4 Strong Cation-Exchange (SCX) and hydrophilic interaction liquid chromatography (HILIC) fractionation and TiO2 and IMAC enrichment of phosphopeptides
SCX and HILIC peptide fractionation was performed on a Waters 1525 HPLC system equipped with a 9.4 × 250 mm Polysulfoethyl A 5 μm column (PolyLC) and 4.6 × 250 mm TSKgel Amide-80 HR 5 μm particle column (Tosoh Bioscience), respectively [14, 15]. For SCX the plasma samples (1000 μL of plasma equivalents) were loaded in 100 % solvent A (7 mM KH2PO4 pH 2.7, 30 % acetonitrile) and eluted with the following gradient: 0 % B for 2 min followed by 0 % B to 25 % B in 33 min, and then 100% B in 1 min for 15 min at 3 mL min−1. Solvent B consisted of 7 mM KH2PO4 pH 2.7, 350 mM KCl, and 30% acetonitrile. For HILIC the plasma samples (500 μL of plasma equivalents) were loaded in 80% solvent B (98% acetonitrile, 0.1% TFA) and eluted with the following gradient: 80 % B for 5 min followed by 80 % B to 60 % B in 40 min, and then 0% B in 15 min at 0.5 mL min−1. Solvent A consisted of 98% HPLC grade water (Honeywell) and 0.1% TFA. Ten SCX fractions were collected and desalted using Sep-Pak 500 mg tC18 cartridges (Waters). Twelve HILIC fractions were collected and each enriched for phosphopeptides after reducing their volume to 50 μL using a SpeedVac concentrator (Savant, Thermo Scientific). Phosphopeptides were enriched using titanium dioxide (TiO2) chromatography according to the manufacturer's instructions (Titanosphere Phos-TiO kit, 200 μL columns, GL Sciences). For IMAC enrichment 5 mg of Fe-NTA (Sigma-Aldrich) was used and the samples were incubated in 250 mM CH3COOH/30% acetonitrile. The same buffer was used to wash the gel 3 times and phosphopeptides were eluted sequentially with 50 mM K2HPO4/NH4OH pH 10 and 150 mM NH4OH/25% acetonitrile (100 μL each) and acidified immediately with 20% TFA. The samples were desalted using Oasis HLB μElution 96-well plate (Waters). After removal of organic solvent using a SpeedVac concentrator, the phosphopeptide samples were suspended in 0.1% formic acid and subsequently analyzed by LC-MS/MS. For discovery experiments, fractionated plasma samples were pooled from 15 individual donors (Fig. 1). For MRM and SWATH MS2 analyses individual donor plasma (500 μL and 200 μL PE, respectively) was processed and analyzed in duplicate. HILIC fractions from 29-60 min were combined, as they contained >95% of all phosphopeptides, yielding a total of 1 fraction. Heavy phosphopeptide standards were spiked in at 150 fmol each and the samples were enriched using TiO2 (Fig. 3).
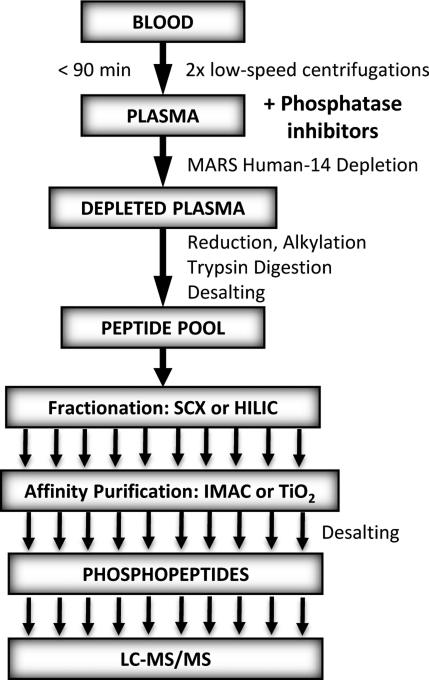
Fig. 1.
The experimental workflow developed for preparation of phosphopeptides from plasma for discovery experiments.
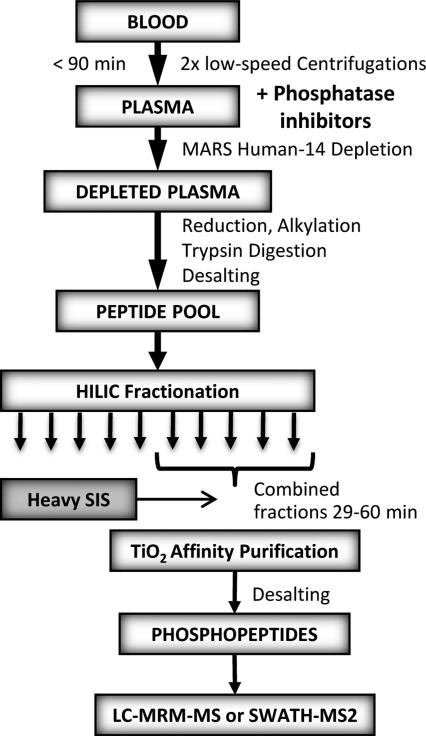
Fig. 3.
The experimental workflow developed for preparation of phosphopeptides from plasma for quantitative mass spectrometric measurements.
2.5.1 Nano-LC-ESI-MS/MS analyses, data dependent acquisitions
The peptide mixtures obtained after tryptic digestion, SCX or HILIC fractionation, TiO2 enrichment, and desalting were analyzed by reversed-phase nano-HPLC-ESI-MS/MS using an Eksigent nano-LC 2D HPLC system (Eksigent, Dublin, CA), which was directly connected to a quadrupole time-of-flight (QqTOF) QSTAR Elite mass spectrometer (AB SCIEX, Concord, CA) in data dependent mode.
2.5.2 Mass spectrometric database searches
Mass spectrometric data were analyzed using two separate bioinformatics database search engine systems, ProteinPilot™ (AB SCIEX) version 4.0.8085 (revision 148085) [16] using the Paragon Algorithm 4.0.0.0, 148083 [17] and an in-house Mascot 2.3 server (Matrix Science) [18] All data were searched using a publicly available human SwissProt UniProt release 2011_05 database of 20,239 protein sequences. For ProteinPilot searches, the following parameters were used: trypsin enzyme specificity, carbamidomethyl (Cys) as a fixed modification, special factors including phosphorylation emphasis and urea denaturation, and thorough search effort setting allowing for biological modifications [17]. For database searches, a ProteinPilot ‘peptide confidence’ cut-off value of 95 was chosen, yielding a peptide level local FDR of <3%. For Mascot searches, the following parameters were used: trypsin enzyme specificity, carbamidomethyl (Cys) as a fixed modification, and the following variable modifications: phosphorylation at Ser, Thr, and Tyr, deamidation of asparagine and glutamine residues, oxidization of methionine, acetylation at the protein N-terminus, cyclization of N-terminal glutamine, and a maximum of three missed tryptic cleavages. For QSTAR Elite data a mass tolerance of 100 ppm (MS1) and 0.4 Da (MS2) was set for the precursor and product ions, respectively. Mascot peptide-spectral matches with significance threshold p < 0.05 were accepted. FDR analysis was performed using the Mascot automatic decoy search. In all cases, the peptide false-positive identification rate was < 3%. Due to the phosphopeptide-centric approach, protein identifications were made based only on the identified phosphopeptides and thus single phosphopeptide identifications were allowed.
2.6.1 MRM transition selection
MS/MS data from the pooled plasma samples obtained during discovery experiments were used to build spectral libraries in Skyline [19]. Initially, two MRM-MS precursor-to-product ion transitions per phosphopeptide were designed in Skyline for all phosphopeptides in the library. The preliminary MRM-MS analysis was performed using the QTRAP 5500 hybrid triple quadrupole/linear ion trap mass spectrometer (AB SCIEX, Foster City, CA) that is capable of high-sensitivity and multiplexed MRM acquistions, and the data was uploaded and analyzed in Skyline. Phosphopeptides with both transitions present and free of coeluting ions were selected for further assay development. The total of 3-4 transitions per each target phosphopeptide were designed and the list was split into 3 separate methods each containing <100 transitions for final sample analysis.
2.6.2 LC-MRM/MS
For selected reaction monitoring (MRM), samples were analyzed by nano-LC-MRM-MS on a QTRAP 5500 mass spectrometer. Chromatography was performed on a NanoLC-Ultra 2D LC system (Eksigent, Dublin, CA) with buffer A (0.1% (vol/vol) formic acid) and buffer B (90% acetonitrile in 0.1% formic acid). Digests were separated on a with a 75 μm inner diameter Integrafrit analytical column (New Objective, Woburn, MA) packed in-house with 10-12 cm of ReproSil-Pur C18-AQ 3 μm reversed phase resin (Dr. Maisch GmbH, Germany) at a flow rate 300 nL/min. Gradient was 3% B from 0-5 min, increased to 7% B over 3 min, increased to 25% B over the next 27 min and increased to 40% B over the next 7 mins. Peptides were ionized using a PicoTip emitter (20 μm, 10 μm tip, New Objective, Woburn, MA). Data acquisition was performed using Analyst 1.5.1 (AB SCIEX) with an ion spray voltage of 2300 V, curtain gas of 20 psi, nebulizer gas of 15psi, and interface heater temperature of 150°C.
The transitions, dwell times, and collision energy are listed in supporting Table 2. Four transitions were assayed per peptide. Values of 100 and 40 were used as the declustering potential and collision cell exit potentials, respectively, for all transitions. MRM transitions were acquired at unit resolution both in the first and third quadrupoles (Q1 and Q3). Samples were processed and analyzed in duplicate with 150 fmol of each heavy phosphopeptide spiked in before TiO2 enrichment and 125 μL PE was injected on the column. Standard curves were performed in duplicate by spiking in the stable isotope-labeled phosphopeptides to determine the linear range in a background matrix of 250 ng of a predigested six protein mix (Michrom). Reproducibility of MRM measurements was tested using 4 replicates of pooled plasma samples and was consistent with previous experience where the QTRAP 5500 was a part of recent multi-site study that assessed system suitability and reproducibilty for MRM-MS analysis [20].
2.6.3 Quantitative MRM Data Analysis
Skyline post-acquisition software was used to process all MRM-MS data [19]. Each transition was individually integrated to generate peak areas and the peak area of the most intense transition ion was used for analysis. All native phosphopeptide data was normalized to the average of the most intense transition ions of 3 heavy phosphopeptide standards (y9-98 from AAIpSGENAGLVR, y8-98 from ISApSAEELR, and y9 from LPTDpSELAPR). If multiple precursor ions were detected for the same phosphopeptide, only the one with the most intense signal was included.
2.7.1 SWATH-MS2
Data acquisitions of 9 plasma samples were performed by reverse-phase nano-HPLC-ESI-MS/MS using an Ultra Plus NanoLC 2D HPLC system (Eksigent, Dublin, CA) which was directly connected to a new generation quadrupole time-of-flight (QqTOF) Triple TOF 5600 mass spectrometer (AB SCIEX, Concord, CAN)_that became available near the end of this study and was capable of SWATH-MS2 acquisitions. Samples were initially re-analyzed in data-dependent mode to obtain MS/MS spectra for the 30 most abundant parent ions following each survey MS1 scan to build spectral libraries. Additional data sets were recorded in data-independent mode using SWATH-MS2 acquisitions. In the SWATH-MS2 acquisition, instead of the Q1 quadrupole transmitting a narrow mass range through to the collision cell, a wider window of ~25 Da is passed in incremental steps over the full mass range from 400-1000. To increase overall efficiencies, two injection replicate SWATH MS2 experiments were performed per sample. The amount of sample injected on the column equaled to 55 μL PE. Additional details describing mass spectrometric instrument parameters and settings and all chromatographic setups and gradient conditions are found in supplemental Methods S1. Lastly, it should be noted that the high reproducibility of the TripleTOF 5600 was assessed in previous studies using new algorithms that examined instrument stability with statistical metrics [21] and where the MS1 signal intensities were examined for repeatability and reproducibility over extended time period [22, 23].
2.7.2 Quantitative SWATH MS2 Data Analysis
Data sets from SWATH-MS2 acquisitions were processed using the full scan MS/MS filtering module for data-independent acquisition within Skyline. The top 8 fragment ions were extracted from SWATH-MS2 acquisitions within Skyline using a fragment ion resolution setting of 10,000. The peak area of the most intense fragment ion was used for quantitative analysis. All native phosphopeptide data was normalized to the average of the most intense transition ions of 2 spiked heavy phosphopeptide standards (y8-98 from ISApSAEELR and y9 from LPTDpSELAPR).
2.8 Statistical analysis
To assess the sample processing and instrument reproducibility, the coefficient of variation (CV) between two process replicates per sample (MRM-MS) or two process replicates with two technical replicates each per sample (SWATH-MS2) were determined for each fragment ion. Phosphopeptides were considered as reproducibly quantifiable only when process replicate CV was <30% for at least 4 out of 6 samples analyzed by MRM in duplicate and at least 6 out of 9 samples analyzed by SWATH MS2. Next, CVinter for all 8 MRM-MS and 9 SWATH-MS2 sample acquisitions was calculated to assess biological variation of phosphopeptides within the human population. Interpersonal variation was considered low when CVinter<30% and high when CVinter>30%.
2.9 Data Accession
All raw data associated with this manuscript may be downloaded from the Buck Institute ftp site at ftp://sftp.buckinstitute.org:251. All confidently identified phosphorylated peptides were transferred to the data-sharing Panorama server [24], allowing for interactive web-based spectral viewing of all PTM-containing peptides in this study (at 95 % confidence). The spectral viewer can be accessed at http://proteome.gs.washington.edu/software/panorama/PlasmaPhosphoproteome.html
3 Results and Discussion
3.1 Workflow development and discovery analysis of phosphopeptides from plasma
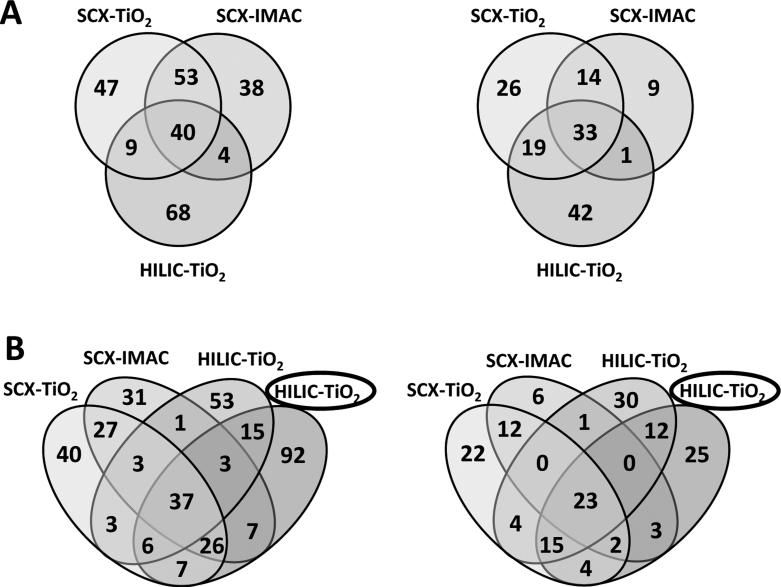
In preparation for MRM-MS assay development, a discovery workflow for phosphopeptide enrichment and subsequent data-dependent analysis (DDA) from plasma was developed (Fig. 1). Pooled plasma samples were subjected to this workflow using larger volumes than we would subsequently employ for our analytical workflow, allowing for a more in depth analysis of the plasma phopshoproteome while building spectral libraries that would be needed later. In this discovery workflow, a previous reported CPTAC blood collection protocol was modified so that phosphatase and protease inhibitors could be added at a very early stage after initial blood draw of the platelet-depleted plasma to preserve the phosphorylation status of the plasma proteome and limit contamination from platelets and other cell types [3]. Since the analysis and quantification of plasma and serum proteins are challenging due to the complexity and large dynamic range of the plasma proteome, a combination of analytical approaches was employed. After immunodepletion of the 14 most abundant proteins and trypsin digestion, the peptides were fractionated by off-line SCX or HILIC chromatography and enriched for phosphoproteins by TiO2 and IMAC. The LC-MS/MS analysis on the QSTAR Elite of 1 mL of plasma equivalents (PE) allowed for identification of over 250 unique phosphorylation sites with 95% confidence on 134 phosphoproteins (Fig. 2A and supporting Table 1). Because MARS Hu-14 immunodepletion removes >97% of the 14 highest abundance proteins, phosphopeptides originating from residual amounts of these proteins were not considered in our data set (APOA1, APOA2, CO3, FIBA). Interestingly, only 78 of over 250 unique phosphosites we identified had been previously reported [5, 6]. However, a more recent study by Jaros et al. [7] reported >500 phosphoproteins in human plasma, of which 58 are common with our discovery results. The analysis of plasma samples prepared using three combinations of fractionation and enrichment methods gave complementary sets of phosphosites with the most identified following SCX fractionation. However, the highest number of phosphoproteins was identified from plasma prepared by the HILIC-TiO2 workflow. Given that HILIC chromatography, unlike SCX fractionation, does not require subsequent desalting, we choose HILIC for our fractionation method in our analytical quantitative assays workflow for MRM-MS and SWATH-MS2 as described below (Fig. 3).
Fig. 2.
Numbers of unique phosphosites (left) and phosphoproteins (right) identified in healthy human plasma A) after fractionation and phosphopeptide enrichment of 1 ml of plasma using SCX-TiO2, SCX-IMAC, and HILIC-TiO2. B) Comparison of the numbers of phosphosite (left) and phosphoprotein (right) identifications obtained during discovery experiments with the results obtained for 200 μl of plasma processed with HILIC-TiO2 and analyzed by data-dependent acquisitions on TripleTOF 5600 (encircled).
3.2 MRM-MS analysis
All phosphopeptides identified in our discovery workflow were then used to build spectral libraries in Skyline [19] for subsequent MRM-MS assay development. A set of preliminary experiments was employed to develop an analytical workflow that was better optimized for sample preparation using smaller plasma volumes that would be more typically available for biomarker studies. The final sample preparation protocol included HILIC fractionation of 500 μL PE followed by TiO2 enrichment (Fig. 3). Heavy phosphopeptide standards were spiked-in at 150 fmol per injection prior to TiO2 enrichment to partially control for technical variation in the phosphopeptide-specific enrichment steps. MRM transitions were refined only for phosphopeptides that were detectable and final methods included 3-4 MRMMS transitions per peptide, with a total of ~500 transitions. MRM data were imported into Skyline and the most intense transitions were used for quantification. Peak areas were normalized to the mean of the 3 heavy peptide most intense transitions. To test sample processing and instrument reproducibility, a set of 4 individually prepared replicate pooled plasma samples was analyzed and compared in Skyline. The coefficients of variation (CV) for all spiked-in heavy peptide transitions was <20 % indicating very good sample processing and instrument reproducibility (Supporting Fig. 1).
Using this analytical workflow, six plasma samples were then processed in duplicate and two with one replicate each, for a total of 8 samples. To select reproducible data for native phosphopeptides, CVs for each sample process replicate were calculated and only phosphopeptides with CVs less than 30% were considered for further study. The MRM analysis yielded quantitative data for 98 unique phosphopeptide sequences corresponding to 90 phosphosites from 58 phosphoproteins (Supporting Table 3).
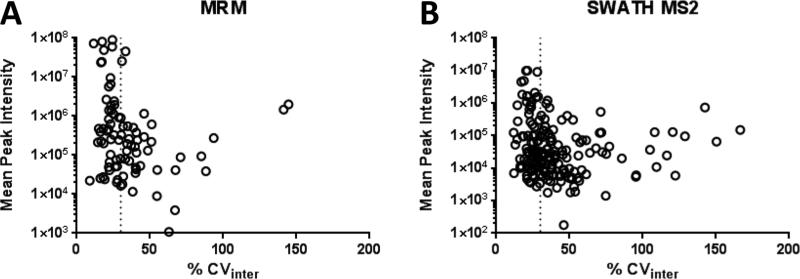
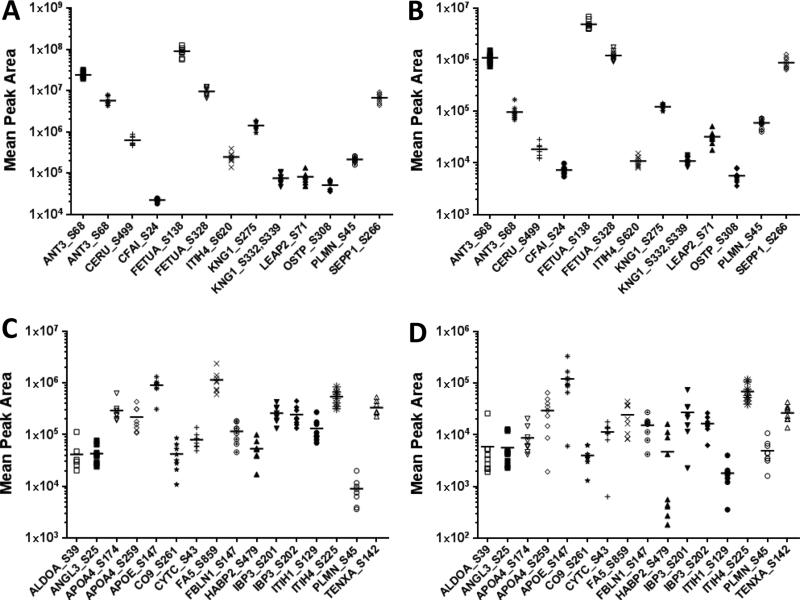
Next, the biological variation in these phosphopeptides levels among the 8 individuals was assessed based on calculating the CVinter. About half of the phosphosites showed relatively low interpersonal variation below CVinter of 30% (Fig. 4A). The most abundant phosphopeptides examined were derived from alpha-2-HS-glycoprotein (FETUA), kininogen 1 (KNG1), antithrombin-III (ANT3), and secreted phosphoprotein 1/osteopontin (OSTP). Phosphopetides from 7 out of 11 phosphoprotein members of the complement and coagulation cascades quantified by MRM showed low biological variation. The other four, including a phosphopeptide from coagulation factor FA5, for which its concentration is known to increase with age [25] had a relatively high biological variation. Phosphopeptides from proteins with function in focal adhesion and ECM-receptor interactions, including integrin 5 (ITA5), osteopontin (OSTP), tenaxin (TENXA), vitronectin (VTNC) showed low biological variation, whereas pS1454 from filamin A (FLNA) had higher variation. Phosphopeptides from protease inhibitor inter-alpha globulin inhibitor H4 (ITHI4), which is involved in acute inflammatory response showed low variation, while phosphopeptides from ITHI1 and ITHI2 were considerably higher. Other phosphosites with high biological variation were derived from lipid carrier proteins apolipoproteins APOL1 and APOA4 and IGF-binding proteins 3 and 5 (IBP3 and IBP5) (Supporting Table 3 and Fig. 5A, C).
Fig. 4.
The dependence of biological variability (CVinter) on the abundance of total peak areas of transition ions of all plasma donor samples in A) MRM and B) SWATH-MS2 measurements for 86 phosphopeptides in MRM and 187 in SWATH-MS2 analysis. Vertical dotted line represents CVinter=30%.
Fig 5.
Mean peak area of the selected product ions of phosphopeptides in plasma from healthy donors analyzed by A) MRM and B) SWATH MS2 with low biological variation (CVinter<30%), and by C) MRM and D) SWATH MS2 with high biological variation (CVinter>30%) consistent between the two methods. The lines represent averages of all individual plasma samples. Corresponding phosphopeptide sequences and transition ions used for quantitation are described in detail in Table 1.
As little is known about absolute concentrations of native phosphopeptides, we used a SID MRM-MS approach to target 2 phosphopeptides derived from APOA4 and ITIH1 for which heavy standard peptides were available. Average concentration of ISApSAEELR phosphopeptide was 226 nmol/L of plasma and 203 nmol/L for AAIpSGENAGLVR (Supplemental Fig. S2). The reported concentration of these proteins in plasma is 3-6 and 2-4 μmol/L, respectively [26]. Therefore, an estimated level of phosphorylation at these two phosphosites (S259 and S129) ranged from 4-7 %.
3.3 Data-dependent analysis prior to SWATH-MS2
The initial DDA analysis using the TripleTOF 5600 mass spectrometer that was used to build the spectral libraries consumed less than half of the plasma volume than what was used for MRM-MS analysis, yet still allowed for confident identification of 193 phosphosites from 84 phosphoproteins (Supporting Table 4). When comparing discovery data obtained from the QStar Elite mass spectrometer, these numbers are higher than obtained using fractionated samples (Fig. 2B). Significantly, an additional 92 phosphosites from 25 phosphoproteins were identified that were not detected in original QSTAR Elite discovery experiments (Fig. 2B). This dramatic improvement can be attributed to increased sensitivity and scanning efficiency in this next generation TripleTOF 5600 instrument.
3.4 SWATH MS2 analysis
SWATH-MS2 analysis was performed in attempt to minimize sample volume and assay development time and maximize numbers of quantifiable phosphopeptides. Data independent SWATH-MS2 acquisitions were obtained in duplicate for 9 plasma samples prepared as two process replicates each. SWATH-MS2 data were analyzed in Skyline and the most intense transition ions were used for quantification. Peak areas were normalized to the mean of the most intense transitions of 2 spiked-in heavy peptides. To select reproducible data, CVs for native phosphopeptides of each sample process replicate were calculated and only phosphopeptides with process replicate CVs of less than 30% were considered. The SWATH MS2 analysis yielded 178 unique phosphopeptide sequences corresponding to 139 phosphosites from 68 phosphoproteins (Supporting Table 4).
Interpersonal variability was evaluated based on calculated CVinter for all 9 plasma donor samples. Similar to the MRM-MS assay results, about half of the phosphopeptides had low biological variation with CVinter < 30% (Fig. 4B). The most abundant phosphopeptides were from alpha-2-HS-glycoprotein (FETUA), kininogen 1 (KNG1), antithrombin-III (ANT3), and fibronectin (FINC). Most of the phosphopeptides from 14 phosphoprotein components of the complement and coagulation pathways that were quantified by SWATH-MS2 had low variability, while phosphopeptides from 5 phosphoproteins (CBPB2, CFAH, CO9, F13B, and FA5) showed high biological variation (Supplemental Table 4). Also most of the phosphopeptides representing proteins involved in focal adhesion and ECM-receptor interactions had low biological variability with the exception for CD44 and collagen alpha 1(IV), and a doubly phosphorylated phosphopeptide FRIpSHELDSASpSEVN from osteopontin, not included in MRM-MS measurements.
Phosphopeptides from additional 29 phosphoproteins not included in the MRM assays were quantified by SWATH-MS2. Some interesting examples of phosphopeptides with high biological variation that may be linked to various disease states included IGF-1, known to be connected with diabetes-associated cancers [27], CD44 receptor for extracellular proteins involved in cell migration and tumor growth and progression [28], vitamin D-binding protein (VTDB) known to affect inflammation and cell proliferation in cancer and cardiovascular disease [29], , adenyl-cyclase associated protein 1 (CAP1) implicated in cell motility and tumor invasiveness [30], and SPARK-like protein 1 (SPRL1) with tumor suppressor function [31]. Interestingly, in addition to the highly variable pS92 on SPRL1, another phosphosite pS295 on SPRL1 had lower biological variability (Supporting Table 4).
3.5 Comparison between MRM and SWATH MS2 quantification
Phosphopeptides quantified with high reproducibility by both MRM-MS and SWATH-MS2 were compared (Table 1). Thirteen phosphosites from 10 proteins showed low interpersonal variation (CVinter<30%) (Fig. 5A-B) and 16 phosphosites from 14 proteins revealed high biological variation ((CVinter>30%) (Fig. 5C-D) in both methods. However, 14 phosphosites from 12 proteins had noticeably different CVinter values as measured by MRM-MS and SWATH-MS2. These larger discrepancies were primarily linked to phosphopeptides that contained multiple phosphorylation sites that were prone to missed cleavage by trypsin, including phosphopeptides from CERU, IBP3, IBP5, KNG1, and ZPI (Table 1). In some cases phosphosites represented by several phosphopeptide forms, for example pS45 from plasminogen (PLMN), varied in their % CVinter which appeared to be reflective of trypsin cleavage efficiency. Considering variability of the most abundant phosphopeptide form, this phosphosite can be regarded as having low biological variation (Fig. 5). Lastly, phosphopeptides from 19 proteins were quantified only by MRM-MS and from 29 proteins only by SWATH-MS2 (Supporting Tables 3 and 4).
Table 1.
Quantitative results of MRM and SWATH-MS2 assays for phosphopeptides in human plasma.
| Protein Name |
Acc. No. | Modified Sequence | Phospho Sites |
Ascore (%) |
Transition | Precursor m/z |
Product m/z |
Prec. Charge |
Product Charge |
Mean ALL | SD ALL | CVinter (%) |
Mean ALL | SD ALL | CVinter (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANT3 | P01008 | ATEDEGpSEQKIPEATNRR | S68 | 100 | y15 | 704.32 | 856.42 | 3 | 2 | 5685867 | 1241637 | 21.8 | 96919 | 28883 | 29.8 |
| ANT3 | P01008 | KATEDEGpSEQKIPEATNR | S68 | 100 | y6 | 694.98 | 687.34 | 3 | 1 | 23877574 | 3929423 | 16.5 | 1097017 | 227005 | 20.7 |
| CERU | P00450 | NNEGTYYSPNYNPQpSR | S499 | 100 | y4 | 661.93 | 469.25 | 3 | 1 | 626291 | 144126 | 23.0 | 18404 | 4935 | 26.8 |
| CFAI | P05156 | VTYTpSQEDLVEK | S24 | 100 | y9 | 746.33 | 1030.51 | 2 | 1 | 22365 | 1965 | 8.8 | 7299 | 1161 | 15.9 |
| FETUA | P02765 | CDSSPDpSAEDVR | S138 | 100 | y8 | 709.25 | 870.40 | 2 | 1 | 90313746 | 21888971 | 24.2 | 4895471 | 924230 | 18.9 |
| FETUA | P02765 | HTFoxMGVVSLGpSPSGEVSHPR | S328 | y10 | 726.33 | 1132.48 | 3 | 1 | 9542050 | 2193977 | 23.0 | 1207999 | 263532 | 21.8 | |
| ITIH4 | Q14624 | NVHpSGSTFFK | S620 | y8 | 602.26 | 495.71 | 2 | 2 | 245101 | 70531 | 28.8 | 10867 | 2290 | 21.1 | |
| KNG1 | P01042 | DIPTNpSPELEETLTHTITK | S275 | 100 | y17 | 740.35 | 995.97 | 3 | 2 | 1420609 | 311680 | 21.9 | 123281 | 14637 | 11.9 |
| KNG1 | P01042 | ETTCSKEpSNEELTEpSCETKK | S332,S339 | y8 | 850.66 | 964.44 | 3 | 1 | 75127 | 20199 | 26.9 | 10917 | 1851 | 17.0 | |
| LEAP2 | Q969E1 | RCpSLSVAQE | S71 | 100 | b7 | 565.24 | 854.36 | 2 | 1 | 81320 | 24228 | 29.8 | 32159 | 8795 | 27.3 |
| OSTP | P10451 | ISHELDpSASSEVN/ISHELDSApSSEVN | S308 | 98 | b8 | 734.30 | 835.39 | 2 | 1 | 51070 | 13382 | 26.2 | 5620 | 1393 | 24.8 |
| PLMN | P00747 | KQLGAGpSIEECAAK | S45 | 100 | y5 | 514.57 | 578.26 | 3 | 1 | 213933 | 30720 | 14.4 | 59995 | 10828 | 18.0 |
| SEPP1 | P49908 | DoxMPApSEDLQDLQK | S266 | 100 | y11 | 793.33 | 662.30 | 2 | 2 | 6670889 | 1546702 | 23.2 | 881271 | 205365 | 23.3 |
| ALDOA | P04075 | GILAADESTGpSIAK | S39 | 100 | y9 | 706.84 | 987.40 | 2 | 1 | 41608 | 28035 | 67.4 | 5895 | 7210 | 122.3 |
| ANGL3 | Q9Y5C1 | IDQDNSpSFDSLSPEPK | S25 | 99 | y4 | 929.89 | 470.26 | 2 | 1 | 43089 | 17460 | 40.5 | 5642 | 3692 | 65.4 |
| APOA4 | P06727 | ENADpSLQASLRPHADELK | S174 | 100 | y15 | 691.99 | 831.43 | 3 | 2 | 294168 | 135256 | 46.0 | 8755 | 5105 | 58.3 |
| APOA4 | P06727 | ISApSAEELR | S259 | 100 | y8 | 528.24 | 844.42 | 2 | 1 | 219702 | 112030 | 51.0 | 29841 | 19085 | 64.0 |
| APOE | P02649 | GEVQAMLGQpSTEELR | S147 | 100 | y8 | 864.39 | 999.41 | 2 | 1 | 907851 | 272929 | 30.1 | 121904 | 86126 | 70.7 |
| CO9 | P02748 | AEQCCEETASpSISLHGK | S261 | y8 | 662.94 | 810.45 | 3 | 1 | 42002 | 23074 | 54.9 | 3978 | 1470 | 37.0 | |
| CYTC | P01034 | LVGGPoxMDApSVEEEGVRR | S43 | 100 | y15 | 632.96 | 793.87 | 3 | 2 | 80008 | 26561 | 33.2 | 11423 | 4639 | 40.6 |
| FA5 | P12259 | LLSLGAGEFKpSQEHAK | S859 | 100 | y14 | 598.96 | 735.87 | 3 | 2 | 1158214 | 532591 | 46.0 | 24453 | 13703 | 56.0 |
| FBLN1 | P23142 | pSQETGDLDVGGLQETDK | S147 | 100 | y8 | 936.40 | 847.42 | 2 | 1 | 116382 | 47351 | 40.7 | 15316 | 6365 | 41.6 |
| HABP2 | Q14520 | LIANTLCNpSR | S479 | 100 | y8 | 621.29 | 1015.40 | 2 | 1 | 53525 | 23249 | 43.4 | 4718 | 5664 | 120.1 |
| IBP3 | P17936 | VDYESQSTDTQNFpSSESKR | S201 | 97 | y15 | 763.32 | 842.38 | 3 | 2 | 261577 | 93275 | 35.7 | 27365 | 20553 | 75.1 |
| IBP3 | P17936 | YKVDYESQSTDTQNFSpSESKR | S202 | y7 | 860.37 | 822.41 | 3 | 1 | 245031 | 101355 | 41.4 | 16588 | 5277 | 31.8 | |
| ITIH1 | P19827 | AAIpSGENAGLVR | S129 | 100 | y9 | 619.30 | 982.44 | 2 | 1 | 131817 | 63980 | 48.5 | 1812 | 916 | 50.6 |
| ITIH4 | Q14624 | pSPEQQETVLDGNLIIR | S225 | 100 | y7 | 631.31 | 800.46 | 3 | 1 | 550666 | 173907 | 31.6 | 69041 | 25343 | 36.7 |
| PLMN | P00747 | QLGAGpSIEECAAKCEEDEEFTCR | S45 | 100 | y19 | 923.70 | 1151.46 | 3 | 2 | 8937 | 4886 | 54.7 | 4925 | 2545 | 51.7 |
| TENXA | Q16473 | LGPLSAEGTTGLAPAGQTSEEpSRPR | S142 | y18 | 854.74 | 947.94 | 3 | 2 | 334359 | 101094 | 30.2 | 26656 | 8903 | 33.4 | |
| CBPB2 | Q96IY4 | SKDHEELSLVApSEAVR | S338 | 100 | y6 | 617.29 | 614.33 | 3 | 1 | 930831 | 265946 | 28.6 | 85372 | 31925 | 37.4 |
| CERU | P00450 | QSEDpSTFYLGER | S725 | 99 | y4 | 756.31 | 474.27 | 2 | 1 | 419925 | 86006 | 20.5 | 13412 | 4121 | 30.7 |
| CERU | P00450 | RQpSEDSTFYLGER/RQSEDpSTFYLGER | S722 | 100 | y4 | 556.57 | 474.27 | 3 | 1 | 194139 | 66831 | 34.4 | 28447 | 8274 | 29.1 |
| CLUS | P10909 | VTTVASHTSDpSDVPSGVTEVVVK | S396 | 100 | y10 | 798.72 | 507.80 | 3 | 2 | 2425944 | 617629 | 25.5 | 155873 | 52970 | 34.0 |
| F13B | P05160 | SGYLLHGpSNEITCNR | S373 | 100 | y4 | 600.93 | 550.24 | 3 | 1 | 1142968 | 285790 | 25.0 | 100126 | 36162 | 36.1 |
| FETUB | Q9UGM5 | GSVQYLPDLDDKNpSQEK | S315 | 100 | y5 | 672.64 | 685.26 | 3 | 1 | 383067 | 155362 | 40.6 | 26081 | 4340 | 16.6 |
| HEP2 | P05546 | GGETAQpSADPQWEQLNNK | S37 | 97 | y7 | 684.96 | 931.46 | 3 | 1 | 616946 | 315367 | 51.1 | 77401 | 11229 | 14.5 |
| IBP3 | P17936 | YKVDYESQSTDTQNFpSSESKR/YKVDYESQSTDTQNFSSEpSKR | S201 | 97 | y19 | 860.37 | 1095.48 | 3 | 2 | 165684 | 65781 | 39.7 | 34288 | 7408 | 21.6 |
| IBP5 | P24593 | IERDpSREHEEPTTSEMAEETYSPK/IERDSREHEEPTTpSEMAEETYSPK | S116 | 98 | y7 | 733.56 | 853.39 | 4 | 1 | 507364 | 195981 | 38.6 | 67677 | 18906 | 27.9 |
| KNG1 | P01042 | EpSNEELTESCETK | S332 | 100 | y8 | 818.31 | 967.44 | 2 | 1 | 1971125 | 506039 | 25.7 | 215318 | 93055 | 43.2 |
| KNG1 | P01042 | EpSNEELTESCETKK | S332 | 100 | y6 | 588.57 | 752.36 | 3 | 1 | 458478 | 99579 | 21.7 | 142726 | 45640 | 32.0 |
| PCSK9 | Q8NBP7 | HLAQApSQELQ | S688 | 100 | b9 | 602.77 | 960.49 | 2 | 1 | 1886731 | 440623 | 23.4 | 307729 | 161316 | 52.4 |
| SEPP1 | P49908 | LPTDpSELAPR | S293 | 100 | y9 | 589.78 | 533.23 | 2 | 2 | 23347 | 6506 | 27.9 | 9394 | 2870 | 30.6 |
| ZPI | Q9UK55 | VVQAPKEEEEDEQEASEEKApSEEEK/VVQAPKEEEEDEQEApSEEKASEEEK | S61 | 100 | y23 | 739.82 | 920.04 | 4 | 3 | 165424 | 42048 | 25.4 | 62784 | 35862 | 57.1 |
3.6 Relevance of phosphorylated proteins to disease
Many phosphoproteins for which phosphopeptides were quantified are known to be involved in processes and pathways associated with human diseases, including cancer, and some of them have been proposed as biomarkers [32]. For example, several phosphopeptides from phosphoproteins involved in the plasminogen activator/plasmin system were quantified, which is one of the major protease systems involved in tumor metastasis. In addition to plasminogen itself, phosphopeptides from proteins involved in down-regulation of the plasminogen activator/plasmin system and thus suppression of cell motility (alpha-2-antiplasmin, A2AP) [33] and from proteins known to stimulate cell migration and tumor invasiveness, like Src/Rac-stimulating vitronectin (VTNC) and integrin alpha 5 (ITA5) [34], were quantified and shown to have low biological variability in the healthy population sampled here. A phosphopeptide from collagen-interacting tenascin-X was among ones with higher biological variation and has been previously associated with breast cancer proteome [35].
Several phosphopeptides identified in our recent study examining phosphoproteins in conditioned media of breast cancer cell lines [3] were also targeted here. Among these were phosphopeptides from CYTC and IBP5 specific for less aggressive luminal type tumors that showed high and low interpersonal variability, respectively, by both MRM and SWATH MS2 (Table 1). In addition, phosphopeptides that were characterized as basal breast cancer tumor specific from OSTP, CD44, and IBP3 were quantified. While the phosphopeptide from OSTP with either phosphorylation at S308 or S310 showed low biological variability, phosphopeptides from CD44 and IBP3 had higher interpersonal variation within healthy population.
4 Concluding remarks
Both MRM-MS and SWATH MS2 methods for phosphopeptide quantification in human plasma were employed in this current study to evaluate phosphopeptide variation among healthy individuals. The two methods produced reproducible relative phosphopeptide quantification data for partially overlapping sets of phosphopeptides, with two thirds showing biological variability of abundance consistent between MRM and SWATH-MS2. Differences in interpersonal phosphorylation variability were shown for phosphopeptides derived from proteins involved in maintaining hemostasis, immune response, cell surface interactions, diabetes pathways, metabolism of lipids, and cell signaling pathways. Our results provide data that will be critical to first establishing the base levels of protein phosphorylation variation within healthy human population. Such data will be critical for future studies that might compare plasma samples among cancer patients. For example, changes in phosphorylation of proteins involved in signaling networks and cell-to-cell interactions may enable disease diagnosis and monitoring progression of treatment. The developed phosphopeptide targets can also complement already developed MRM-MS assays for nonphosphorylated plasma peptides [36, 37] and constitute a reference for selection of biomarker candidates. Indeed, the total number of phosphoproteins and defined phosphosites that have been identified in human plasma to date has increased significantly just in the last year, including novel phosphosites identified in this current study [3, 7].
Our results also suggest that SWATH-MS2 analysis combined with phosphopeptide enrichment can provide reproducible and sensitive quantification of plasma phosphosites. Almost twice as many phosphopeptides were quantified from half of the plasma volume compared to MRM-MS analysis. The SWATH-MS2 analysis did not require extensive method development and phosphopeptide targets were not limited to pre-selected ones like in the MRM-MS method. Additional incorporation of heavy phosphopeptide standards, potential sample preparation automation, or introducing a multiplexed SISCAPA approach, would be expected to increase method sensitivity and technical reproducibility even further.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lorri Reinders from the Buck Institute for help with preparation of plasma samples for immunodepletion and Dr. Michael MacMaster from UCSF for help with obtaining plasma samples. This work was supported by grants from the National Cancer Institute, U24 CA126477 (SJF) and a U24 Subcontract (BWG) that are part of the NCI Clinical Proteomic Technologies for Cancer initiative (http://proteomics.cancer.gov). We also acknowledge the support of several instruments from the NCRR shared instrumentation program, including the AB SCIEX 5500 QTRAP 5500 (S10 RR027953; BWG), the QSTAR Elite (S10 RR024615; BWG) and the TripleTOF 5600 (1S10 OD016281; BWG).
Abbreviations
- MRM
multiple reaction monitoring
- SCX
strong cation exchange
Footnotes
CONFLICT OF INTEREST STATEMENT
Authors declare no conflicts of interest.
SUPPORTING INFORMATION
This article contains supporting Figs. 1 and 2, Tables 1 to 4, and Methods 1.
References
- 1.Blume-Jensen P, Hunter T. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 2.Anderson NL, Anderson NG. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Zawadzka AM, Schilling B, Cusack MP, Sahu AK, Drake P, Fisher SJ, Benz CC, Gibson BW. Mol Cell Proteomics. 2014;13:1034–1049. doi: 10.1074/mcp.M113.035485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly-Spratt KS, Pitteri SJ, Gurley KE, Liggitt D, Chin A, Kennedy J, Wong CH, Zhang Q, Buson TB, Wang H, Hanash SM, Kemp CJ. PLoS One. 2011;6:e19721. doi: 10.1371/journal.pone.0019721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrascal M, Gay M, Ovelleiro D, Casas V, Gelpi E, Abian J. J Proteome Res. 2010;9:876–884. doi: 10.1021/pr900780s. [DOI] [PubMed] [Google Scholar]
- 6.Zhou W, Ross MM, Tessitore A, Ornstein D, Vanmeter A, Liotta LA, Petricoin EF., 3rd J Proteome Res. 2009;8:5523–5531. doi: 10.1021/pr900603n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaros JA, Guest PC, Ramoune H, Rothermundt M, Leweke FM, Martins-de-Souza D, Bahn S. J Proteomics. 2012;76:36–42. doi: 10.1016/j.jprot.2012.02.015. Spec No. [DOI] [PubMed] [Google Scholar]
- 8.Jaros JA, Martins-de-Souza D, Rahmoune H, Rothermundt M, Leweke FM, Guest PC, Bahn S. J Proteomics. 2012;76:43–55. doi: 10.1016/j.jprot.2012.05.027. Spec No. [DOI] [PubMed] [Google Scholar]
- 9.Picotti P, Aebersold R. Nat Methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 10.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R. Mol Cell Proteomics. 2012;11:O111 016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Huttenhain R, Surinova S, Gillet LC, Mouritsen J, Brunner R, Navarro P, Aebersold R. Proteomics. 2013;13:1247–1256. doi: 10.1002/pmic.201200417. [DOI] [PubMed] [Google Scholar]
- 12.Held JM, Schilling B, D'Souza AK, Srinivasan T, Behring JB, Sorensen DJ, Benz CC, Gibson BW. Int J Proteomics. 2013;2013:791985. doi: 10.1155/2013/791985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNulty DE, Annan RS. Mol Cell Proteomics. 2008;7:971–980. doi: 10.1074/mcp.M700543-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Villen J, Gygi SP. Nat Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pappin DJ, Hojrup P, Bleasby AJ. Curr Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 17.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 19.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abbatiello SE, Mani DR, Schilling B, Maclean B, Zimmerman LJ, Feng X, Cusack MP, Sedransk N, Hall SC, Addona T, Allen S, Dodder NG, Ghosh M, Held JM, Hedrick V, Inerowicz HD, Jackson A, Keshishian H, Kim JW, Lyssand JS, Riley CP, Rudnick P, Sadowski P, Shaddox K, Smith D, Tomazela D, Wahlander A, Waldemarson S, Whitwell CA, You J, Zhang S, Kinsinger CR, Mesri M, Rodriguez H, Borchers CH, Buck C, Fisher SJ, Gibson BW, Liebler D, Maccoss M, Neubert TA, Paulovich A, Regnier F, Skates SJ, Tempst P, Wang M, Carr SA. Mol Cell Proteomics. 2013;12:2623–2639. doi: 10.1074/mcp.M112.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma ZQ, Polzin KO, Dasari S, Chambers MC, Schilling B, Gibson BW, Tran BQ, Vega-Montoto L, Liebler DC, Tabb DL. Anal Chem. 2012;84:5845–5850. doi: 10.1021/ac300629p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham AJ, Bunk DM, Kilpatrick LE, Billheimer DD, Blackman RK, Cardasis HL, Carr SA, Clauser KR, Jaffe JD, Kowalski KA, Neubert TA, Regnier FE, Schilling B, Tegeler TJ, Wang M, Wang P, Whiteaker JR, Zimmerman LJ, Fisher SJ, Gibson BW, Kinsinger CR, Mesri M, Rodriguez H, Stein SE, Tempst P, Paulovich AG, Liebler DC, Spiegelman C. J Proteome Res. 2010;9:761–776. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, Verdin E, Kahn CR, Maccoss MJ, Gibson BW. Mol Cell Proteomics. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma V, M., B., Eckels J, Stergachis AB, Jaffe JD, MacCoss MJ. 60th Annual ASMS Conference on Mass Spectrometry & Allied Topics; Vancouver, CAN. May 20-24, 2012. [Google Scholar]
- 25.Silliman CC, Dzieciatkowska M, Moore EE, Kelher MR, Banerjee A, Liang X, Land KJ, Hansen KC. Transfusion. 2012;52:417–424. doi: 10.1111/j.1537-2995.2011.03316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hortin GL, Sviridov D, Anderson NL. Clin Chem. 2008;54:1608–1616. doi: 10.1373/clinchem.2008.108175. [DOI] [PubMed] [Google Scholar]
- 27.De Pergola G, Silvestris F. J Obes. 2013;2013:291546. doi: 10.1155/2013/291546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louderbough JM, Schroeder JA. Mol Cancer Res. 2011;9:1573–1586. doi: 10.1158/1541-7786.MCR-11-0156. [DOI] [PubMed] [Google Scholar]
- 29.Malik S, Fu L, Juras DJ, Karmali M, Wong BY, Gozdzik A, Cole DE. Crit Rev Clin Lab Sci. 2013;50:1–22. doi: 10.3109/10408363.2012.750262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou GL, Zhang H, Field J. Cell Adh Migr. 2013:8. doi: 10.4161/cam.27479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan MM, Sage EH. Int J Biochem Cell Biol. 2004;36:991–996. doi: 10.1016/j.biocel.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 32.Polanski M, Anderson N. Leigh, Biomarker Insights. 2006;2:1–48. [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashido Y, Hamana T, Ishida Y, Shintani T, Koizumi K, Okamoto T. Oncol Rep. 2007;17:417–423. [PubMed] [Google Scholar]
- 34.Noh H, Hong S, Huang S. Theranostics. 2013;3:487–495. doi: 10.7150/thno.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng Z, Hincapie M, Pitteri SJ, Hanash S, Schalkwijk J, Hogan JM, Wang H, Hancock WS. Anal Chem. 2011;83:4845–4854. doi: 10.1021/ac2002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson L, Hunter CL. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Kuzyk MA, Smith D, Yang J, Cross TJ, Jackson AM, Hardie DB, Anderson NL, Borchers CH. Mol Cell Proteomics. 2009;8:1860–1877. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.