Abstract
Intrauterine growth restriction leads to the development of adult onset obesity/metabolic syndrome, diabetes mellitus, cardiovascular disease, hypertension, stroke, dyslipidemia, and non-alcoholic fatty liver disease/steatohepatitis. Continued postnatal growth restriction has been shown to ameliorate many of these sequelae. To further our understanding of the mechanism of how intrauterine and early postnatal growth affects adult health we have employed Affymetrix microarray-based expression profiling to characterize hepatic gene expression of male offspring in a rat model of maternal nutrient restriction in early and late life. At day 21 of life (p21) combined intrauterine and postnatal calorie restriction treatment led to expression changes in circadian, metabolic, and insulin-like growth factor genes as part of a larger transcriptional response that encompasses 144 genes. Independent and controlled experiments at p21 confirm the early life circadian, metabolic, and growth factor perturbations. In contrast to the p21 transcriptional response, at day 450 of life (d450) only seven genes, largely uncharacterized, were differentially expressed. This lack of a transcriptional response identifies non-transcriptional mechanisms mediating the adult sequelae of intrauterine growth restriction. Independent experiments at d450 identify a circadian defect as well as validate expression changes to four of the genes identified by the microarray screen which have a novel association with growth restriction. Emerging from this rich dataset is a portrait of how the liver responds to growth restriction through circadian dysregulation, energy/substrate management, and growth factor modulation.
Keywords: Intrauterine growth restriction (IUGR), liver, microarray, transcriptome, developmental origins of health and disease, obesity, circadian
INTRODUCTION
Aside from the perinatal complications associated with low birth weight, individuals born with intrauterine growth restriction suffer from chronic diseases late in life that ultimately lead to a shortened lifespan. The late David Barker elegantly described this association through epidemiologic analysis [Barker, 1988], and directed animal studies confirm and mechanistically describe the epigenetic processes operative [Seki et al., 2012]. The aging associated metabolic sequelae of low birth weight include obesity and metabolic syndrome [Nobili et al., 2008], diabetes mellitus [Eriksson et al., 2006], cardiovascular disease [Kajantie et al., 2005], hypertension [Barker et al., 1990], stroke [Martyn et al., 1998], dyslipidemia [Sohi et al., 2011b], and non-alcoholic fatty liver disease/steatohepatitis [Alisi et al., 2011].
Animal models of differing technique and species offer mechanistic inquiry into the pathophysiology of intrauterine growth restriction. Specific methodology to induce growth restriction include in utero maternal calorie restriction or protein malnourishment, uterine artery ligation, nicotine exposure, and hypoxia using various animal species [Vuguin, 2007]. Our laboratory has employed rodent models of maternal nutrient restriction during the perinatal period to induce growth restriction to characterize the somatic changes to liver, skeletal muscle, heart, pancreas, lung, hypothalamus, and the brain. Our recent study has further characterized the metabolic profile consisting of serum concentrations of glucose, lipids and insulin in these animals [Garg et al., 2013]. Interestingly, our studies validate those of others that revealed intrauterine growth restricted offspring exposed to continued restriction in the early neonatal period results in overall diminution of adult body size, however with amelioration of the later metabolic sequelae [Dai et al., 2012; Garg et al., 2012; Jimenez-Chillaron and Patti, 2007]. Thus fetal programming induced in utero may be modified by early postnatal growth. The nascence of late-life adult disease in the fetal and postnatal periods is termed the developmental origins of health and disease [Barker, 2004]. Scientific inquiry of this hypothesis has therefore evolved into life course study.
New genetic technologies offer the possibility to evaluate genome-wide changes in gene expression. Excepting placental studies and one recent report examining multiple organs and employing a low protein diet to induce intrauterine growth restriction [Vaiman et al., 2011], few transcriptome studies examining organ systems exist in intrauterine growth restriction. We therefore sought to characterize the liver transcriptome in intrauterine growth restricted rat offspring during the suckling transition (day 21 after parturition, p21) and in the aging adult (day 450 of adult life, d450) through microarray-based expression profiling using a maternal nutrient restriction model. The studies described herein test the hypothesis that the hepatic transcriptional profile may identify mechanisms operative behind the aging associated sequelae of intrauterine growth restriction and may further define the amelioration seen by delayed re-feeding during the critical neonatal growth period. These discovery driven studies identify a transcriptional response at p21 that is not found at d450. Validation experiments performed at p21 designed to interrogate circadian influence confirmed the changes to circadian, metabolic, and growth factor genes. In contrast to the early life transcriptome changes, validation experiments at d450 confirm only a few novel genes to change expression despite persistent circadian dysregulation. Emerging from this rich dataset is a comprehensive portrait of the transcriptional response of the liver to perinatal calorie restriction in early and late life, pointing to the importance of metabolic and growth factor genes.
MATERIALS AND METHODS
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Animal Research Committee of the University of California, Los Angeles (Permit Number: 1999-104-42). Deep anesthesia was achieved with administration of isoflurane, and euthanasia with pentobarbital (100 mg/kg, intraperitoneal); all efforts were made to minimize suffering.
Animals
Pregnant Sprague-Dawley rats (8–10 weeks old, 225–250 g; Charles River Laboratories, Hollister, CA) were housed in individual cages with ad libitum access to water, exposed to 12-h light/dark cycles at 21–23°C, and fed with standard rat chow (NIH-31 Modified Open Formula Mouse/Rat Sterilizable Diet composed of 63.9% carbohydrate, 6.25% fat, and 18.6% protein; product number 7013, Harlan Industries, Indianapolis, IN). Upon birth, neonatal gender was identified and litters culled to 6 male newborn pups with litter birth weights closest to the median weight. Maternal cross-fostering was employed in all experimental groups.
Prenatal and postnatal nutrition management and the definition of experimental groups
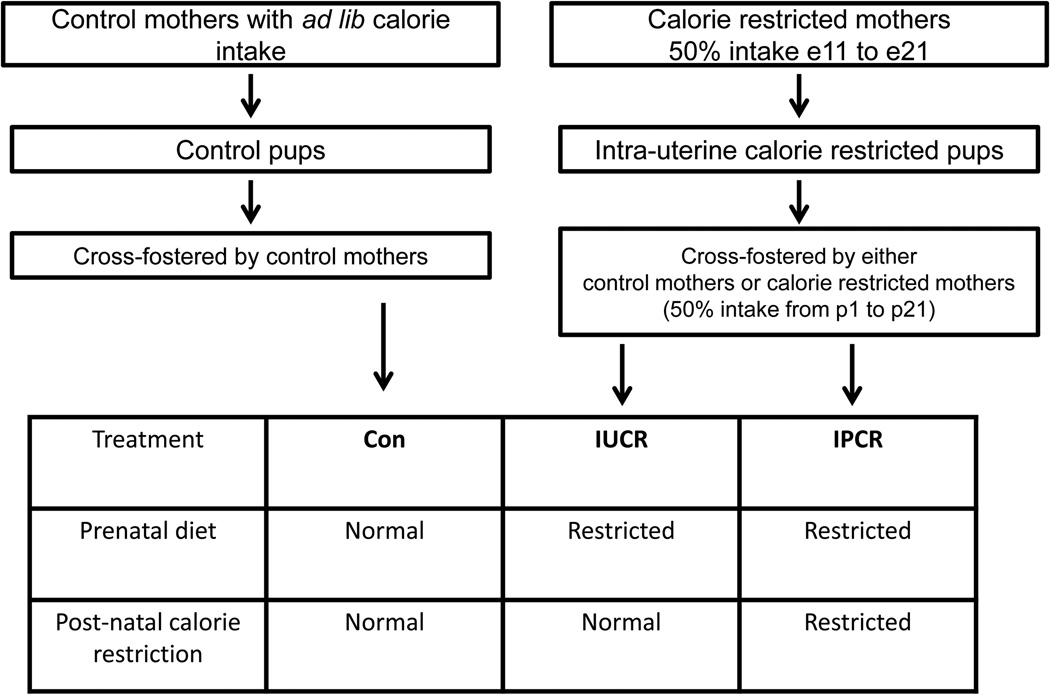
Figure 1 describes the three experimental groups employed in these studies. Control-fed rats (Con) were allowed ad libitum access to feeding throughout gestation, and when appropriate for age at tissue collection, through lactation, and adulthood. Control offspring were cross fostered by mothers who received ad libitum feeding throughout gestation and lactation, and upon weaning at parturition day 21 (p21), received ad libitum feeding as juveniles and adults. The rats exposed to intrauterine calorie restriction (IUCR) were born from mothers who had 50% calorie restriction (11 g/day) from embryonic day 11 (e11) through birth (e21), and owing to the maternal nutrient restriction, were born with intrauterine growth restriction. IUCR offspring, were cross-fostered by ad libitum fed mothers during gestation and lactation, and when appropriate for age at collection, were also fed ad-libitum as juveniles and adults. The last group was subjected to combined intrauterine and postnatal calorie restriction (IPCR), and was born from mothers subjected to 50% calorie restriction from e11 to e21 and cross-fostered by mothers subjected to intrauterine calorie restriction and maintained at 50% calorie limitation during lactation (20 g/day). If appropriate for age at tissue collection, the IPCR group received ad libitum feeding as juveniles and adults beginning at post-weaning (p22). Legacy studies performed in our laboratory have included a group of animals which received postnatal calorie restriction only. Owing to the similar phenotype and hormonal profiles of the IPCR and postnatal calorie restriction groups [Garg et al., 2013; Garg et al., 2012] we have elected not to study this group in these experiments. Previously studied parameters found to be similar in these two groups include body weights, body fat percentages, intravenous glucose tolerance testing, and serum levels of glucose, leptin, and adiponectin. Excluding this group in these studies minimizes the number of animals used and allows for timely collection of tissues at multiple points in the day.
Figure 1.
Microarray-based expression profiling of liver tissues
Liver tissue was dissected and snap-frozen in liquid nitrogen for subsequent processing. Total RNA was extracted using Trizol (InVitrogen; Carlsbad, CA) with RNA cleanup using the mini RNEASY column (Qiagen; Valencia, CA). RNA quality was assured by spectrophotometric absorption at 260nm/280nm, as well as by the Agilent Bioanalyzer which assured integrity of the small and large ribosomal subunits and lack of degradation (Agilent; Santa Clara, CA). One µg of total RNA was used to generate microarray probes by standard Affymetrix protocol (Enzo Diagnostics; Farmingdale, NY) which were hybridized to the Affymetrix Rat 1.0 gene arrays (Affymetrix; Santa Clara, CA). Each treatment group was interrogated by microarray analysis from three different animals. The data was normalized using the Robust Multichip Average algorithm with the Affymetrix software Expression Console. The normalized data files (cel) and corresponding text files were uploaded into the dCHIP program [Li and Wong, 2001] for pair-wise comparisons. Probesets were filtered to exclude low expressed genes having a mean expression value less than 64 microarray units (non-log transformed) in the p21 control samples; a value at or below this threshold is near the background level of the microarray thereby being biologically irrelevant and/or spuriously assayed. The microarray data has been deposited into the Gene Expression Omnibus (accession number: GSE41709)
Validation of microarray studies through independent experiments
The differentially expressed genes identified in the microarray screen were validated with independent experiments. To validate the p21 findings, independent litters of Con and IPCR treatments were examined in the morning and evening to capture representative diurnal and nocturnal time points using a feeding schedule designed to minimize food entrainment (see below). To validate the d450 microarray results additional historical banked samples from independent experiments previously performed were evaluated. The total number of d450 samples studied (including the samples evaluated by microarray) are Con (n=8), IUCR (n=7), and IPCR (n=8).
Feeding time and minimization of food entrainment
As per vivarium protocol, light entrainment occurs on a 12 hour schedule. Customary with our established protocol of maternal calorie restriction, the IUCR and IPCR groups were fed at zeitgeber time 4 (ZT4), with lights on set at ZT0. The initial microarray experiment employed animals cared for by this standard protocol. To confirm the early life microarray results independent and controlled experiments were designed to incorporate a rolling feeding schedule which offset feeding time by 2 hours per day from ZT0 to ZT12, the times which we are allowed vivarium access. The rolling feeding schedule achieved a balance of limiting a strong and uniform food entrainment cue while minimizing prolonged periods of food restriction. Another significant modification to the standard collection protocol was an additional collection of tissue at ZT16 designed to collect a representative nocturnal time point. To make the morning and evening tissue collections comparable, fasting time before tissue collection was set at exactly 16 hours. These experiments employed two mothers and their individual litters of six for each treatment group and time point.
Confirmatory quantitative reverse-transcription polymerase chain reaction assays
Differentially expressed genes were confirmed by reverse transcription and Taqman based quantitative polymerase chain reaction (RT-qPCR) (Table 1). For a gene to be considered adequately validated three criteria were met i. RT-qPCR amplification was linear as evaluated by standard curve analysis ii. Using the RNA originally employed in the microarray analysis there was correlation with the microarray signal changes to the calculated RQ obtained by RT-qPCR iii. The direction of expression changes obtained by RT-qPCR were concordant in the microarray samples and the validation samples. 56 genes were screened. Sequences of the primers and probes reported are found in Supplementary Table S1. One µg of RNA was used as a template for reverse transcription using Super Script III in conjunction with an oligo-dT primer according to manufacturer’s protocol (Invitrogen). This cDNA was used as a template for RT-qPCR using the Step One Plus Real Time PCR System thermocycler (Applied Biosystems, Foster City, CA) using the following parameters: 50°C for 2 minutes, 95°C for 20 seconds, then 40 cycles of 95°C for 15 seconds and 60°C for 20 seconds. Data were normalized to the amplification of either 18S, Rpl13a, or Rn45s (selected for the optimal performing standard curve) and relative quantification to the control sample was made using the ΔΔCt method [Livak and Schmittgen, 2001]. Data is displayed as relative quotient (RQ) with error bars representing the standard error of the mean calculated from the power base 2 transformed variance of the ΔΔCt.
Table 1.
Genes examined by RT-qPCR
| Gene | Abbreviation |
|---|---|
| Circadian locomotor output cycles kaput | Clock |
| Aryl hydrocarbon receptor nuclear transporter-like | Arntl |
| Neuronal PAS domain protein 2 | Npas2 |
| Period 2 | Per2 |
| Insulin-like growth factor binding protein 2 | Igfbp2 |
| Serine dehydratase | Sds |
| Cytoplasmic aspartate aminotransferase | Got1 |
| Aldo-keto reductase, family 1 member b7 | Akr1b7 |
| Acyl-CoA synthetase short-chain family member 2 | Acss2 |
| Zinc finger protein 189 | Zfp189 |
| Apolipoprotein L3 | Apol3 |
| Westmead DMBA8 nonmetastatic cDNA 1 | Wdnm1 |
| Small nucleolar RNA, H/ACA box 5C | Snora5 |
Data analysis
Statistical significance was determined by a Fisher’s two way analysis of variance for data sampled at two time points in a day using SigmaStat (Systat Software, San Jose, CA). If this data did not possess a normal distribution or equal variance, a non-parametric variant of the Fisher’s two way analysis of variance was performed through a rank transformation. Pair-wise comparisons of treatments and time points were evaluated by the Holm Sidak method. For data sampled at only one time point in a day (the d450 data), RT-qPCR results are presented as a mean and standard error of the mean of the RQ summarized from all of the individual experiments examined. A Fisher’s one way analysis of variance and post hoc Fisher’s protected least significant difference test were performed using Statview (SAS Institute, Cary, NC). The threshold of significance was set at 0.05.
Western blot analysis
Western blot analysis was performed as previously described using specific primary antibodies directed against Stat5 and Phospho-Stat5 (Tyr694) (Cell Signaling Technology) at a 1:1000 dilution with an over-night hybridization [Oak et al., 2006; Shin et al., 2012]. The quantification of protein bands was performed by densitometry using ImageQuant software (GE Healthcare). The optical density was corrected for inter-lane loading variability using an internal control, vinculin (from Sigma Chemical Co. St. Louis, MO). Statistical significance was determined by a Fisher’s two way analysis of variance.
RESULTS AND DISCUSSION
Male rats subjected to intrauterine growth restriction develop obesity which is prevented by postnatal calorie restriction
Body weights were affected by the experimental manipulation of maternal nutrient restriction (Table 2). Birth weight as recorded at p2 revealed that the average pup weight was reduced by 21% in the IUCR group as compared to the ad libitum fed control animals (Con) (p<0.004). At p21, the IUCR group had undergone partial “catch-up growth”, however there was continued diminution in size with a 14% decrease in body weight as compared to the normally fed control animals (p<0.0001). At p21 the IPCR group was markedly smaller with a 68% decrease in body weight as compared to the normally fed control animals (p<0.0001). At d450 the IUCR group had now exceeded the weight of the control fed animals by 11% (p<0.05), and the IPCR group remained 15% lighter than the normally fed control animals (p<0.05) [Garg et al., 2013]. Other phenotypic characterization has been previously reported by our laboratory at p21 and d450 [Garg et al., 2012] [Garg et al., 2013] [Dai et al., 2012] [Shin et al., 2012].
Table 2.
Body weights at p2, p21 and d450
| Con (g) | IUCR (g) | IPCR (g) | |
|---|---|---|---|
| Body weight, p2 | 8.0 +/− 0.2 | 6.3 +/− 0.2a | N/A |
| Body weight, p21 | 70.0 +/− 1.3 | 60.5 +/−1.2b | 22.5 +/−1.0b |
| Body weight, d450 [21] | 895.7 +/−36.9 | 989.8 +/− 34.3c | 756.3 +/−28.8c,d |
g=grams,
p<0.004 vs Con,
p<0.0001 vs Con,
p<0.05 vs Con,
p<0.05 vs IUCR
Microarray-based expression profiling of the liver of growth restricted male offspring in early life identifies a robust transcriptional response that is characterized by circadian, metabolic, and insulin-like growth factor binding genes
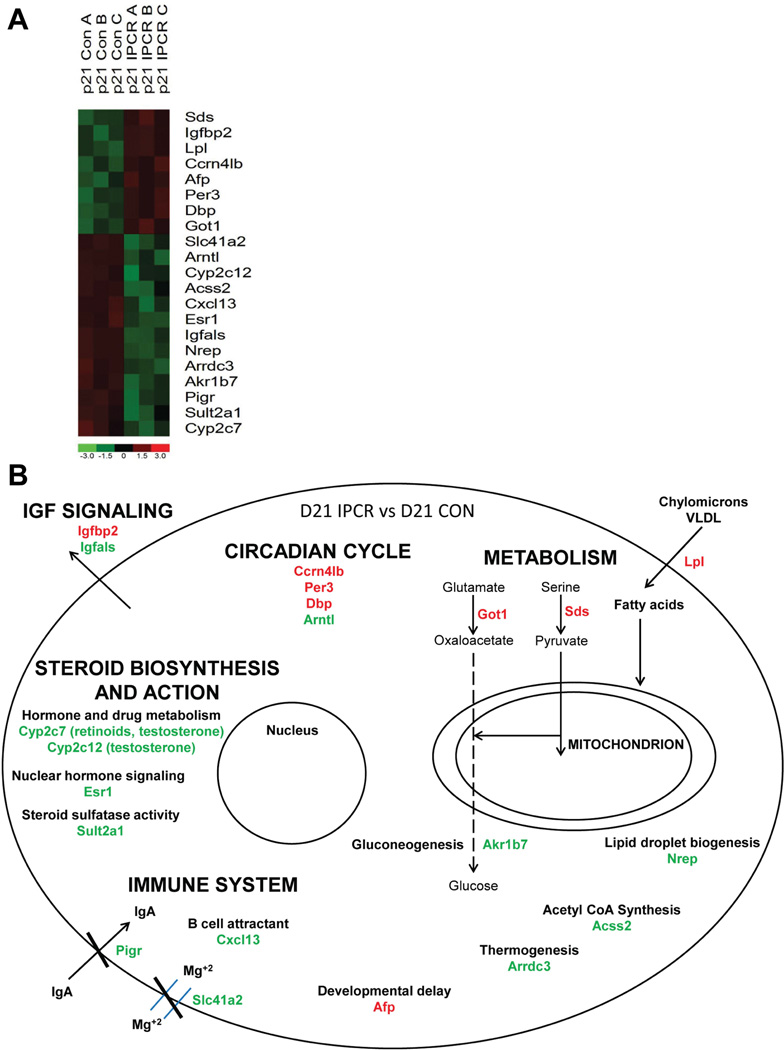
To identify the hepatic genes most reliably changed by perinatal calorie restriction, microarray-based expression profiling was performed in ad libitum fed controls and calorie restricted offspring at p21 with three independent liver samples profiled per group. At p21 the samples studied include the Con and IPCR groups. We purposely excluded the IUCR group at the p21 time-point as these animals were in a dynamic state of undergoing ‘catch-up’ growth and therefore would not provide a clear transcriptional profile. In addition, the marked phenotypic differences between control and IUCR animals is not apparent until later in life. A comparison was made between Con liver samples (n=3) to all IPCR samples (n=3) at p21 with the following comparison criteria: a minimum of two-fold or greater difference and a p-value less than 0.05 using a Welch modified two sample t-test. This identified 144 probesets to be differentially expressed; 103 of these probesets are down-regulated and 41 are up-regulated (Supplementary Table S2). The 144 stringently identified probesets were evaluated with the online software Database for Annotation, Visualization and Integrated Discovery (DAVID) of the National Institutes of Allergy and Infectious Diseases, software which identifies groups of genes that are over-represented using an unbiased algorithm based upon the Gene Ontology classification [Huang da et al., 2009]. This analysis identified the terms circadian rhythm (p<7.5 × 10−6), energy metabolism (amine catabolic process (p<0.00013), lipid biosynthetic process (p<0.00024), starch, sucrose and metabolism (p<0.0067)), insulin-like growth factor binding (p<0.017), steroid hormone biosynthesis (p<7.8 × 10−5), and immune response (p<0.04) to be highly over-represented. The expression profile and cellular location and function of the most highly differentially expressed genes are displayed in Figure 2. These four-fold or greater or lesser differentially expressed genes include 8 up-regulated genes and 13 down-regulated genes, and incorporate representative examples of the major gene ontology over-represented groups.
Figure 2.
Perinatal calorie restriction alters circadian gene expression in early life
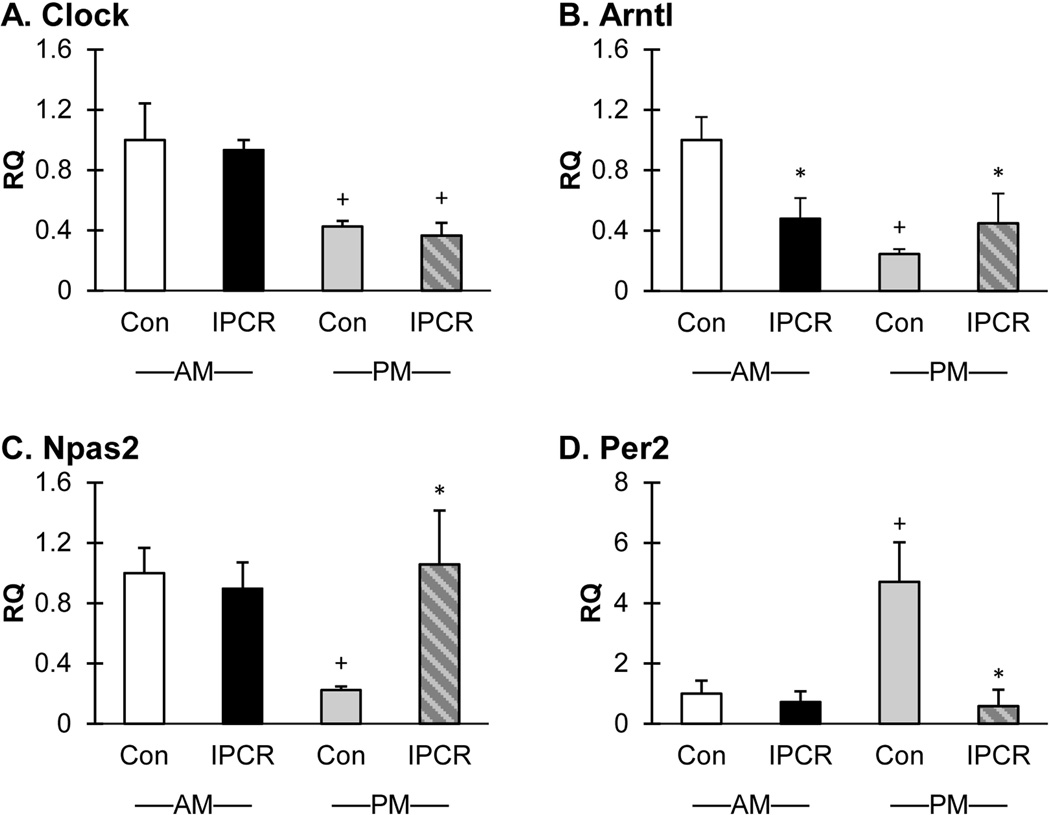
We surmise that Arntl is the most significant circadian gene to change expression in response to perinatal calorie restriction and make this conclusion while considering the results of the microarray screen, the validation experiments at p21, and the central placement of Arntl within the circadian regulatory system.
The circadian regulatory system is comprised of a series of proteins which are sequentially produced and degraded through both transcriptional and post-transcriptional mechanisms [Ko and Takahashi, 2006]. The microarray data at p21 reveals seven circadian genes at every level of the regulatory system to be significantly differentially expressed at a cutoff of a 50% change or greater (Supplementary Figure S1: carbon catabolite repression 4-like b (Ccrn4lb, also known as nocturnin); the period homologs 3, 1, and 2 (Per3, Per1, Per2); nuclear receptor subfamily 1, group D, member 1 (Nr1d1); Npas2; and Arntl).
As a broad survey of the circadian system, RT-qPCR of Clock, Arntl, Npas2, and Per2 was performed in the validation experiments (Figure 3) using 6 independent liver samples from each treatment group and time point. Consistent with the microarray findings, the expression of Arntl is decreased in the ZT4 (morning) time-point in the IPCR group (RQ 0.5; p<0.004). The RQ of Arntl at the ZT16 (evening) time-point is 0.2 and 0.4 for the Con and IPCR groups, respectively (Con AM vs Con PM, p<0.001; Con PM vs IPCR PM, p<0.049). The validation experiment did not confirm the morning expression changes for Clock, Npas2, and Per2 in the IPCR group but did however identify an increase in expression at the night sampling time for Npas2 and Per2. Clock gene expression varied by time of day with increased expression at ZT4 in both the Con and IPCR groups (the RQ in the AM Con and IPCR groups is 1.0 and 0.9, respectively; the RQ in the PM Con and IPCR groups is 0.4; Con AM vs Con PM, p<0.001; IPCR AM vs IPCR PM, p<0.001). The expression of Npas2 was found to be decreased in the evening IPCR group (RQ 0.2; p<0.002). The expression of Per2 was found to be increased in the evening Con group (RQ 4.7; Con AM vs Con PM, p<0.011; Con PM vs IPCR PM, p<0.008). Discordance of the morning expression of Clock, Npas2, and Per2 expression in the microarray screen and validation experiments may be explained by the methodologic differences between the experiments and likely highlights the importance of timing of last meal in relation to tissue collection.
Figure 3.
To summarize, circadian genes are the most significant over-represented functional group identified to change expression in response to perinatal calorie restriction in early life. Further, the validation experiments reveal maximal discordance between the normally fed controls and the calorie restricted group in the evening sampling point, and that Arntl is the most robust and centrally acting circadian associated transcription factor to change expression. Interestingly, for numerous circadian associated genes such as Per3 and Nocturnin, there are ascribed metabolic functions which highlight the indivisibility of circadian time keeping with metabolic regulation[Pendergast et al., 2012] [Costa et al., 2011] [Dallmann and Weaver, 2010] [Wang et al., 2001] [Green et al., 2007] [Kawai et al., 2010b] [Kawai et al., 2010a].
Perintatal calorie restriction alters metabolic gene expression in early life
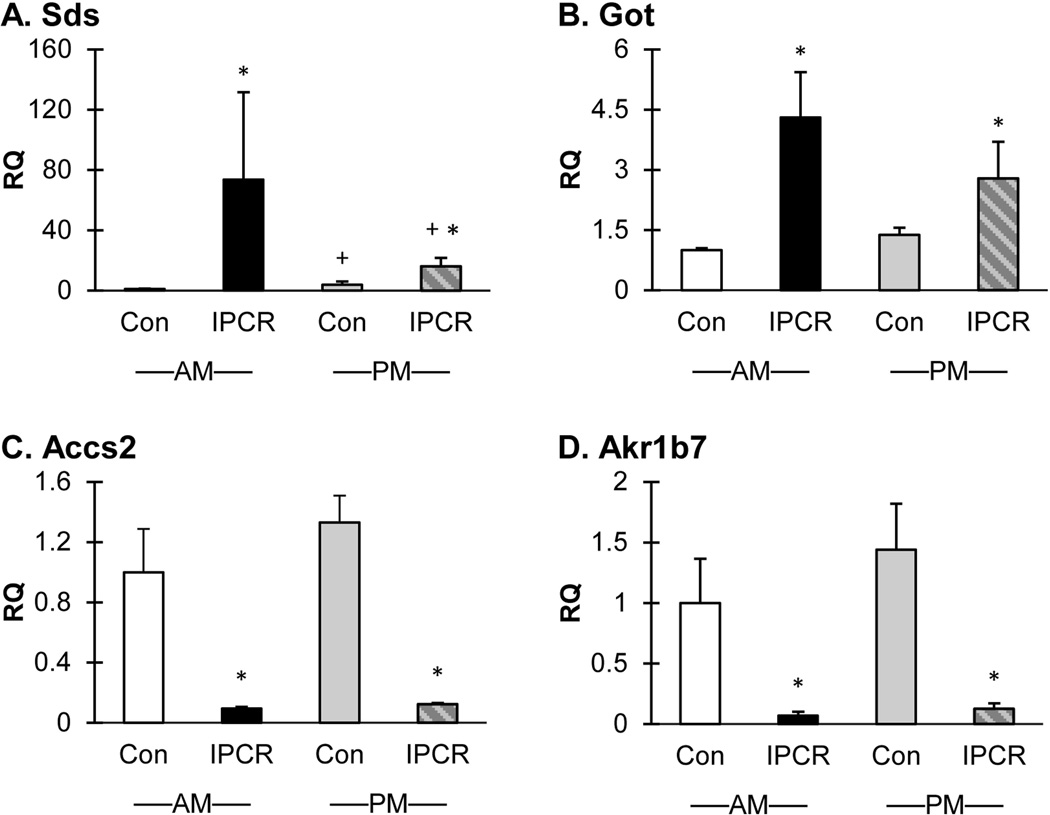
As a broad survey of metabolism, RT-qPCR of Sds, Got1, Accs2, and Akr1b7 (Figure 4) and Lpl (Supplementary Figure S2) was performed in the validation experiments using 6 independent liver samples from each treatment group and time point. The expression changes for each of these genes were uniformly validated. The up-regulated genes Sds and Got1 highlight the importance of carbon sources which may be consumed in the tricarboxylic cycle or used as substrate for gluconeogenesis. Additionally, the action of Lpl, also highly up-regulated, provides saturated fats to be delivered to the liver. The down-regulated genes Accs2 and Akr1b7 are involved in acetyl-CoA generation and gluconeogenesis, respectively.
Figure 4.
Sds is the highest up-regulated gene identified in the microarray screen and the profound increase in expression is validated in the confirmatory experiments (Figure 4, Panel A). The RQ of Sds in the IPCR group is 73.7 (Con AM vs IPCR AM, p<0.001) and 16.1 (IPCR AM vs IPCR PM, p<0.019; Con PM vs IPCR PM, p<0.004) for the morning and evening samples respectively. The RQ of SDS in the evening Con group is 3.8 (Con AM vs Con PM, p<0.019).Owing to the large RQ, the RT-qPCR results were confirmed in an independent re-analysis and a representative experiment displayed. Rat Sds is able to remove the nitrogen moiety from serine and threonine through a pyridoxol-5’-phosphate requiring catalysis resulting in the release of pyruvate and a-ketobutyrate, respectively [Ogawa et al., 1989].
The aminotransferase Got1 serves the same purpose as Sds to mobilize and transfer carbon skeletons within the cell from amino acid precursors. In the validation experiments the RQ of Got1 in the IPCR group is 4.3 (Con AM vs IPCR AM, p<0.001) and 2.8 (Con PM vs IPCR PM, p<0.011) for the morning and evening samples respectively (Figure 4, Panel B). In the Con treatment group, Sds and Got1 expression reach a zenith in the evening; in the IPCR treatment group, Sds and Got1 expression reach a zenith in the morning.
Acss2 is a cytosolic enzyme responsible for the activation of acetyl-coenzyme A from acetate. The RQ of Acss2 in the IPCR group is 0.1 (p<0.001) for both the morning and evening samples (Figure 4, Panel C). Time of day does not affect the expression of Acss2 in both normally fed animals and calorie restricted groups. Acetyl Co-A is of primary importance for both metabolic control and gene expression regulation. Placed at the intersection of gluconeogenesis and fatty acid oxidation, Acetyl-CoA may be utilized for different purposes depending upon energy status, and may be directed to either a cytosolic or mitochondrial location. During times of energy abundance, Acetyl-CoA is formed from glucose, and cytosolic levels are plentiful. During fasting conditions, cytosolic Acetyl Co-A levels are low, and available Acetyl-CoA remains sequestered within the mitochondrion. In addition to the metabolic function, Acetyl-CoA provides a regulatory signal through protein acetylation, most simplistically and well known by the control of gene expression through histone acetylation and de-acetylation. Therefore, the profound down-regulation of Acss2 is of significant importance from both a gene regulatory and metabolic perspective.
Akr1b7 is highly down-regulated in response to maternal perinatal calorie restriction. The RQ of Akr1b7 in the IPCR group is 0.1 for both the morning (p<0.001) and evening (p<0.001) samples (Figure 4, Panel D). There are no differences in expression of Akr1b7 within a treatment group between morning and evening samples. Directed studies have revealed that Akr1b7 is responsible for metabolism of bile through aldo-keto reduction and is transcriptionally regulated by FXR binding to bile acids [Schmidt et al., 2011]. Targeted over-expression of Akr1b7 leads to decreased gluconeogenesis, blood glucose levels, and hepatic lipid accumulation in diabetic mice [Ge et al., 2011]. The observed down-regulation of Akr1b7 in response to perinatal calorie restriction is therefore consistent with the drive for increased gluconeogenesis imparted by calorie restriction.
Lipoprotein lipase (Lpl), the third most highly up-regulated gene, mobilizes the energy stored in fats by catalyzing the hydrolysis of lipoprotein triglycerides, thereby generating free fatty acids for either fuel consumption or energy storage [Davies et al., 2012]. Perinatal calorie restriction induces a 12 and 6.5 fold increase in Lpl expression in the morning and evening, respectively. Time of day does affect the expression of Lpl in the IPCR group (p<0.036, Supplementary Figure S2).
To summarize, metabolic genes are prominently altered at p21 in response to perinatal calorie restriction. Alternative fuels such as fatty acids and amino acid derived carbon skeletons likely provide much needed resources for cellular maintenance and growth during periods of scarcity. Specifically, fatty acids are obtained through the action of Lpl from circulating lipoprotein triglycerides, and carbon skeletons commandeered through the deamination reaction of Sds and the transaminase reaction of Got1. The regulatory role of Akr1b7 in controlling gluconeogenesis, and the decreased production of acetyl-CoA through the down-regulated Acss2, are key effects of perinatal calorie restriction.
Perinatal calorie restriction suppresses growth hormone/insulin-like growth factor 1 signaling in early life
The finding of Igfbp2 as the second highest up-regulated gene in the microarray screen denotes the importance of the endocrine somatotrophic axis in our calorie restriction model of IUGR (Figure 2). A survey of the microarray data for genes known to be involved in the growth hormone/insulin-like growth factor axis identified a profound down-regulation of Ghr, Stat 5b (the second messenger of Ghr), insulin-like growth factor 1 (Igf1), and insulin-like growth factor binding protein, acid labile subunit (Igfals) (Supplementary Figure S3). The reciprocal expression of the binding proteins Igfbp2 and Igfals result in profound suppression of the growth effect of Igf1. Igfbp2 is known to sequester Igf1 [Hoeflich et al., 2001] and therefore prevent a local tissue growth effect. Igfals, a binding protein produced by the liver in response to growth hormone signaling, is known to extend the circulating half-life of insulin-like growth factors by creating a vascular tripartite protein complex containing the growth factor, an additional Igfbp, and itself [Ooi et al., 1998].
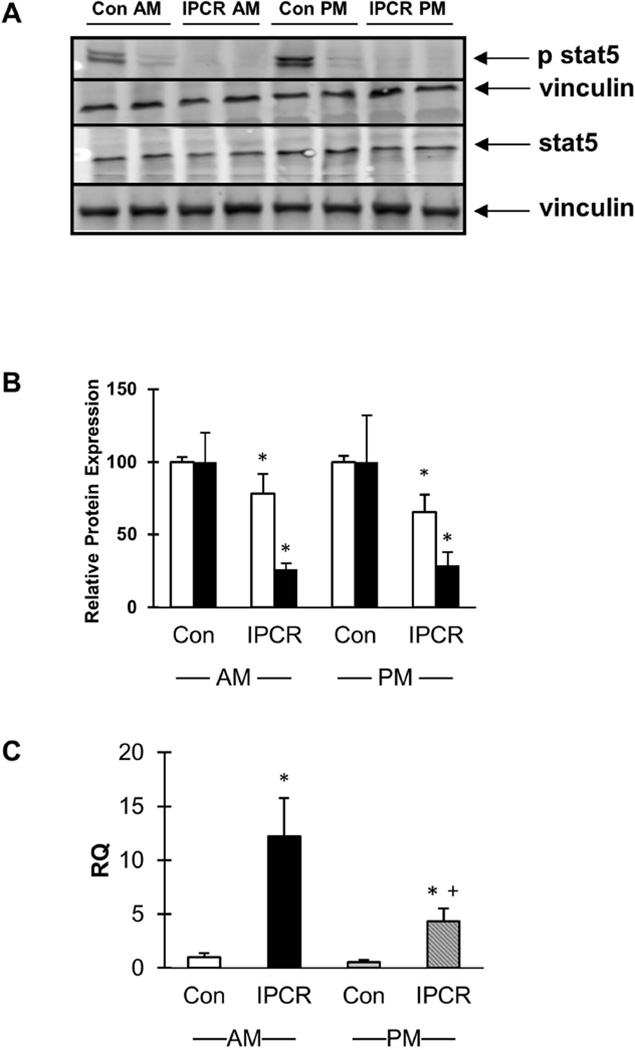
Western blot analysis of liver obtained from p21 males subjected to IPCR validate a decrease in total Stat5 protein (Figure 5, panels A and B) between 22% (p<0.016) and 35% (p<0.008) in the morning and evening, respectively. phospho-stat5 protein levels were decreased by 74% (p<0.005) to 72% (p<0.025) in the morning and evening, respectively. RT-qPCR for Igfbp2 confirmed up-regulation in the IPCR liver in both morning and evening (Figure 5, panel C; RQ of the IPCR group is 12.2 (p<0.001) and 4.4 (p<0.001) for the morning and evening samples, respectively).
Figure 5.
To summarize, the microarray screen has identified profound suppression of the somatotrophic axis at multiple levels of the growth hormone/insulin-like growth factor cascade. Growth hormone secretion is known to follow gender specific patterns, and induces epigenetic changes to loci located throughout the genome that affects many genes not directly involved in growth. Cytochrome P450, family 2, subfamily c, polypeptide 12 (Cyp2c12), the most highly down-regulated gene identified in the microarray screen (Figure 2), is one such gene expressed in a gender specific pattern [Kamataki et al., 1983] for which growth hormone is permissive [Endo et al., 2005]. These data support the somatotrophic axis as an important determinant of fetal growth [D'Ercole et al., 1980; Gluckman and Pinal, 2003].
Microarray-based expression profiling of the liver of growth restricted male offspring at d450 identifies a few genes to change their expression and fails to identify a transcriptional response despite the development of obesity
Using the d450 Con group (n=3) as a reference, the microarray analysis failed to identify any significantly differentially expressed genes in the d450 IUCR group (n=3). When a relaxed metric cutoff was used a small number of uncharacterized or heterogeneous genes were identified (Supplementary Table S3). Although the aggregate action of these few genes with subtle expression differences may have biologic importance, the validation of any one specific gene would be difficult owing to the low fold change difference. Alternatively, the subtle expression changes may be the result of stochastic sampling or quantification error. Interestingly, the microarray analysis did identify a small number of genes to be significantly differentially expressed in the d450 IPCR group (n=3) when using the d450 Con samples (n=3) as a reference (Supplementary Table S4). This important observation indicates that post natal calorie restriction imparts a transcriptional change in adulthood that overshadows that imparted by intrauterine calorie restriction.
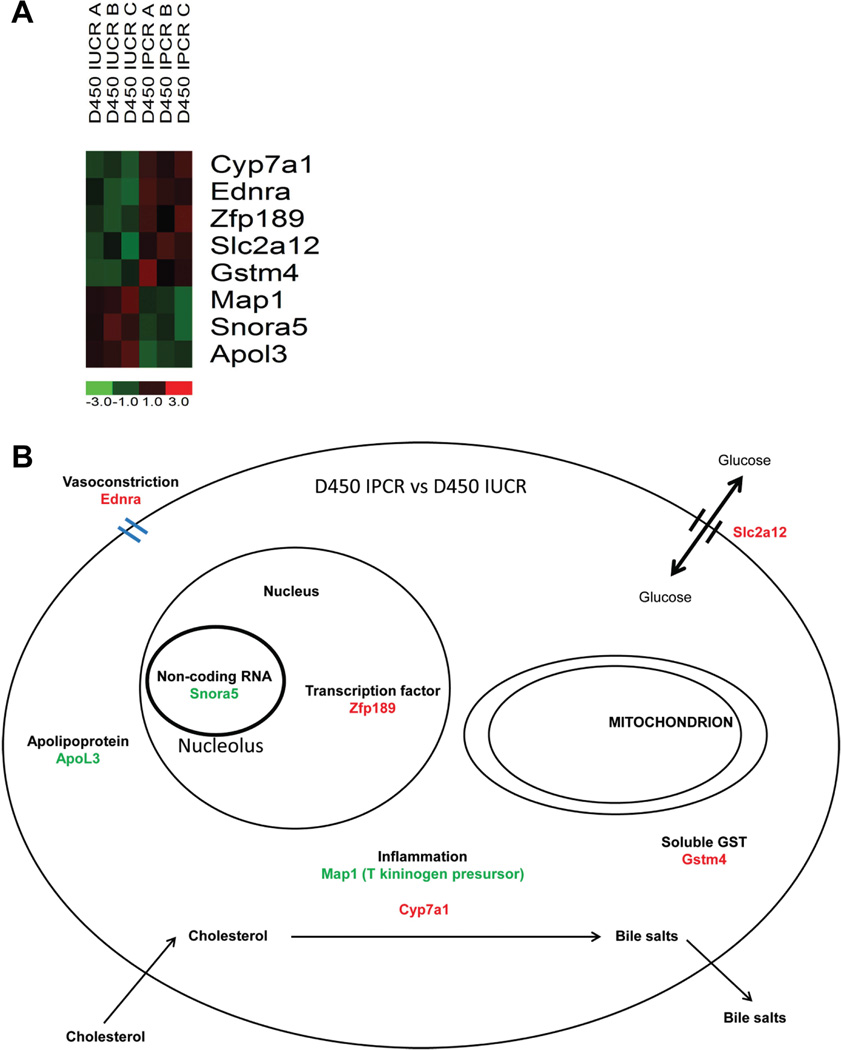
To discern the changes induced by intrauterine calorie restriction that are modified by postnatal calorie restriction, a comparison of the d450 IPCR samples (n=3) to the d450 IUCR samples (n=3) provides an interesting gene list with significant expression fold differences. This comparison elucidates the protective effect of postnatal calorie restriction on intrauterine calorie restriction. With a comparison metric of two-fold with p-value testing, 8 genes were found to be differentially expressed; 3 are down-regulated and 5 are up-regulated. The expression profile and cellular location and function of these most highly differentially expressed genes are displayed in Figure 6. As the most significant fold changes at d450 are found in the comparison between the IUCR and IPCR groups we validate the mismatch hypothesis [Gluckman et al., 2005]. The mismatch hypothesis states that the greater the degree of mismatch between the developmental and mature environments the greater the risk for adult onset disease. The addition of postnatal calorie restriction serves to impart the greatest change to the hepatic transcriptome at d450.
Figure 6.
The finding of up-regulation of 7 alpha hydroxylase (Cyp7a1) in the IPCR group is significant in several respects. Cyp7a1 is the rate limiting step of hepatic excretion of cholesterol and is known to be epigenetically regulated by early life nutritional cues and expressed later in life in a gender specific pattern. The conversion of cholesterol into bile acids occurs in the liver and is catalyzed at the rate limiting step by Cyp7a1 which hydroxylates cholesterol in the 7 alpha position [Jelinek et al., 1990]. Transcriptional regulation of Cyp7a1 occurs through the liver × receptor [Repa and Mangelsdorf, 2000]. Directed studies in a maternal low protein model of intrauterine growth restriction has identified that circulating cholesterol levels are higher in 130 day old male rats and that the observed gender difference is correlated with differential expression of Cyp7a1 protein thought to be regulated at the histone level through increased tri-methyl and decreased acetylation of H3K9 [Sohi et al., 2011a]. The finding of Cyp7a1 within the late adult life microarray screen importantly serves to validate our experimental findings by recapitulating these prior published studies.
Hepatic circadian dysregulation resolves in IPCR and develops in IUCR at d450
Prompted by the observation that circadian genes are the most significant over-represented functional group of genes identified in the microarray screen in early life, we sought to characterize their expression at d450. Interrogation of the d450 microarray data revealed a trend of three circadian genes to be differentially expressed by perinatal calorie restriction using a cutoff of a 50% change or greater (Arntl, Npas2, and Nocturnin), however these genes did not pass significance by p-value testing (Supplementary Figure S4). Therefore, using additional banked samples, RT-q-PCR of Clock, Arntl, Npas2, and Per2 was examined in normally fed (Con, n=8) and perinatally restricted male rat offspring (IUCR, n=7; IPCR, n=7) at d450 (Figure 7). The expression of Arntl (RQ=4.6) and Npas2 (RQ=4.5) are increased in IUCR offspring at d450 when compared to either the Con or IPCR groups (Arntl: one way ANOVA, p<0.0096, F=5.99; Con vs IUCR, p<0.006; IUCR vs IPCR, p<0.0086; Npas2 one way ANOVA, p<0.0056, F=6.903; Con vs IUCR, p<0.038; IUCR vs IPCR, p<0.005). Expression of Clock and Per2 were unchanged by perinatal calorie restriction at d450 (Clock one way ANOVA p<0.7062, F=0.354; Per2 one way ANOVA P<0.2722, F=1.394). These data demonstrate that the circadian alteration seen at p21 in the IPCR group has resolved by d450 (Figures 3 and 7). We conclude that active calorie restriction at p21 in the IPCR group leads to transient circadian gene expression changes which subsequently normalize with restitution of a normal feeding schedule. Interestingly, at d450 the IUCR group has now acquired a circadian defect despite ad libitum feeding. The exact temporal acquisition of the circadian defect seen in IUCR is not addressed by these experiments and further directed studies are required. Interestingly, whether in the IPCR group at p21 or the IUCR group at d450, Arntl is the most significant circadian gene altered by perinatal calorie restriction.
Figure 7.
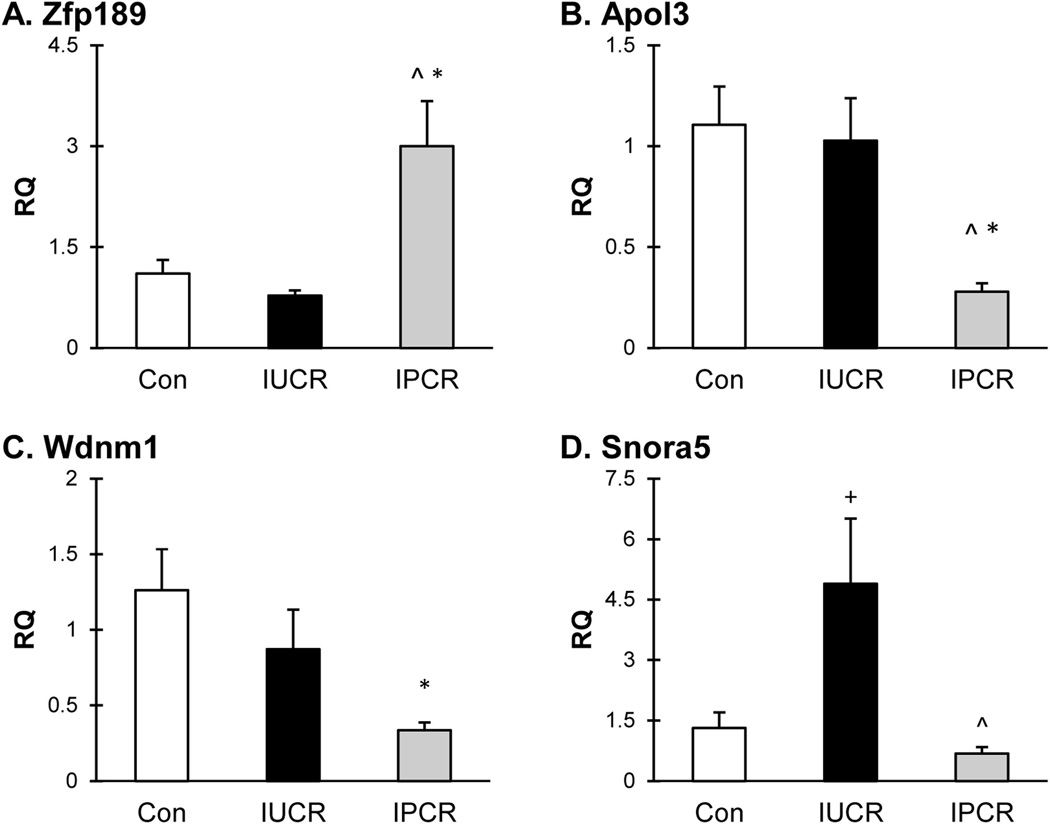
RT-qPCR in independent experiments confirms that the late adult life hepatic expression of the novel genes Zfp189, Apol3, Wdnm1, and Snora5 is altered by maternal perinatal calorie restriction in male offspring
We have selected to validate Zfp189, Apol3, Wdnm1, and Snora5 based upon their high fold change differences within the microarray screen and their novel association with growth restriction.
Zfp189 is a scantily described transcription factor with a human homolog, Znf189, of which the maternal genotype is associated with isolated cleft lip [Jugessur et al., 2010]. RT-qPCR confirmed up-regulation of Zfp189 in the IPCR group (Figure 7, panel A; RQ=3.0; Con vs IPCR, p<0.0042; IUCR vs IPCR, p<0.0016; one way ANOVA overall p<0.0027, F=8.071). The RQ of the Con and IUCR groups are 1.1 and 0.8, respectively. The mechansim by which Zfp189 expression is regulated is unknown, however Genomatix promoter analysis identifies several Stat, p53, and cAMP response sites within 500 bp of the transcription start site.
Apol3 is a cytoplasmic protein thought to affect the movement of lipids or the binding of lipids to organelles [Page et al., 2001] and whose expression is responsive to tumor necrosis factor alpha (TNFa) [Horrevoets et al., 1999]. RT-qPCR confirmed down-regulation of Apol3 in the IPCR group (Figure 7, panel B; RQ=0.3; Con vs IPCR, p<0.0013; IUCR vs IPCR, p<0.0039; one way ANOVA overall p<0.0023, F=8.329). The RQ of the Con and IUCR groups are 1.1 and 1.0, respectively.
Wdnm1 is a member of the four disulfide core family of proteins that was found to have anti-metastatic activity in a mammary adenocarcinoma cell line. The mechanism by which Wdnm1 exerts it’s anti-metastatic activity is by functioning as a secreted proteinase inhibitor that is regulated by TNFa [Kho et al., 2008]. RT-qPCR confirmed down-regulation of Wdnm1 in the IPCR group (Figure 7, panel C; RQ=0.3; Con vs IPCR, p<0.0057; IUCR vs IPCR, p<0.0098; one way ANOVA overall p<0.019, F=4.840). The RQ of the Con and IUCR groups are 1.3 and 0.9, respectively.
Snora5 is a ribosomal protein thought to be required for proper pseudo-uridinylation incorporation of the 18S ribosomal subunit and therefore may affect protein translation [Kiss et al., 2004]. RT-qPCR confirmed up-regulation of Snora5 in the IUCR group (Figure 7, panel C; RQ=4.9; Con vs IUCR, p<0.018; IUCR vs IPCR, p<0.0035; one way ANOVA overall p<0.0079, F=6.230). The RQ of the Con and IPCR groups are 1.3 and 0.7, respectively.
Owing to the paucity of ascribed functions and disparate nature of these novel genes it is hard to draw general conclusions of their mechanistic action in the phenotype of the growth restricted aging adult. However, two of these genes are known to be regulated by TNFa: whether their response is specific to developmental cues or secondary to generalized obesity is unknown. Further directed studies will elucidate the role that these candidate genes may play in the pathophysiology associated with abnormal developmental programming.
Conclusions
Microarray based expression profiling has been used to interrogate the hepatic transcriptome in a rat model employing maternal nutrient restriction. These data reveal significant early life gene expression changes involving a unique combination of circadian, metabolic, and growth hormone/insulin-like growth factor genes which collectively represent the response of the neonatal liver to maternal nutrient limitation. Interestingly, late in life with aging, the hepatic transcriptome is remarkably similar to the ad libitum fed littermates indicating that the maturity associated metabolic sequelae of growth restriction is mediated by non-transcriptional mechanisms. As the p21 microarray data largely implicate a profound decrease in IGF signaling we surmise that the protective effect of postnatal calorie restriction on the intrauterine growth restriction phenotype is mediated through a postnatal IGF regulated process. Future directed studies examining and manipulating the IGF tone in the postnatal period will further define the protective effect of postnatal growth restriction on intrauterine growth restriction. Furthermore, despite a lifetime of ad libitum feeding, IUCR offspring develop abnormal circadian gene expression.
The mechanism by which nutrient restriction affects circadian timekeeping in growth restriction is unknown. Our data suggest that the most likely transcriptional target affected by nutritional cues is the PAS (Period/Arntl/Single minded) domain containing protein Arntl which heterodimerizes with the transcriptionally unchanged Clock protein. Additionally, it is unknown if this circadian alteration is a result of central modification working through the suprachiasmatic nucleus of the hypothalamus or alternatively mediated through peripheral circadian timekeeping within the liver.
The circadian system harmonizes cellular growth, cell cycle control, anabolism, catabolism, and nutrient disposition to synchronize sleep/wake schedules, feeding behavior, physical activity, thermoregulation, and growth. Disruption of the circadian cycle, as found in shift workers, can lead to obesity and coronary artery disease, and highlights the connection between circadian timekeeping function and metabolism [Knutsson, 1989]. The experiments described herein illustrate the importance of the circadian system in metabolic disease: guided future studies examining the circadian regulation of metabolism may lead to chronotherapeutic approaches to combat the obesity phenotype of perinatal growth restriction.
Supplementary Material
ACKNOWLEDGEMENTS
WAF is grateful for the support of Professor Gautam Chaudhuri, M.D., Ph.D. and the Department of Obstetrics and Gynecology. We thank Yun Dai for help in obtaining the initial samples employed in the microarray analysis. This work was supported by grants HD41230 and HD25024 (to S.V.D.) from the National Institutes of Health.
Footnotes
Gene Expression Omnibus accession number: GSE41709
REFERENCES
- Alisi A, Panera N, Agostoni C, Nobili V. Intrauterine growth retardation and nonalcoholic Fatty liver disease in children. Int J Endocrinol. 2011;2011:269853. doi: 10.1155/2011/269853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ. Childhood causes of adult diseases. Arch Dis Child. 1988;63:867–869. doi: 10.1136/adc.63.7.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ. Developmental origins of adult health and disease. J Epidemiol Community Health. 2004;58:114–115. doi: 10.1136/jech.58.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Bull AR, Osmond C, Simmonds SJ. Fetal and placental size and risk of hypertension in adult life. BMJ. 1990;301:259–262. doi: 10.1136/bmj.301.6746.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MJ, So AY, Kaasik K, Krueger KC, Pillsbury ML, Fu YH, Ptacek LJ, Yamamoto KR, Feldman BJ. Circadian rhythm gene period 3 is an inhibitor of the adipocyte cell fate. J Biol Chem. 2011;286:9063–9070. doi: 10.1074/jbc.M110.164558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ercole AJ, Applewhite GT, Underwood LE. Evidence that somatomedin is synthesized by multiple tissues in the fetus. Dev Biol. 1980;75:315–328. doi: 10.1016/0012-1606(80)90166-9. [DOI] [PubMed] [Google Scholar]
- Dai Y, Thamotharan S, Garg M, Shin BC, Devaskar SU. Superimposition of postnatal calorie restriction protects the aging male intrauterine growth- restricted offspring from metabolic maladaptations. Endocrinology. 2012;153:4216–4226. doi: 10.1210/en.2012-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann R, Weaver DR. Altered body mass regulation in male mPeriod mutant mice on high-fat diet. Chronobiol Int. 2010;27:1317–1328. doi: 10.3109/07420528.2010.489166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BS, Beigneux AP, Fong LG, Young SG. New wrinkles in lipoprotein lipase biology. Curr Opin Lipidol. 2012;23:35–42. doi: 10.1097/MOL.0b013e32834d0b33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Takahashi Y, Sasaki Y, Saito T, Kamataki T. Novel gender-related regulation of CYP2C12 gene expression in rats. Mol Endocrinol. 2005;19:1181–1190. doi: 10.1210/me.2004-0063. [DOI] [PubMed] [Google Scholar]
- Eriksson JG, Osmond C, Kajantie E, Forsen TJ, Barker DJ. Patterns of growth among children who later develop type 2 diabetes or its risk factors. Diabetologia. 2006;49:2853–2858. doi: 10.1007/s00125-006-0459-1. [DOI] [PubMed] [Google Scholar]
- Garg M, Thamotharan M, Dai Y, Lagishetty V, Matveyenko AV, Lee WN, Devaskar SU. Glucose intolerance and lipid metabolic adaptations in response to intrauterine and postnatal calorie restriction in male adult rats. Endocrinology. 2013;154:102–113. doi: 10.1210/en.2012-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg M, Thamotharan M, Dai Y, Thamotharan S, Shin BC, Stout D, Devaskar SU. Early postnatal caloric restriction protects adult male intrauterine growth-restricted offspring from obesity. Diabetes. 2012;61:1391–1398. doi: 10.2337/db11-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Yin L, Ma H, Li T, Chiang JY, Zhang Y. Aldo-keto reductase 1B7 is a target gene of FXR and regulates lipid and glucose homeostasis. J Lipid Res. 2011;52:1561–1568. doi: 10.1194/jlr.M015859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Morton SM, Pinal CS. Life-long echoes--a critical analysis of the developmental origins of adult disease model. Biol Neonate. 2005;87:127–139. doi: 10.1159/000082311. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Pinal CS. Regulation of fetal growth by the somatotrophic axis. J Nutr. 2003;133:1741S–1746S. doi: 10.1093/jn/133.5.1741S. [DOI] [PubMed] [Google Scholar]
- Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller SR, Besharse JC. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci U S A. 2007;104:9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich A, Nedbal S, Blum WF, Erhard M, Lahm H, Brem G, Kolb HJ, Wanke R, Wolf E. Growth inhibition in giant growth hormone transgenic mice by overexpression of insulin-like growth factor-binding protein-2. Endocrinology. 2001;142:1889–1898. doi: 10.1210/endo.142.5.8149. [DOI] [PubMed] [Google Scholar]
- Horrevoets AJ, Fontijn RD, van Zonneveld AJ, de Vries CJ, ten Cate JW, Pannekoek H. Vascular endothelial genes that are responsive to tumor necrosis factor-alpha in vitro are expressed in atherosclerotic lesions, including inhibitor of apoptosis protein-1, stannin, and two novel genes. Blood. 1999;93:3418–3431. [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jelinek DF, Andersson S, Slaughter CA, Russell DW. Cloning and regulation of cholesterol 7 alpha-hydroxylase, the rate-limiting enzyme in bile acid biosynthesis. J Biol Chem. 1990;265:8190–8197. [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Chillaron JC, Patti ME. To catch up or not to catch up: is this the question? Lessons from animal models. Curr Opin Endocrinol Diabetes Obes. 2007;14:23–29. doi: 10.1097/MED.0b013e328013da8e. [DOI] [PubMed] [Google Scholar]
- Jugessur A, Shi M, Gjessing HK, Lie RT, Wilcox AJ, Weinberg CR, Christensen K, Boyles AL, Daack-Hirsch S, Nguyen TT, Christiansen L, Lidral AC, Murray JC. Maternal genes and facial clefts in offspring: a comprehensive search for genetic associations in two population-based cleft studies from Scandinavia. PLoS One. 2010;5:e11493. doi: 10.1371/journal.pone.0011493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajantie E, Osmond C, Barker DJ, Forsen T, Phillips DI, Eriksson JG. Size at birth as a predictor of mortality in adulthood: a follow-up of 350 000 person-years. Int J Epidemiol. 2005;34:655–663. doi: 10.1093/ije/dyi048. [DOI] [PubMed] [Google Scholar]
- Kamataki T, Maeda K, Yamazoe Y, Nagai T, Kato R. Sex difference of cytochrome P-450 in the rat: purification, characterization, and quantitation of constitutive forms of cytochrome P-450 from liver microsomes of male and female rats. Arch Biochem Biophys. 1983;225:758–770. doi: 10.1016/0003-9861(83)90087-5. [DOI] [PubMed] [Google Scholar]
- Kawai M, Green CB, Horowitz M, Ackert-Bicknell C, Lecka-Czernik B, Rosen CJ. Nocturnin: a circadian target of Pparg-induced adipogenesis. Ann N Y Acad Sci. 2010a;1192:131–138. doi: 10.1111/j.1749-6632.2009.05221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Green CB, Lecka-Czernik B, Douris N, Gilbert MR, Kojima S, Ackert-Bicknell C, Garg N, Horowitz MC, Adamo ML, Clemmons DR, Rosen CJ. A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPAR-gamma nuclear translocation. Proc Natl Acad Sci U S A. 2010b;107:10508–10513. doi: 10.1073/pnas.1000788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho Y, Kim S, Yoon BS, Moon JH, Kwak S, Park G, Woo J, Oh S, Hong K, Kim H, You S, Choi Y. WDNM1 is associated with differentiation and apoptosis of mammary epithelial cells. Anim Biotechnol. 2008;19:89–103. doi: 10.1080/10495390801887361. [DOI] [PubMed] [Google Scholar]
- Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol. 2004;24:5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsson A. Shift work and coronary heart disease. Scand J Soc Med Suppl. 1989;44:1–36. [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(ΔDelta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martyn CN, Gale CR, Jespersen S, Sherriff SB. Impaired fetal growth and atherosclerosis of carotid and peripheral arteries. Lancet. 1998;352:173–178. doi: 10.1016/S0140-6736(97)10404-4. [DOI] [PubMed] [Google Scholar]
- Nobili V, Alisi A, Panera N, Agostoni C. Low birth weight and catch-up-growth associated with metabolic syndrome: a ten year systematic review. Pediatr Endocrinol Rev. 2008;6:241–247. [PubMed] [Google Scholar]
- Oak SA, Tran C, Pan G, Thamotharan M, Devaskar SU. Perturbed skeletal muscle insulin signaling in the adult female intrauterine growth-restricted rat. Am J Physiol Endocrinol Metab. 2006;290:E1321–E1330. doi: 10.1152/ajpendo.00437.2005. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Gomi T, Konishi K, Date T, Nakashima H, Nose K, Matsuda Y, Peraino C, Pitot HC, Fujioka M. Human liver serine dehydratase. cDNA cloning and sequence homology with hydroxyamino acid dehydratases from other sources. J Biol Chem. 1989;264:15818–15823. [PubMed] [Google Scholar]
- Ooi GT, Hurst KR, Poy MN, Rechler MM, Boisclair YR. Binding of STAT5a and STAT5b to a single element resembling a gamma-interferon-activated sequence mediates the growth hormone induction of the mouse acid-labile subunit promoter in liver cells. Mol Endocrinol. 1998;12:675–687. doi: 10.1210/mend.12.5.0115. [DOI] [PubMed] [Google Scholar]
- Page NM, Butlin DJ, Lomthaisong K, Lowry PJ. The human apolipoprotein L gene cluster: identification, classification, and sites of distribution. Genomics. 2001;74:71–78. doi: 10.1006/geno.2001.6534. [DOI] [PubMed] [Google Scholar]
- Pendergast JS, Niswender KD, Yamazaki S. Tissue-specific function of Period3 in circadian rhythmicity. PLoS One. 2012;7:e30254. doi: 10.1371/journal.pone.0030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa JJ, Mangelsdorf DJ. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annu Rev Cell Dev Biol. 2000;16:459–481. doi: 10.1146/annurev.cellbio.16.1.459. [DOI] [PubMed] [Google Scholar]
- Schmidt DR, Schmidt S, Holmstrom SR, Makishima M, Yu RT, Cummins CL, Mangelsdorf DJ, Kliewer SA. AKR1B7 is induced by the farnesoid × receptor and metabolizes bile acids. J Biol Chem. 2011;286:2425–2432. doi: 10.1074/jbc.M110.181230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Williams L, Vuguin PM, Charron MJ. Minireview: Epigenetic programming of diabetes and obesity: animal models. Endocrinology. 2012;153:1031–1038. doi: 10.1210/en.2011-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin BC, Dai Y, Thamotharan M, Gibson LC, Devaskar SU. Pre- and postnatal calorie restriction perturbs early hypothalamic neuropeptide and energy balance. J Neurosci Res. 2012;90:1169–1182. doi: 10.1002/jnr.23013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohi G, Marchand K, Revesz A, Arany E, Hardy DB. Maternal protein restriction elevates cholesterol in adult rat offspring due to repressive changes in histone modifications at the cholesterol 7alpha-hydroxylase promoter. Mol Endocrinol. 2011a;25:785–798. doi: 10.1210/me.2010-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohi G, Revesz A, Hardy DB. Permanent implications of intrauterine growth restriction on cholesterol homeostasis. Semin Reprod Med. 2011b;29:246–256. doi: 10.1055/s-0031-1275523. [DOI] [PubMed] [Google Scholar]
- Vaiman D, Gascoin-Lachambre G, Boubred F, Mondon F, Feuerstein JM, Ligi I, Grandvuillemin I, Barbaux S, Ghigo E, Achard V, Simeoni U, Buffat C. The intensity of IUGR-induced transcriptome deregulations is inversely correlated with the onset of organ function in a rat model. PLoS One. 2011;6:e21222. doi: 10.1371/journal.pone.0021222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuguin PM. Animal models for small for gestational age and fetal programming of adult disease. Horm Res. 2007;68:113–123. doi: 10.1159/000100545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Osterbur DL, Megaw PL, Tosini G, Fukuhara C, Green CB, Besharse JC. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev Biol. 2001;1:9. doi: 10.1186/1471-213X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.