Abstract
Background: Dual-specificity phosphatase 6 (DUSP6) is a negative feedback mechanism of the mitogen-activated protein (MAP) kinase superfamily (MAPK/ERK, SAPK/JNK, p38), that is associated with cellular proliferation and differentiation. It has been reported that the expression of DUSP6 in different types of breast cancer is diverse and therefore it has altered functions in various types of breast cancer. Our aim was to explore the exact function of DUSP6 in triple-negative breast cancer cells (MDA-MB-231 cell) and to determine whether the suppression of DUSP6 by small interfering RNA (siRNA) and mircroRNA (miRNA) inhibits the growth of human MDA-MB-231 breast cancer cells. Methods: DUSP6-siRNA was used to inhibit the expression of DUSP6 directly and miR-145 to inhibit the expression of DUSP6 either in MDA-MB-231 breast cancer cells and successful transfection being confirmed by Real-time PCR and Western Blotting. Down regulation of DUSP6 in MDA-MB-231 cells suppressed the cell proliferation as investigated by MTT assay and colony form assay. Transwell test and Scratch assay were conducted to investigate the migration and invasion of MDA-MB-231 cells. T-test (two-tailed) was used to compare differences between groups, and the significance level was set at P<0.05. Results: DUSP6 mRNA expression and protein expression were reduced after transfection with DUSP6-siRNA directly and similar trend with transfection with miR-145. The treated group with DUSP6-siRNA or miR-145 suppressed MDA-MB-231 cells proliferation, migration and invasion, and meanwhile the cells were arrested at G0/G1 phase. Conclusions: DUSP6 plays a role in triple-negative breast cancer cells that might promote growth in MDA-MB-231 triple-negative breast cancer cells.
Keywords: siRNA, miR-145, DUSP6, breast cancer, MDA-MB-231 cells
Introduction
Breast cancer is the most common form of cancer among women in the USA, where it is estimated that breast cancer will account for 207,090 new cancer cases every year and it is predicted to cause 39,840 deaths annually, which was ranked second among women in the USA in 2010 [1]. Small interfering RNA (siRNA) is a kind of small molecular RNA (21-25 nucleotides), as a member of the siRISC it arouses the complementary target mRNA silencing directly [2]. MicroRNAs (miRNAs) are a kind of small non-coding RNAs that control gene expressions by targeting mRNAs for translational repression or cleavage [3]. Both are small (~22nt) noncoding RNAs that once into the cyto RNA-induced Complex (RISC), bind to their targeting mRNA and impair translation, have functions in regulating cellular differentiation, proliferation, and apoptosis [4].
Dual specificity phosphatase 6 (DUSP6) is one member of the family of mitogen-activated protein kinase (MAPK) phosphatase (MKPs) or cysteine-dependent DUSPs that specially dephosphorylates extracellular-signal-regulated kinase (ERK), so functioned as a negative regulator of ERK, working as a key effector of MAPK signal pathway [5]. In different cancer types, the expression of DUSP6 is different, what that means is that the function of DUSP6 is not fixed [6]. For instances, previous study has reported that in Myeloma, Melanoma, Glioma and some other kinds of cancer, the expression of DUSP6 is increased, which indicated that DUSP6 may be a tumor promotor [6], while in pancreatic invasive cancer, primary lung cancer, and ovarian cancer, the expression of DUSP6 is decreased, meaning DUSP6 may be a tumor suppressor [7-9].
It was found that in estrogen receptor-negative breast cancer, the expression of DUSP6 is high while in estrogen receptor-positive breast cancer, the expression is lost [10]. What is exactly the function of DUSP6 in breast cancer is still not clear, so in this experiment, we explored its’ function in triple-negative breast cancer, knockdown by RNAi and miRNA so that it can be a new treatment to triple-negative breast cancer with the help of technology of RNA delivery.
Materials and methods
Cell lines, culture and transfection
Human breast cancer cell line MDA-MB-231 was purchased from Chinese Academy of Sciences (Shanghai, China). The MDA-MB-231 cells were cultured in DMEM/high glucose medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA, USA), penicillin (100 U/ml) and streptomycin (100 µg/ml) (Enpromise, China). Cells were incubated at 37°C in a humidified chamber supplemented with 5% CO2. The MDA-MB-231 cells were cultured to 30~40% confluence in 6-well plates and were transfected with siRNA of DUSP6 or miR-145 mimics (Genepharma Co, Ltd. Shanghai, China) at working concentrations using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), in accordance with the manufacturer’s instructions. SiRNA- and miR- negative control (NC) were used as negative controls. After 48 h of incubation, cells were harvested for further analysis. All transfections were performed in triplicates.
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
For detection of mRNA of DUSP6 expression, primer design and qRT-PCR were carried out. cDNA was generated by reverse transcription using the PrimeScript™ RT-PCR kit in accordance with the manufacturer’s instructions (Takara, Tokyo, Japan). Real time PCR was performed on a 7900HT Fast RT-PCR instrument (Applied Biosystems, Singapore). The amplification procedure was as follows: 5 min at 95°C, followed by 40 cycles at 95°C for 30 sec and 65°C for 45 sec. Expression of mRNA was assessed by evaluating threshold cycle (CT) values. The CT values of the DUSP6 were normalized with the expression level of GAPDH. The primer sequences were as follows: 5’-AAC AGG GTT CCA GCA CAG CAG-3’ (forward) and 5’-GGC CAG ACA CAT TCC AGC AA-3’ (reverse) for DUS-P6; 5’-CATGAGAAGTATGACAACAGCCT-3’ (forward) and 5’-AGTCCTTCCACGATACCAAAGT-3’ (reverse) for GAPDH. The quantification experiment was done in triplicate for every sample.
Cell proliferation assay
Cell proliferation was determined using an MTT assay kit (Sigma, Santa Clara, CA, USA) in accordance with the manufacturer’s instructions. Briefly, cells (2×103 cells/well) that transfected with miR- or siRNA- mimics or NC mimics were seeded into 96-well culture plates (BD Biosciences, Franklin Lakes, NJ, USA) and incubated overnight at 37°C in 5% CO2. Cell proliferation was assessed at 24, 48, 72 and 96 hours, following addition of 0.5 mg/ml MTT (Sigma) solution. After a 4-hr incubation, the medium was replaced with 150 µl dimethylsulfoxide (DMSO; Sigma) and vortexed for 10 min. The optical density (OD) of each well was measured using a microplate reader at 490 nm. Each experiment was performed in triplicate.
Colony formation assay
SiRNA or miRNA mimics groups and control are the same with MTT assay. After 24 hours. Transfection, MDA-MB-231 cells were digested with trypsin and suspended into a single cell status. 500 cells from each group were cultured in the 6-well plate for 14 days. When the colonies were visible by eye, the culture was terminated by removing the medium and washing cells twice with phosphate-buffered saline (PBS). The colonies were fixed with 95% ethanol for 10 min, dried and stained with 0.1% Crystal Violet solution for 10 min, and the plate was washed three times with water. Colonies less than 2 mm in diameter and faintly stained were ignored. Each treatment was performed in triplicate.
Scratch assay
The “scratch” assay, which used for evaluating cell migration, was also called the wound healing assay. MDA-MB-231 cells were transfected with siDUSP6 mimics (100 nM) or NC mimics, and when cells reached 90% confluence, a scratch was made through each well using a sterile pipette tip. Cells were monitored under the microscope (magnification, ×50) for 0, 12, 24 and 36 h after wounding. Images of cells were captured at the same position before and after incubation to document the repair process. The experiments were repeated three times.
Transwell invasion and migration assay
The transwell invasion assay was performed to evaluate cell invasion ability. The filters (Corning, Lowell, MA, USA) were washed with serum-free DMEM, and placed into a 24-well plate. The lower chambers contained DMEM with 10% FBS. For the upper chambers, 2×104 cells, re-suspended in 200 µl DMEM with 0.1% BSA, were plated in the top chamber of Transwells (Millipore) with a Matrigel (2 mg/ml)-coated membrane containing 8-mm diameter pores. Plates were then incubated at 37°C in 5% CO2. After 18 hours, cells remaining on the upper membrane surface were removed by cotton swab scrubbing; cells on the lower surface of the membrane were fixed in 10% formalin at room temperature for 30 min and stained with 0.5% crystal violet. Images of six randomly selected fields-of-view were captured, and the cells were counted. For the migration assays, the infected cells (2×104 cells/Transwell) were plated in the top chamber with no Materiel. After 18 hours, the number of migrated cells was counted as described above. The stained cells were dissolved in glacial acetic acid, and solutions were transferred to a 96-well culture plate for colorimetric reading of OD at 560 nm. The OD value represents the invasive ability. Each experiment was carried out in triplicate.
Dual-luciferase reporter assay
HEK293T cells were seeded in 12-well plates (BD, USA) and cultured until the cells reached 80-90% confluence. The 3’-UTR segments of the DUSP6 mRNA sequence containing the predicted miR-145 binding sites were amplified by PCR in a total volume of 50 µl using the Primer Star kit (Takara, Tokyo, Japan) in accordance with the manufacturer’s instructions. The corresponding mutant constructs were created by mutating the seed regions of the miR-145-binding sites (5’-AACUGGAA-3’ to 5’-UUGACCUU-3’). The mutant constructs were generated by mutation. The primers used in the reaction were (Forward: 5’-AGTAATTCTAGGCGATCGCTCGAGAAGAAA-3’; Reverse: 5’-GATATTTTATTGCGGCCAGCGGCCGCTAAA-3’). Fragments were sub cloned into the XhoI site in the 3’-UTR of Renillaluciferase of the psiCHECK-2 reporter vector. 293T cells were transiently co-transfected with 0.2 µg psiCHECK-2/DUSP6 3’-UTR or psiCHECK-2/DUSP6 3’-UTR mutant reporter plasmids and together with 100 nmol/l miR-145 or miR-NC using Lipofectamine™ 2000 (Invitrogen), according to the manufacturer’s instructions. After 48 hours, firefly and Renillaluciferase activities were measured by using a Dual Luciferase Assay (Promega, Madison, WI, USA). Firefly luciferase values were normalized to Renilla, and the ratio of Firefly/Renilla was presented.
Western blot analysis
Cells were lysed in lysis buffer (10 mmol/L Tris-HCl, pH 7.4, 1% NP-40, 0.1% deoxycholic acid, 0.1% SDS, 150 mmol/L NaCl, 1 mM EDTA and 1% protease inhibitor cocktail) (Sigma). The protein concentrations were quantified using a BCA protein assay kit (Pierce). Protein was separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (Beyotime Institute of Biotechnology, Jiangsu, China). The membrane was immunoblotted overnight at 4°C with primary antibodies against DUSP6 (1:1,000, Abcam, USA) and β-tubulin (1:2,000, Cwbio, Jiangsu, China), as a loading control. Horseradish peroxidase-conjugated secondary antibodies were incubated with the membrane for 30 minutes at 37°C after three washes with TBST. Immunoreactive protein bands were detected with an Odyssey Scanning system.
Statistical analysis
GraphPad Prism version 6.0 (GraphPad, San Diego, CA, USA) was used for all statistical analyses. Data was presented as the means ± standard deviation from at least three separate experiments. The t-test (two-tailed) was used to draw a comparison between groups, and the significance level was set at P<0.05.
Results
Expression of DUSP6 directly downregulated by DUSP6-siRNA and the growth of MDA-MB-231 cells inhibited
The siRNA of DUSP6 was transferred into MDA-MB-231 breast cancer cells with lipofectamine 2000. After 36 hours. Real time PCR was conducted to investigate the gene level of DUSP6 and after 60 hours, western blot experiment was conducted to investigate the protein level of DUSP6 that was affected by DUSP6-siRNA. As is shown in Figure 1A and 1B, the expression of DUSP6 was down regulated with siDUSP6, compared with the siRNA-NC group both at gene or protein level. When transferred with siDUSP6, the proliferation of MDA-MB-231 breast cancer cells was directly inhibited, compared with NC group from 48 hours to 120 hours, at 50 nm or 100 nm concentration, in a time- and concentration-dependent manner (Figure 1C), detected by MTT assays that optical density (OD) values at 490 nm. Colony formation assays showed much less colony formation in the group transfected with 100 nM siDUSP6 compared with the NC group (64 ± 4 siDUSP6 group vs. 112 ± 7 NC group, **P<0.01) (Figure 1D, 1E). These results indicated that DUSP6-siRNA suppressed MDA-MB-231 cellular proliferation.
Figure 1.
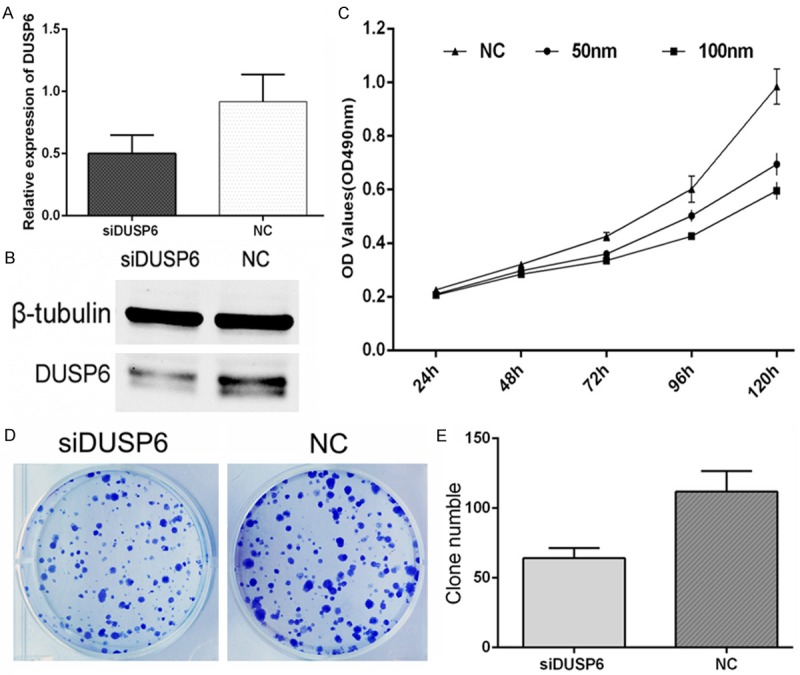
Down regulation of DUSP6 inhibited the growth of MDA-MB-231 cells. A and B. siRNA of DUSP6 (siDUSP6) down regulated the expression of DUSP6 in mRNA and protein; C. The proliferation of cells transfected with siDUSP6 mimics was inhibited by MTT cell proliferation assays; D and E. Colony formation assays showed Crystal Violet staining of the siDUSP6-transfected group and NC-transfected group (Comparison was draw by the t-test (two-tailed). Data represent means ± SD; *P<0.05).
DUSP6-siRNA directly inhibited MDA-MB-231 cells migration and invasion
To investigate how siDUSP6 affects cellular migration and invasion, we performed wound healing assays and transwell assays with MDA-MB-231 cells transfected with siDUSP6 mimics (100 nM) or NC mimics. The wound healing assay results showed that the migration ability of the siDUSP6 mimic groups was lower than the NC group. As is shown in Figure 2, the cell-free area of the siDUSP6 group was significantly wider than the NC group at 24 hours after drawing the “scratch” line on the monolayer cells. While the NC group filled in the gap at 36 hours. The monolayer of siDUSP6 group-transfected cells still showed a clear gap in the scratched region. In transwell invasion assays, the number of invaded cells stained with Crystal Violet was significantly less in the siDUSP6 (100 nM) group (Figure 3A). Absorbance at 573 nm showed that breast cancer cells invaded through matrigel in 100 nM DUSP6-siRNA group (0.38 ± 0.0118) was statistically reduced than NC group (0.56 ± 0.0321) in MDA-MB-231 cells (Figure 3B) and confirmed the results observed by inverted microscope (Figure 3C). These results showed that siRNA of DUSP6 can suppress cellular migration and invasion ability.
Figure 2.
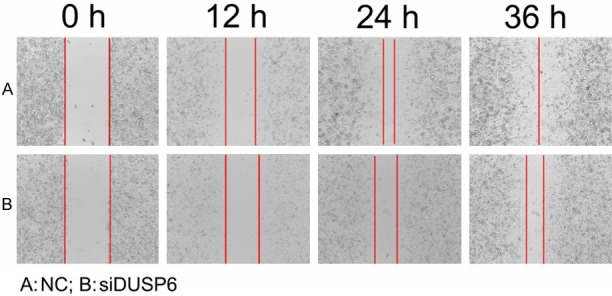
siDUSP6 showed impaired migration in wound healing assays. A. Images showed the gap of the scratched region of the NC group cells; B. Image showed the region of the siDUSP6 group cells.
Figure 3.
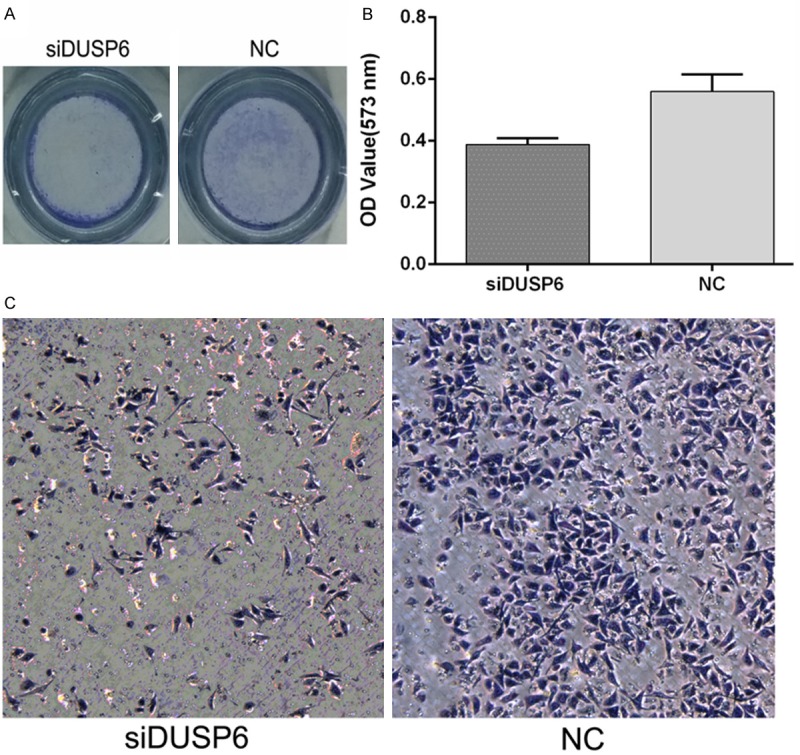
siDUSP6 affected the invasion of MDA-MB-231 cells. A and C. Overall pictures and pictures on an inverted microscope with ×50 magnification; B. Invasion rates were determined by solubilization of crystal violet and spectrophotometric reading at OD 573 nm (Comparison was draw by the t-test (two-tailed). Data represent means ± SD; *P<0.05).
Downregulation of DUSP6 by miR-145 suppressed the growth, migration, invasion of MDA-MB-231 cells
To further study the function of DUSP6, we decided to use miR-145 to inhibit the expression of DUSP6, and explore the function of miR-145, partially through down regulation of DUSP6. To investigate whether DUSP6 is a potential targets of miR-145, we searched several online databases, including targetscan, miRanda and miRBase. All databases showed that miR-145 had a binding site in the 3’-UTR of DUSP6 mRNA. To verify miR-145 binding to this predicted site, we performed a luciferase reporter assay in the 293T cell line. We cloned the 3’-UTR of DUSP6 containing the putative miR-145 binding site into a luciferase reporter construct, in addition to a mutated DUSP6 3’-UTR (Figure 4A). The results (Figure 4C) showed that the luciferase activity significantly decreased after co-transfection with psiCHECK-2/DUSP6 3’-UTR and miR-145 mimics in comparison with control cells, demonstrated that miR-145 specifically binds to the 3’-UTR of DUSP6 mRNA. Additionally, western blot analysis indicated that DUSP6 protein levels were lower in the miR-145-overexpressing group compared with the NC group (Figure 4B). MiR-145 was transferred into MDA-MB-231 cells with 100 nM, and significantly inhibited the growth of MDA-MB-231 cells (Figure 4D-F). The ability of migration (Figure 5A, 5C) and invasion (Figure 5B, 5D) of MDA-MB-231 cells were also suppressed by miR-145. All these data’s indicated that miR-145 partially targeted DUSP6 to play a role in MDA-MB-231 cells and further confirmed that the DUSP6 promoted the growth and ability of migration and invasion in MDA-MB-231 cells.
Figure 4.
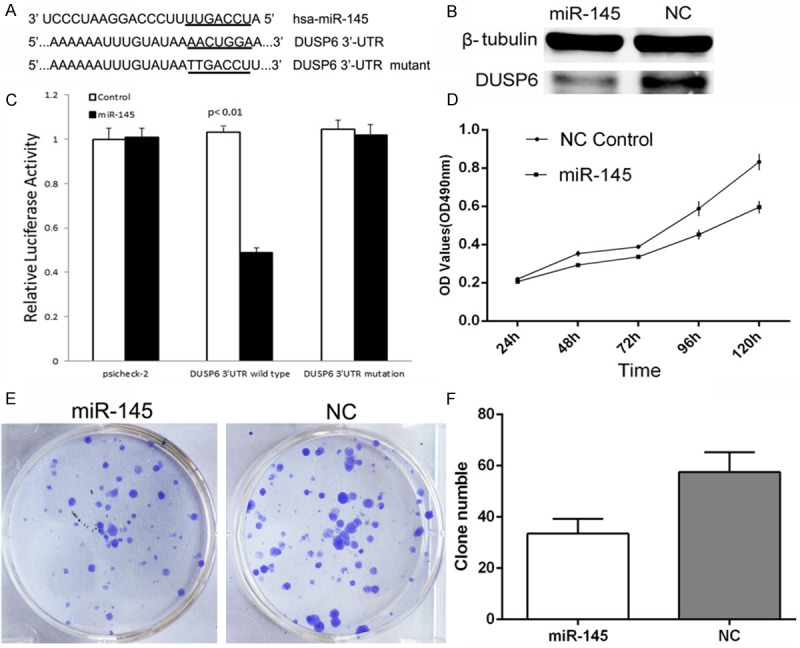
DUSP6 is a direct target of miR-145 and miR-145 inhibited the growth of MDA-MB-231 cells. A. The binding site for miR-145 in the 3’-UTR of DUSP6 mRNA; B. Western blot analysis was used to detect DUSP6 protein expression levels; C. The relative luciferase activity (Renilla/firefly) was measured in HEK293T cells after co-transfection of the DUSP6 3’-UTR or DUSP6 3’-UTR mutant luciferase construct with either miR-145 mimics or miR-NC; D-F. MTT and Colony formation assays showed Crystal Violet staining of the miR-145-transfected group and NC-transfected group (Comparison was draw by the t-test (two-tailed). Data represent means ± SD; *P<0.05).
Figure 5.
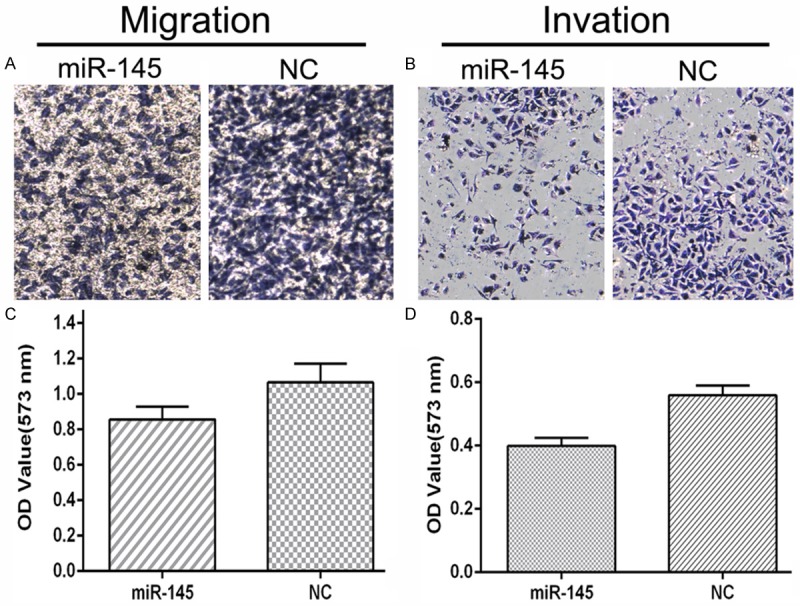
Overexpression of miR-145 inhibited cell migration and invasion ability. A and B. pictures of migration and invasion on an inverted microscope with ×50 magnification; C and D. Migration and invasion rates were determined by solubilization of crystal violet and spectrophotometric reading at OD 573 nm (Comparison was draw by the t-test (two-tailed). Data represent means ± SD; *P<0.05).
Downregulation of DUSP6 by siRNA or miRNA arrests the cell cycle of MDA-MB-231 cells
The effect of DSUP6 on the cell cycle progression was analyzed by flow cytometry by using both DUSP6-siRNA and miR-145 mimics in MDA-MB-231 cells compared with related NC mimics. The FACS results indicated that DUSP6-siRNA significantly increased the percentage of cells in G0/G1 compared to siRNA-NC group (Figure 6A). When transferred with miR-145 mimics, the similar results were gotten (Figure 6B). These results indicated that the down regulation of DUSP6, directly by siRNA-DUSP6 or by miR-145 could impact cell cycle progression in MDA-MB-231 cells.
Figure 6.
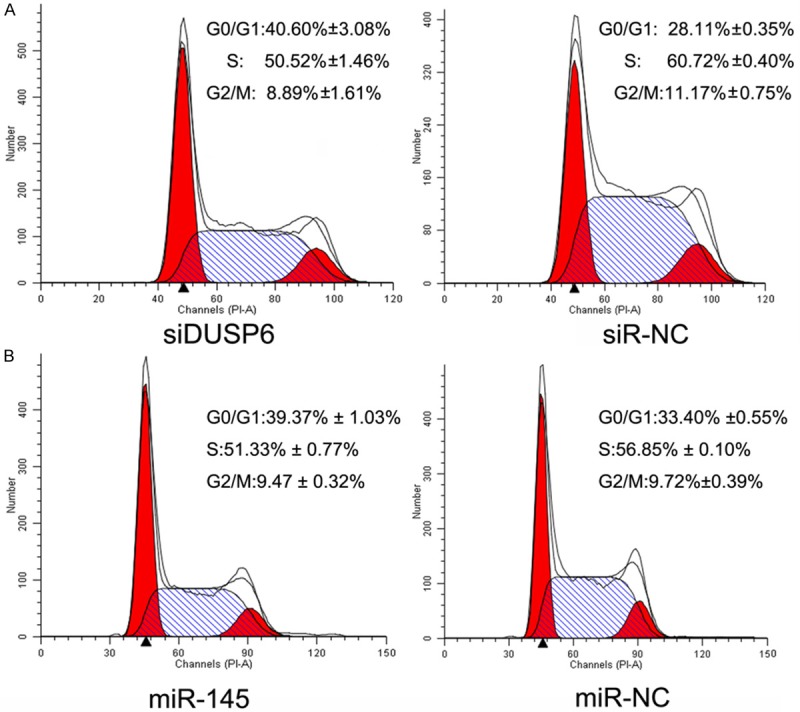
DUSP6 affected cell cycle distribution. A and B. Cell distribution was analyzed by flow cytometry 36 h after transfection of MDA-MB-231 cells with siDUSP6 or miR-145 mimics compared with NC group. SiDUSP6 and miR-145 arrested cells mostly in G0/G1 phase (Comparison was draw by the t-test (two-tailed). Data represent means ± SD; *P<0.05).
Discussion
The selective targeting of cancer cells would enable treatments to inhibit tumor growth and invasion while sparing the surrounding normal cells, reducing the adverse side effects of conventional chemotherapies. Therefore, therapeutic molecules that specially target cancer cells are the key to the development of patient-tailored cancer treatment [11]. Due to the versatility of the RNAi process, RNA-based gene therapy is considered as a potential approach to target various human diseases, effectively and safely [12]. Therefore, Using RNAi to inhibit the action of genes relevant for cancer cell survival and progression may be preferred than other inhibitors such as small molecules, peptides and antibodies, since the elevated target specificity is coupled to low toxicity [13]. For RNAi therapy, the alternative effective strategies for siRNA or miRNA delivery are essential and the ideal delivery system should be nontoxic, non-immunogenic, and sufficient for siRNA protection, to reach the target cell and facilitate cell uptake, and to release siRNA into the cytoplasm to achieve gene silencing [14]. Luckily various carriers have been reported to provide promising application in siRNA delivery in vitro and in vivo, such as liposomes, nanoparticles and inorganic materials, with the of progression of nanotechnology [14,15].
Since the ERK1/2 pathway mediates mitogenic signaling amongst other responses, DUSP6 has been proposed as a tumor suppressor, whose inactivation is frequently associated with cancer [16]. While in Manzano’s findings, their microarrays, showed that the expression of DUSP6 is just found in estrogen receptor-negative breast cancer, and not in estrogen receptor-positive breast cancer [10]. In our experiment, we found that in triple-negative MDA-MB-231 breast cancer cells, the expression of DUSP6, both in mRNA or protein levels, was significantly decreased with the transferation of siRNA of DUSP6. The proliferation of MDA-MB-231 cells was suppressed by the siRNA, as measured by MTT and colony formation assays, respectively. The migration and invasion of cells were also inhibited. Additionally, compared with the NC control group, flow cytometry showed that when transferred with siRNA of DUSP6, downregulation of DUSP6 by the siRNA arrested cells in the G0/G1 phase. We found that DUSP6 may act as a tumor promotor and siRNA of DUSP6 could inhibit the growth of MDA-MB-231 cells.
MiRNA-145, as reported in many other articles, downregulates the expression of cyclin-dependent kinase 6 in human cervical carcinoma cells to suppress the proliferation of Hela cells [17]. It also suppresses thyroid cancer growth and metastasis by targeting AKT3 [18]. In our study, we found that overexpression of miR-145 could significantly suppress the growth of MDA-MB-231 cells and the migration and invasion of cells were also inhibited by overexpression of miR-145, which was consistent with the finding studied by others [19,20]. What’s the difference is that their results showed that miR-145 regulated the cells’ growth by targeting ERBB3 and ARF6 while in our study miR-145 suppressed the MDA-MB-231 cells’ proliferation through targeting DUSP6. Based on the database of miRwalk, miRbase and Target scan, we constructed a psiCHECK-2 plasmid containing the 3’-UTR of DUSP6 (psiCHECK-2/DUSP6 3’-UTR). The dual luciferase assays confirmed that DUSP6 is a direct target gene of miR-145. The protein level of DUSP6 was downregulated by miR-145 and the flow cytometry also showed that overexpression of miR-145 could arrest the cells in G0/G1 phase. In the light of these results, we could get miR-145 to inhibit the growth of MDA-MB-231, at least partially through DUSP6.
As previously known that siRNAs complement their target mRNA, induce gene silencing through a sequence-specific cleavage of the target mRNA, whereas microRNAs shows a partial complement to target mRNA, mediate translational repression or transcript degradation [13]. Taking all these into consideration, we came to the conclusion that DUSP6 is a cancer gene in triple-negative breast cancer. Therefore, RNAi mediated silencing of the DUSP6 gene has inhibitory effects on cells’ proliferation, migration and invasion. Gene therapy, especially for RNAi to special gene, will play an important role in cancer therapy in near future with the progress got in research of gene carriers and nanotechnology.
Acknowledgements
This research was made possible with financial support from the National Natural sciences Foundation of China, for the project 81272240, and the Shanghai Science Committee Foundation (to L.F.) (no. STCSM 10411964700).
Disclosure of conflict of interest
None.
Abbreviations
- siRNA
Small interfering RNA
- miRNA
microRNA
- NC
Negative Control
- qPCR
quantitative polymerase chain reaction
- nm
nmol/l
- DUSP6
Dual specificity phosphatase 6
- FBS
Fetal bovine serum
- DMEM
Dulbecco’s modified Eagle’s medium
- RL
Renilla luciferase
- FL
Firefly luciferase
References
- 1.Cheng YC, Ueno NT. Improvement of survival and prospect of cure in patients with metastatic breast cancer. Breast Cancer. 2012;19:191–9. doi: 10.1007/s12282-011-0276-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li XY, Luo QF, Wei CK, Li DF, Fang L. siRNA-mediated silencing of CDK8 inhibits proliferation and growth in breast cancer cells. Int J Clin Exp Pathol. 2013;7:92–100. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Li D, Chen L, Hu Z, Li H, Li J, Wei C, Huang Y, Song H, Fang L. Alterations of microRNAs are associated with impaired growth of MCF-7 breast cancer cells induced by inhibition of casein kinase 2. Int J Clin Exp Pathol. 2014;7:4008–4015. [PMC free article] [PubMed] [Google Scholar]
- 4.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piya S, Kim JY, Bae J, Seol DW, Moon AR, Kim TH. DUSP6 is a novel transcriptional target of p53 and regulates p53-mediated apoptosis by modulating expression levels of Bcl-2 family proteins. FEBS Lett. 2012;586:4233–4240. doi: 10.1016/j.febslet.2012.10.031. [DOI] [PubMed] [Google Scholar]
- 6.Bermudez O, Pagès G, Gimond C. The dual-specificity MAP kinase phosphatases: critical roles in development and cancer. Am J Physiol Cell Physiol. 2010;299:C189–202. doi: 10.1152/ajpcell.00347.2009. [DOI] [PubMed] [Google Scholar]
- 7.Furukawa T, Fujisaki R, Yoshida Y, Kanai N, Sunamura M, Abe T, Takeda K, Matsuno S, Horii A. Distinct progression pathways involving the dysfunction of DUSP6/MKP-3 in pancreatic intraepithelial neoplasia and intraductal papillary-mucinous neoplasms of the pancreas. Mod Pathol. 2005;18:1034–1042. doi: 10.1038/modpathol.3800383. [DOI] [PubMed] [Google Scholar]
- 8.Chan DW, Liu VW, Tsao GS, Yao KM, Furukawa T, Chan KK, Ngan HY. Loss of MKP3 mediated by oxidative stress enhances tumorigenicity and chemoresistance of ovarian cancer cells. Carcinogenesis. 2008;29:1742–1750. doi: 10.1093/carcin/bgn167. [DOI] [PubMed] [Google Scholar]
- 9.Okudela K, Yazawa T, Woo T, Sakaeda M, Ishii J, Mitsui H, Shimoyamada H, Sato H, Tajiri M, Ogawa N, Masuda M, Takahashi T, Sugimura H, Kitamura H. Down-regulation of DUSP6 expression in lung cancer: its mechanism and potential role in carcinogenesis. Am J Pathol. 2009;175:867–881. doi: 10.2353/ajpath.2009.080489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manzano RG, Martinez-Navarro EM, Forteza J, Brugarolas A. Microarray phosphatome profiling of breast cancer patients unveils a complex phosphatase regulatory role of the MAPK and PI3K pathways in estrogen receptor-negative breast cancers. Int J Oncol. 2014;45:2250–2266. doi: 10.3892/ijo.2014.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toniatti C, Jones P, Graham H, Pagliara B, Draetta G. Oncology drug discovery: planning a turnaround. Cancer Discov. 2014;4:397–404. doi: 10.1158/2159-8290.CD-13-0452. [DOI] [PubMed] [Google Scholar]
- 12.Uchino K, Ochiya T, Takeshita F. RNAi therapeutics and applications of microRNAs in cancer treatment. Jpn J Clin Oncol. 2013;43:596–607. doi: 10.1093/jjco/hyt052. [DOI] [PubMed] [Google Scholar]
- 13.Esposito CL, Catuogno S, de Franciscis V. Aptamer-mediated selective delivery of short RNA therapeutics in cancer cells. J RNAi Gene Silencing. 2014;10:500–506. [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Li X, Huang L. Non-viral nanocarriers for siRNA delivery in breast cancer. J Control Release. 2014;190:440–450. doi: 10.1016/j.jconrel.2014.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozpolat B, Sood AK, Lopez-Berestein G. Liposomal siRNA nanocarriers for cancer therapy. Adv Drug Deliv Rev. 2014;66:110–116. doi: 10.1016/j.addr.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Díaz-García CV, Agudo-López A, Pérez C, Prieto-García E, Iglesias L, Ponce S, Rodríguez Garzotto A, Rodríguez-Peralto JL, Cortés-Funes H, López-Martín JA, Agulló-Ortuño MT. Prognostic value of dual-specificity phosphatase 6 expression in non-small cell lung cancer. Tumour Biol. 2015;36:199–206. doi: 10.1007/s13277-014-2729-8. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Wang L, Li B, Huo M, Mu M, Liu J, Han J. miR-145 downregulates the expression of cyclin-dependent kinase 6 in human cervical carcinoma cells. Exp Ther Med. 2014;8:591–594. doi: 10.3892/etm.2014.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boufraqech M, Zhang L, Jain M, Patel D, Ellis R, Xiong Y, He M, Nilubol N, Merino MJ, Kebebew E. miR-145 suppresses thyroid cancer growth and metastasis and targets AKT3. Endocr Relat Cancer. 2014;21:517–531. doi: 10.1530/ERC-14-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan X, Chen X, Liang H, Deng T, Chen W, Zhang S, Liu M, Gao X, Liu Y, Zhao C, Wang X, Wang N, Li J, Liu R, Zen K, Zhang CY, Liu B, Ba Y. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol Cancer. 2014;13:220. doi: 10.1186/1476-4598-13-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. lincRNA-RoR and miR-145 Regulate Invasion in Triple-negative Breast Cancer via Targeting ARF6. Mol Cancer Res. 2015;13:330–8. doi: 10.1158/1541-7786.MCR-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]


