Abstract
Cervical cancer is the second most common cancer among women worldwide and is the leading cause of deaths in developing countries. Persistent infections with a subset of HPVs, called “high-risk HPVs”, including HPV16 and HPV18, are the primary cause of cervical cancer. The apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) family of proteins is a group of cellular enzymes that catalyze the deamination of cytidine (C) to uracil (U) in single-stranded DNA/RNA, and functions as antiviral factors in the innate immune system of the host. Recent studies have shown that APOBEC3A could restrict certain DNA viruses, including HPVs. In this study, we confirmed that the expression of APOBEC3A was decreased in cervical cancer tissues. Furthermore, APOBEC3A inhibited the cervical cells proliferation, migration as well as invasion, and promoted apoptosis depend on cytidine deaminase. In addition, APOBEC3A decreased HPV16-E6, HPV16-E7 and HPV18-E6 depend on cytidine deaminase, but no effect on HPV18-E7. Therefore, we believe that, in cervical cancer cells, the expression of APOBEC3A possesses anticancer and antiviral effects by differential inhibition of HPV E6 and E7 expression depend on cytidine deaminase.
Keywords: APOBEC3A, cervical cancer, HPV E6, HPV E7
Introduction
The apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) family of proteins is a group of cellular enzymes that catalyze the deamination of cytidine (C) to uracil (U) in single-stranded DNA/RNA [1,2], and functions as antiviral factors in the innate immune system of the host [3]. In humans, the APOBEC family contains at least 11 members [4,5], including activation-induced cytidine deaminase and APOBEC1, -2, -3A, -3B, -3C, -3DE, -3F, -3G, -3H, and -4 (3). The APOBEC proteins target not only viral DNA/RNA but also the host genomic DNA, generating enriched clusters of C-to-T conversions in the genome [6-8]. Of these APOBEC3A (A3A) family members, hA3A, hA3B, and hA3H genes are expressed in skin keratinocytes, the primary host cell for HPV infection [9]. Recent studies have shown that APOBEC3A could restrict certain DNA viruses, including adeno-associated viruses and other parvoviruses [10-12]. In addition, other groups found evidence for editing of HPV DNA in precancerous lesions and cell lines [13]. The latter study (13) proved that APOBEC3A functions as a restriction factor of human papilloma viruses (HPVs). HPVs are small, nonenveloped DNA viruses known as one of the most prevalent sexually transmitted pathogens. Persistent infections with a subset of HPVs, called “high-risk HPVs”, including HPV16 and HPV18, are the primary cause of cervical cancer and some head-and-neck cancers [14-16]. HPV-induced carcinogenesis requires two viral oncoproteins, E6 and E7, which bind to and degrade p53 and Rb, respectively, thereby inhibiting these tumor suppressors and contributing significantly to the development of malignant cancer phenotypes [14,17]. However, there is currently no evidence that APOBEC3A proteins could directly interfere with the cervical cells proliferation, migration or invasion.
APOBEC3A cytidine deaminase has a canonical deaminase domain (H-x-E-x24-28-P-C-x-x-C) in which the glutamate has a role in proton transfer and the histidine and two cysteines coordinate a zinc cofactor [11,12]. To investigate the mechanism by which APOBEC3A regulates HPV infection, we tested the role of cytidine deaminase activity using the additional point mutations at conserved residues (H70 and F75) [11,12,18] which we denied mut-APOBEC3A.
In the present study, we discovered the expression of APOBEC3A between the normal cervical lesions and HPV-positive cervical lesions from early disease stages to cancer. Then we further explored the anticancer effects of APOBEC3A on cervical cancer cells. To ascertain whether cytidine deaminase activity is required for inhibition of cervical cancer development, we also analyze the properties of mut-APOBEC3A.
Materials and methods
Patient samples
48 paraffin-embedded specimens of cervical specimens were collected from 2008 to 2012 at the department of Pathology, The Six Affiliated Hospital Of Sun Yat-Sen University. Clinical stages were determined by two certified gynecologists according to a modified International Federation of Gynecology Obstetrics (FIGO) staging system for cervical cancer, which was published in 2009 [19]. The study was approved by the Institutional Research Ethics Committee of The Third Affiliated Hospital Of Sun Yat-Sen University. Consent was obtained from the patients and the clinical research materials were collected from the patients prior to clinical treatment. The samples included 22 cases of normal cervical tissues and 26 cases of cervical cancer. The normal cervical tissues were obtained from patients who underwent a hysterectomy for reasons other than neoplasia of the cervix without a history of CIN or abnormal liquid-based cytological test (LCT) smears. The cervical neoplasm specimens were obtained from patients who underwent an operation or cervical biopsy. Patients with acute pelvic infection or who had received a cytology examination within six months, cervical therapy or preoperative treatment (such as radiotherapy and chemotherapy) were excluded.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections of tissue sections were deparaffinized and rehydrated, and antigen activity was retrieved by microwaving the samples in 10 mM citrate buffer (pH 6.0) at 95°C for 15 min. After quenching endogenous peroxidase activity with H2O2 and blocking with normal goat serum, the sections were incubated with monoclonal antibodies against APOBEC3A (1:100; Abcam) according to the standard protocols at 4°C overnight, respectively. Subsequently, the samples were incubated with a biotinylated secondary antibody and visualized with 3- 3’-diaminobenzidine and counterstained with hematoxylin. Evaluations of the stained tissue sections were performed by two independent pathologists who were unaware of the clinical parameters of the patients at the time of evaluation. Immunoreactivity was semiquantitatively evaluated based on staining intensity and distribution using the immunoreactive score: immunoreactive score = intensity score × proportion score. The intensity score was defined as 0, negative; 1, weak; 2, moderate; or 3, strong. The proportion score was defined as 0, 0-25%; 1, 26-50%; 2, 51-75%, or 3, 76-100% positive cells. The total score ranged from 0 to 6. The immunoreactivity was divided to three levels on the basis of the final score: negative immunoreactivity was defined as a total score of 0-1, low immunoreactivity was defined as a total score of 2-3, moderate immunoreactivity was defined as a total score of 4-5, and high immunoreactivity was defined as a total score of 6.
Cell lines and cell culture
A total of 3 cervical cancer cell lines: SiHa (HPV 16-infected cervical cancer cell lines), HeLa (HPV 18-infected cervical cancer cell lines) and C33A (HPV-uninfected cervical cancer cell line) were used in this study. Cells were cultured in DMEM-H medium (from Hyclone) containing 10% fetal bovine serum at 37°C in a humidified atmosphere containing 5% CO2.
APOBEC3A vector
Full-length APOBEC3A coding sequences were PCR amplified and cloned into a pcDNA 3.1 expression vector (Invitrogen, Carlsbad, Califonia, US), following the manufacturer’s suggested protocol. Then mut-A3A using the additional point mutations at conserved residues (H70 and F75) which lost deaminase activity were generated by site-directed mutagenesis was constructed using the primer: Forward: TCTGTGGCTTTTACGGCCGCCGTGCGGAGCTGCGCTTGTTGGACCTGGTTCCTTCTTT; Reverse: AAAGAAGGAACCAGGTCCAACAAGCGCAGCTCCGCACGGCGGCCGTAAAAGCCACAGA.
DNA sequence was used to verify the constructs. The transfection efficiency of the APOBEC3A overexpression vector was verified via RT-PCR. Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, California, US) was used for transfection according to standard protocol.
RNA extraction, reverse transcription and quantitative real-time PCR
Total RNA was extracted using TRIzol (Takara, Japan) according to the manufacturer’s instructions. 1 μg total RNA was reverse transcribed using RT-PCR Kit (Takara, Japan). Quantitative PCR was carried out by the SYBR Green method (Takara, Japan) using a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, California, US). The primers were purchased from TaKaRa (Japan), and 18srRNA was used as an endogenous control. The sequences are shown in Table 1. The fold changes were calculated through relative quantification with 2-ΔΔCt. All of the reactions were performed in a 20 μL reaction volume in triplicate.
Table 1.
Primers used in this study
| Gene name | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| 18srRNA | CCTGGATACCGCAGCTAGGA | GCGGCGCAATACGAATGCCCC |
| APOBEC3A | GGCATTGGAAGGCATAAGAC | CAACTGCAAAGAAGGAACCA |
| HPV16 E6 | TATGCTGTATGTGATAAATG | TTTGTTGTATTGCTGTTC |
| HPV16 E7 | CAGAGGAGGAGGATGAAATAGA | AGCGTAGAGTCACACTTGCA |
| HPV18 E6 | CCGTTGAATCCAGCAGAAAA | TTGTGTTTCTCTGCGTCGTT |
| HPV18 E7 | AGCGACTCAGAGGAAGAAAA | CATTGTGTGACGTTGTGGTT |
MTS assays
For cell proliferation assays, cells transfected with vectors for 48 h were reseeded in 96-well plates at 10,000 cells/well in a final volume of 100 μL and incubated for 24 h, 48 h and 72 h. The effects of APOBEC3A on cell proliferation were determined with an MTS assay according to the standard protocol.
Cell cycle analysis
For cell cycle analysis, we used flow cytometric analysis. For flow cytometric analysis, cells transfected with vectors were plated in 6-well plates at 2×105 per well and incubated at 37°C for 48 h. The cells were then collected, washed with PBS and fixed with cold 70% ethanol at 4°C overnight. The cell cycle distribution was analyzed with propidium iodide (PI, Sigma-Aldrich) staining and flow cytometry.
Propidium iodide and FITC-conjugated annexin V staining
The cell apoptosis rate was detected by Annexin V FITC/Propidium Iodide Assay kit (KeyGEN, Nanjing, China), according to the manufacturer’s instructions. Briefly, cells were harvested by 0.25% Trypsin without EDTA after treatments and washed twice with cold PBS, and stained with Annexin V-FITC/PI staining kit according to manufacturer’s instructions, and then sorted by fluorescence activated cell sorting using a Beckman Coulter instrument.
In vitro migration and invasion assays
For the Transwell migration assays, 1×105 cells were plated in the top chamber of each insert (BD Biosciences, Canada). For the invasion assays, 1×105 cells were plated in the upper chamber of each insert, which had previously been coated with 150 mg of Matrigel (BD Biosciences, Canada). The cells in both assays were trypsinized and resuspended in FBS-free DMEM-H medium, and 600 μL of medium that had been supplemented with 10% FBS was added to the lower chambers in the migration assay wells and 20% FBS medium in the lower chambers in the invasion assay. After 12 hours or 24 hours of incubation at 37°C for migration or invasion assays, respectively, the cells remaining in the top chambers or on the upper membranes of the inserts were removed, and the cells that had migrated or invaded the lower surface were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The number of cells were imaged and counted using an IX71 inverted microscope (Olympus, Tokyo, Japan).
Western blot analysis
Protein lysates were resolved through 10% SDS-PAGE and electrophoretically transferred to a PVDF (polyvinylidene difluoride) membrane (Millipore). Then, the membrane was probed with a mouse monoclonal antibody against human APOBEC3A (1:300 dilution, Abcam) or HPV16 E6+HPV18 E6 (1:500, Abcam), HPV16 E7 (1:400, Abcam), HPV18 E7 (1:1000, Abcam) and β-actin (1:1000, Abcam) as a protein loading control. Horseradish peroxidase-conjugated secondary antibodies (1:10000, southern biotech) and an Immobilon TM Western chemiluminescent ECL kit (Millipore) were used to detect bound antibody. The intensity of the proteins was visualized with Fuji (Japan) SUPER RX-N-C films.
Result
APOBEC3A expression levels were low in the cervical cancer
The APOBEC3A expression was located in the nucleus and cytoplasm (Figure 1). Positive A3A immunoreactivity was detected in 46.15% (12/26) of cervical cancer and 86.36% (19/22) of normal cervical tissues. The A3A positive expression rates were significantly lower in the cancer group than in the normal tissue group (χ2 = 8.423, P<0.01).
Figure 1.
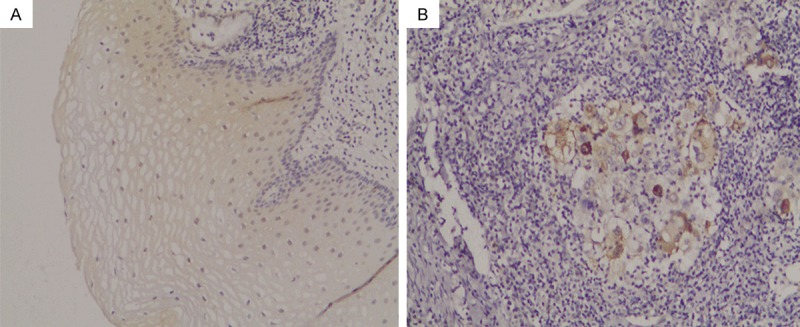
Immunohistochemical staining of APOBEC3A in cervical samples. APOBEC3A staining in normal cervical tissue (A) and cervical cancer (B). Image magnifications were 100×.
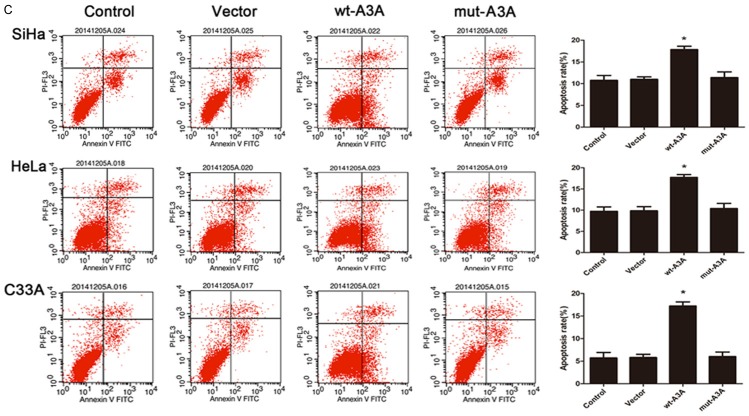
APOBEC3A expression inhibits growth and promotes apoptosis in cervical cancer cells
To determine the functional role of APOBEC3A in proliferation and apoptosis, we up-regulated APOBEC3A expression using an exogenous APOBEC3A expression vector including wt-A3A and mut-A3A in the three cervical cancer cells. The validation of A3A overexpression through plasmid was showed in Figure 2. Forty-eight hours after transfection, we performed a MTS assay to detect the OD values in 4 groups. As shown in Figure 3A, enhanced expression of wt-A3A inhibited cervical cancer cells proliferation. Also, the cell cycle distribution showed that after overexpression of wt-A3A, G1 phase cells proportion was increased and S phase cells proportion was decreased (Figure 3B), indicating that APOBEC3A inhibited cells growth. Meanwhile, the cell apoptosis rate was significantly increased when up-regulation of wt-A3A (Figure 3C). However, mut-A3A had no significant effect on the cell growth and apoptosis.
Figure 2.
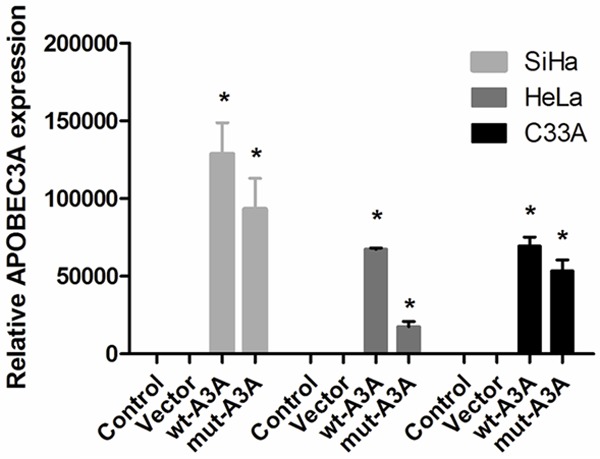
The effect of overexpression of APOBEC3A was validated by qRT-PCR in three cervical cancer cells.
Figure 3.
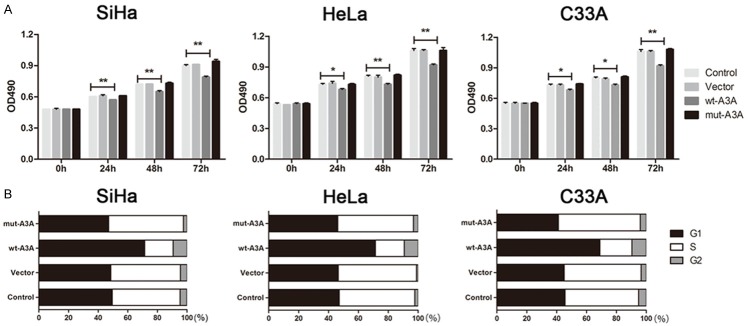
Overexpression of APOBEC3A inhibited growth and promoted apoptosis in SiHa, HeLa and C33A cells. A. Overexpression of wt-A3A inhibited proliferation in three cervical cells, but mut-A3A had no effect on proliferation. B. Overexpression of wt-A3A increased the G1 phase cells proportion and decreased the S phase cells proportion. Overexpression of mut-A3A had no differences. C. Over-expression of wt-A3A promoted the cervical cells apoptosis. *P<0.05, **P<0.01.
APOBEC3A expression attenuates migration and invasion in cervical cancer cells
We also examined whether APOBEC3A inhibited migration and invasion in cervical cancer cells. As shown in Figure 4, enhanced expression of wt-A3A attenuated cell invasion and migration in all the three cervical cancer cells, whereas mut-A3A had no significant effect on migration and invasion.
Figure 4.
APOBEC3A expression attenuated migration and invasion in SiHa, HeLa and C33A cells. A. Exogenous overexpression of wt-A3A attenuated cervical cell migration whereas mut-A3A had no effect. B. Overexpression of wt-A3A suppressed cervical cell invasion. Mut-A3A had no significant differences. *P<0.05, **P<0.01.
APOBEC3A overexpression has effect of HPV expression depend on cytidine deaminase
SiHa was a HPV 16-infected cervical cancer cell line and HeLa was a HPV 18-infected cervical cancer cell line, whereas C33A was a HPV-uninfected cervical cancer cell line. So we just used SiHa and HeLa for further research. As shown in Figure 5, in SiHa cells, wt-A3A reduced the mRNA and protein expression of HPV16-E6 and HPV16-E7. In HeLa cells, wt-A3A inhibited the expression of HPV18-E6 at the levels of both mRNA and protein. However, it had no effect on the expression of HPV18-E7 (Figure 5). Mut-A3A didn’t attenuate either HPV16-E6/E8 or HPV18-E6/E8.
Figure 5.

APOBEC3A overexpression has effect of HPV expression depend on cytidine deaminase. In SiHa cells, wt-A3A reduced the mRNA and protein expression of HPV16-E6 and HPV16-E7, whereas mut-A3A had no effect. In HeLa cells, wt-A3A inhibited the expression of HPV18-E6 at the levels of both mRNA and protein, but no effect on the expression of HPV18-E7. However, mut-A3A didn’t attenuate either HPV18-E6 or HPV18-E8.
Discussion
Cervical cancer is the second most common cancer among women worldwide and is the leading cause of deaths in developing countries [20,21]. Here, we confirmed that APOBEC3A was decreased in cervical cancer tissues compared with the normal tissue. We using overexpression of APOBEC3A in the cervical cancer cells proved that APOBEC3A could inhibit cell growth, migration and invasion, and promote cell apoptosis. What is more, this effect depends on cytidine deaminase. For HPV infection, APOBEC3A decreased HPV16-E6, HPV16-E7 and HPV18-E6 depend on cytidine deaminase, but no effect on HPV18-E7. In this report we provide multiple pieces of evidence of APOBEC3A anticancer and antiviral effects by differential inhibition of HPV E6 and E7 expression on cervical cancer through a cytidine deaminase dependent mechanism.
Constantly expanding knowledge about pathogenesis has shown that persistent infection with high-risk human papillomavirus types is necessary for the transformation of normal cervical epithelium to cervical precancer and invasive cancer in the vast majority of cases [22]. In 2008, Vartanian and colleagues revealed a potential role for APOBEC3A in restricting HPV infection. To link this observation to the APOBEC3A proteins, those authors showed that cotransfection of HPV DNA with APOBEC3A, APOBEC3C, and APOBEC3H expression plasmids resulted in substantial hypermutation [13]. Recently, some studies reported that APOBEC3A could decrease the infectivity of HPV, which is consistent with this study [23,24]. However, they had no deeper exploration about the function of APOBEC3A in cervical cancer cells growth. In this study, we demonstrated that APOBEC3A inhibited cervical cancer cells proliferation, migration and invasion.
APOBEC3A cytidine deaminase has a canonical deaminase domain (H-x-E-x24-28-P-C-x-x-C) in which the glutamate has a role in proton transfer and the histidine and two cysteines coordinate a zinc cofactor. In this study, we used the additional point mutations at conserved residues (H70 and F75) to investigate function of APOBEC3A. We proved that APOBEC3A anticancer effect on cervical cancer depend on cytidine deaminase.
In addition to providing evidence for APOBEC3A inhibition of cervical cancer, our study also offers insights into the HPV infection. The ability of APOBEC3A to restrict HPV is likely deaminase dependent, since catalytically inactive APOBEC3A could not restrict HPV. These results mirror data from previous studies suggesting a deaminase-dependent mechanism for APOBEC3A inhibition of retrotransposons and parvoviruses [10-12,25,26]. In our study, we showed that wt-APOBEC3A could decrease HPV16-E6, HPV16-E7 and HPV18-E6, whereas mut-APOBEC3A had no effect. This was consisted with the previous research. However, no difference was observed in HPV18-E7.
In summary, our study demonstrates that APOBEC3A could inhibit the cervical cancer cell proliferation, migration and invasion, which was deaminase dependent. Moreover, APOBEC3A decreased the expression of HPV-E6, HPV16-E7 and HPV18-E6. Further studies are required to clarify the specific mechanism of how APOBEC3A proteins mediate HPV infectivity defect and prevent cervical cancer development.
Acknowledgements
The work in this article was Supported by Medical Scientific Research Foundation of Guangdong Province, China (No. 2012B031800455).
Disclosure of conflict of interest
None.
References
- 1.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 2.Wijesinghe P, Bhagwat AS. Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic acids res. 2012;40:9206–9217. doi: 10.1093/nar/gks685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wedekind JE, Dance GS, Sowden MP, Smith HC. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 2003;19:207–216. doi: 10.1016/S0168-9525(03)00054-4. [DOI] [PubMed] [Google Scholar]
- 6.Suspene R, Aynaud MM, Guetard D, Henry M, Eckhoff G, Marchio A, Pineau P, Dejean A, Vartanian JP, Wain-Hobson S. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc Natl Acad Sci U S A. 2011;108:4858–4863. doi: 10.1073/pnas.1009687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen BR. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J Virol. 2006;80:1067–1076. doi: 10.1128/JVI.80.3.1067-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bishop KN, Holmes RK, Sheehy AM, Malim MH. APOBEC-mediated editing of viral RNA. Science. 2004;305:645. doi: 10.1126/science.1100658. [DOI] [PubMed] [Google Scholar]
- 9.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van T VL, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitt K, Guo K, Algaier M, Ruiz A, Cheng F, Qiu J, Wissing S, Santiago ML, Stephens EB. Differential virus restriction patterns of rhesus macaque and human APOBEC3A: implications for lentivirus evolution. Virology. 2011;419:24–42. doi: 10.1016/j.virol.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narvaiza I, Linfesty DC, Greener BN, Hakata Y, Pintel DJ, Logue E, Landau NR, Weitzman MD. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. 2009;5:e1000439. doi: 10.1371/journal.ppat.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 13.Vartanian JP, Guetard D, Henry M, Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320:230–233. doi: 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- 14.Zur HH. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev CANCER. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 15.Grce M, Matovina M, Milutin-Gasperov N, Sabol I. Advances in cervical cancer control and future perspectives. Coll Antropol. 2010;34:731–736. [PubMed] [Google Scholar]
- 16.Coquillard G, Palao B, Patterson BK. Quantification of intracellular HPV E6/E7 mRNA expression increases the specificity and positive predictive value of cervical cancer screening compared to HPV DNA. Gynecol Oncol. 2011;120:89–93. doi: 10.1016/j.ygyno.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Yugawa T, Kiyono T. [Molecular basis of cervical carcinogenesis by high-risk human papillomaviruses] . Uirusu. 2008;58:141–154. doi: 10.2222/jsv.58.141. [DOI] [PubMed] [Google Scholar]
- 18.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pecorelli S, Zigliani L, Odicino F. Revised FIGO staging for carcinoma of the cervix. Int J Gynaecol Obstet. 2009;105:107–108. doi: 10.1016/j.ijgo.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 21.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 22.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 23.Warren CJ, Xu T, Guo K, Griffin LM, Westrich JA, Lee D, Lambert PF, Santiago ML, Pyeon D. APOBEC3A functions as a restriction factor of human papillomavirus. J Virol. 2015;89:688–702. doi: 10.1128/JVI.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahasan MM, Wakae K, Wang Z, Kitamura K, Liu G, Koura M, Imayasu M, Sakamoto N, Hanaoka K, Nakamura M, Kyo S, Kondo S, Fujiwara H, Yoshizaki T, Mori S, Kukimoto I, Muramatsu M. APOBEC3A and 3C decrease human papillomavirus 16 pseudovirion infectivity. Biochem Biophys Res Commun. 2015;457:295–299. doi: 10.1016/j.bbrc.2014.12.103. [DOI] [PubMed] [Google Scholar]
- 25.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulliard Y, Narvaiza I, Bertero A, Peddi S, Rohrig UF, Ortiz M, Zoete V, Castro-Diaz N, Turelli P, Telenti A, Michielin O, Weitzman MD, Trono D. Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J Virol. 2011;85:1765–1776. doi: 10.1128/JVI.01651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]