Abstract
Background: Primary angle closure glaucoma (PACG) has been thought to have a significantly genetic basis for a long time, and genome-wide association studies (GWAS) have identified various candidate genes including PCMTD1-ST18 rs1015213 as susceptibility loci. However, different results produced inconsistent results and make the conclusions controversial in some extent. Thus, we carried out a systematic review, attempting to summarize the recent evidence and determine the association of rs1015213 with PACG risk. Methods: A systematic literature search was conducted to identify all published studies on associations of rs1015213 (PCMTD1-ST18) polymorphism and PACG risk up to April 30, 2015. Selection of eligible studies was undertaken by two investigators according to inclusion criteria. Summary odds ratios (ORs) and 95% confidence intervals (95% CIs), as well as the pooled ocular biometric measures in different genotype or allele groups, were collected and analyzed. Heterogeneity was measured using the chi-square-based Q statistic test and I2 metric. Publication bias of the included articles was evaluated using funnel plots. Results: 21 eligible studies were included, among them 15 studies with enough data to estimate OR were included for meta-analysis, with a total of 24764 subjects (4737 PACG patients and 20027 controls), including 19416 Asian subjects (4378 PACG patients and 15038 controls) and 5348 Caucasian subjects (359 PACG patients and 4989 controls). Low heterogeneity was detected among studies (for Asian subgroups P=0.80, I2=0%, for Caucasian subgroups P=0.78, I2=0%, for all groups, P=0.89, I2=0%), thus, only fixed-effects model was used in the meta-analysis. The results showed that the frequencies of the TT genotype of rs1015213 were significant higher in PACG group than the controls in Asians (OR=1.51, 95% CI 1.27-1.79, P<0.01) but not in Caucasians (OR=1.54, 95% CI 0.94-2.54, P=0.09). In sensitivity analysis the significance of the pooled OR remained almost the same when removing studies individually. Visual inspection of the funnel plots revealed no asymmetry. 6 studies were included for evaluating the association between rs1015213 polymorphism with axial length (AL) and anterior chamber depth (ACD), all of them showed that rs1015213 polymorphism is independent with AL (Shi, P=0.528; Day, P=0.74; Nongpiur, pooled P=0.067, respectively). 5 studies showed that rs1015213 polymorphism was significantly associated with a shallow ACD (P<0.05) but the other study did not support this result. Conclusion: Our meta-analysis suggests that rs1015213 (TT genotype) is associated with PACG in Asian populations, but this association is not significant in Caucasian population and need more data. Some literatures also supported that rs1015213 polymorphism was associated with a shallow ACD but not with a short AL, however the evidences are not sufficient yet.
Keywords: Meta-analysis, primary angle closure glaucoma, rs1015213, polymorphism
Introduction
Glaucoma is the second leading cause of blindness worldwide. It contains two main subtypes, primary open angle glaucoma (POAG) and primary angle closure glaucoma (PACG). Epidemiological studies have revealed that POAG is mainly common in populations of western countries, but in populations of Asian countries PACG accounts for more than 50% of all cases of glaucoma. In China there are over 1 million PACG patients and this disease is very common in people over the age of 40 especially for women [1-3].
Ethnicity and family history are thought to be potential risk factors for PACG, which suggests a strong genetic predisposition to this disease. Researches about the PACG genetic basis become hot topic for a long time [4]. With such genetic bases the eye has a higher risk of suspicious anatomical features, such as shallow anterior chamber, narrow angle, thick crystal, short axial length, etc., so that the irido-corneal angle can be obstructed and the aqueous outflow channel is blocked, which lead to a high intraocular pressure (IOP) and finally destruction of the optic nerve progressively with corresponding loss of the peripheral visual field.
The molecular mechanisms of PACG are very complex and the genetic polymorphism is still under investigation. Recently some genetic susceptibility loci for PACG were identified. Jin et al. [5] found that the frequencies of the G allele (P=0.009; OR=0.52) of TMCO1 (rs4656461) and T allele (P=0.03; OR=0.69) of ATOH7 (rs1900004) were higher in cases of PACG group than in controls. Awadalla et al. [6] found that eNOS gene polymorphism was associated with PACG significantly (P=0.003; OR=0.5). Recently, some genome wide association studies (GWAS) was conducted by Vithana et al. [7] in Asian countries and United Kingdom (total sample size is 22322 with 3771 cases and 18551 controls) to determine the potential risk loci for PACG. They found that the minor allele frequency (MAF) of rs11024102 in PLEKHA7 (OR=1.22, P=5.33×10-12), rs3753841 in COL11A1 (OR=1.20, P=9.22×10-10) and rs1015213 in PCMTD1-ST18 (OR=1.50, P=3.29×10-9) were higher in PACG patients than controls. These three susceptibility genes may influence the anatomical features, such as trabecular meshwork, iris, ciliary body and eventually cause PACG. Because PLEKHA7, COL11A1 and PCMTD1 can expressed in tissues form the irido-corneal angle, this maybe the way how the polymorphisms of those three loci associated with PACG. The similar result was obtained by Awadalla et al. [8] from populations of Australia (232 cases and 288 controls) and Nepal (106 cases and 204 controls). But in contrast, Chen et al. [9] failed to get same evidence like the former findings in subsequent groups. Day et al. [10] also found that ocular biometric such as axial length (AL) and keratometry were both not associated with genotypes of those three loci above, only anterior chamber depth (ACD) is associated with rs1015213 genotypes (-0.07 mm, 95% CI -0.01 to -0.14 mm, P=0.028).
These studies produced inconsistent results and make the conclusions discrepant in some extent. Possible reasons for this discrepancy in the reported associations include non-homogeneous populations, different sample sizes, and distinct methodologies. Systematic review and meta-analysis are good ways to assess genetic variants-associated disease risk when individual studies produced inconsistent results. We want to get a summarized evidence of the association between the genetic polymorphisms and PACG risk, then we viewed the literature and found that the association between PACG and both of rs11024102 and rs3753841 were already analyzed by two other systematic reviews [11,12], yet there is no systematic view for the association of rs1015213 (PCMTD1-ST18) polymorphism and PACG risk. Thus in this study, we conducted a systematic review and a quantitative meta-analysis to determine the recent evidence of the association between rs1015213 polymorphism and PACG as well as the association of rs1015213 polymorphism with AL and ACD.
Materials and methods
Search strategy
A systematic literature search was conducted to identify all published studies on associations of rs1015213 (PCMTD1-ST18) polymorphism and PACG.
We carried out systematic literature searches via the database of PubMed, EMbase and Cochrane Library. The cut-off date was April 30, 2015. We used the following search terms including (“PCMTD1-ST18” or “rs1015213”) and “glaucoma” and (“closed” or “closure” or “uncompensative” or “uncompensated” or “narrow”) and (“mutation” or “variation” or “polymorphism” or “diversities” OR “diversity”) in this study and we screened the references cited in the publications matching the key words listed above. The search strategy contains no language, journal, country, age, gender, ethnicity restrictions.
Eligible studies were included if they (1) Using human patients sustaining PACG as cases; (2) The focus was the effects of rs1015213 (PCMTD1-ST18) on the development of PACG; (3) used a case-control, cohort or population-based epidemiological survey design to compare PACG cases and normal controls, or the parameters of AL and ACD of different allele in PACG in defined populations; (4) Supplying sufficient genotype or allele data which could be used to estimate ORs or the value of AL and ACD; and (5) were original research articles, not reviews or comments.
Descriptive research, Abstracts from conferences, full texts without raw data available for retrieval, or incomplete data that cannot be used to estimate the risk of PACG or the value of AL and ACD, republished data, duplicate studies and reviews were excluded.
Data extraction
We collect the Information on first author’s surname, year of publication, study location, ethnicity, sample size of cases and controls, single nucleotide polymorphism (SNP), MAF wherever available, matching variables, assays/platforms used in genotype determination, Hardy-Weinberg equilibrium whenever reported, as well as the data of AL, ACD and per-allele change of AL/ACD. Data were extracted by two investigators (Xiaoling Y, Yanting X) from all eligible articles into data collection forms independently. Disagreements were resolved through discussion between the two investigators and consult to another senior investigator (Ming L) if necessary until a consensus was achieved.
Statistical methods
To assess the strength of association between rs1015213 (PCMTD1-ST18 gene) polymorphism and PACG risk, we estimated crude ORs with its 95% confidence intervals (95% CI). Chisquare-based Q statistic and the I2 metric were used to detect potential heterogeneity across studies. If the P-value was above 0.10 from the Q-test or I2<50%, which indicates an absence of heterogeneity, the pooled ORs were summarized in a fixed-effect model using the Mantel-Haenszel method [13]. Otherwise, the random-effect model was used to analyze the summary Ors [14]. The association of the allele with AL and ACD was assessed under the additive genetic model, dominant model, and recessive model if possible and the mean value were compared by t-test. A sensitivity analysis was conducted by removing one included study at a time to determine whether the results were affected by one specific study. Subgroup analyses were performed according to ethnicity. Publication bias was investigated by funnel plot with obviously visual symmetry suggesting lack of publication bias. All analyses were done with Revman (version 5.3; The Cochrane Collaboration, Copenhagen, Denmark) and SPSS (ver. 18.0; SPSS Inc., Chicago, IL). The values of P<0.05 were considered statistically significant.
Results
Study characteristics
The selection process for the articles in our meta-analysis is described in Figure 1. The initial search strategy yielded 47 articles. Among them, 20 duplicate studies were excluded firstly, 3 were excluded after reviewing the title and abstract, 3 articles were excluded after full-text review according to the exclusion criteria. Among the 21 eligible included studies (note: 11 studies provided by Vithana et al., 4 studies provide by Nongpiur et al., and 2 by Awadalla et al.), 6 population-base survey studies were concerned with the association of rs1015213 with AL and ACD however the data were not sufficient to fulfill meta-analysis because distinct methodologies were performed [10,15,16], 15 case control studies provided a total of 26,365 subjects (4737 cases and 20027 controls) with Asian or Caucasian ancestry for the meta-analysis of rs1015213 variant and PACG risk [7,8,15,17,13], studies (86.7%) were for subjects of Asians and only 2 studies (13.3%) were performed in Caucasians. Different genotyping assays or platforms were selected in genotype determination, including TaqMan SNP assay, Illumina 610K Quad BeadChip assay, TaqMan Genotyping Assay, Autoflex mass spectrometer, and Sequenom MassArray platform. The included studies were similar in matching status, and two studies showed significant Hardy-Weinberg equilibrium deviation [7,15]. All the studies were not family based. The characteristics of these studies are further described in Tables 1, 2.
Figure 1.
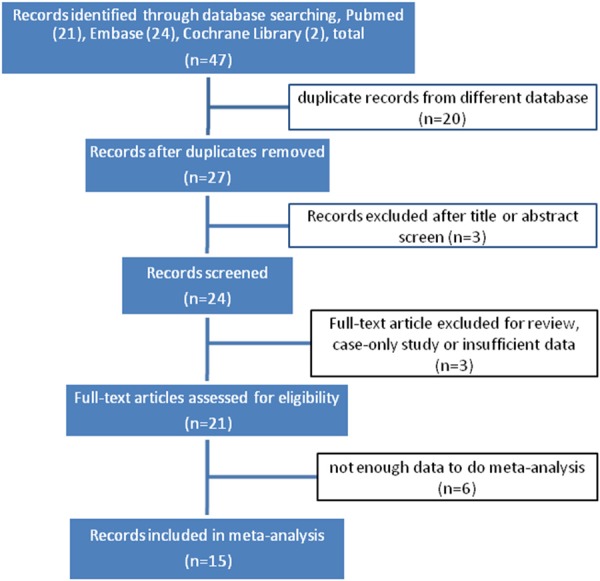
Flow diagram of eligible studies for this systematic review. (※, Three articles included more than one independent study).
Table 1.
Characteristics of the selected studies evaluating PACG risk correlated with PCMTD1-ST18 rs1015213. Vithana’s article included 11 independent studies and Awadalla’s article included 2 independent studies
| Authors | year | country | population | matching variables | genotype | study design | outcomes | case | control | HWE Deviation |
|---|---|---|---|---|---|---|---|---|---|---|
| Awadalla (a) | 2013 | Australian | Caucasian | Gender-, ethnicity-matched | TT/CT+CC | case control | MAF | 232 | 288 | N |
| Awadalla (b) | 2013 | Nepalese | Asian | Gender-, ethnicity-matched | TT/CT+CC | case control | MAF | 106 | 204 | N |
| Duvesh | 2013 | India | Asian | Ethnicity-matched | TT/CT+CC | case control | MAF | 180 | 411 | N |
| Shi | 2013 | China (Jiangsu) | Asian | Age-, gender-matched | TT/CT+CC | case control | MAF | 453 | 599 | N |
| Vithana (a) | 2012 | Singapore (stage 1) | Asian | Genetically-matched | TT/CT+CC | case control | MAF | 983 | 943 | N |
| Vithana (b) | 2012 | Hong Kong (stage 1) | Asian | Genetically-matched | TT/CT+CC | case control | MAF | 297 | 1044 | N |
| Vithana (c) | 2012 | India (stage 1) | Asian | Genetically-matched | TT/CT+CC | case control | MAF | 338 | 2535 | N |
| Vithana (d) | 2012 | Malaysia (stage 1) | Asian | Genetically-matched | TT/CT+CC | case control | MAF | 83 | 3057 | N |
| Vithana (e) | 2012 | Vietnam (stage 1) | Asian | Genetically-matched | TT/CT+CC | case control | MAF | 153 | 2016 | N |
| Vithana (f) | 2012 | China (Beijing, stage 2) | Asian | Genetically-matched | TT/CT+CC | case control | MAF | 992 | 1667 | N |
| Vithana (g) | 2012 | Singapore (stage 2) | Asian | Genetically-matched | TT/CT+CC | case control | MAF | 304 | 1479 | N |
| Vithana (h) | 2012 | China (Shantou, stage 2) | Asian | Genetically-matched | TT/CT+CC | case control | MAF | 245 | 601 | N |
| Vithana (i) | 2012 | India (stage 2) | Asian | Genetically-matched | TT/CT+CC | case control | MAF | 80 | 308 | N |
| Vithana (j) | 2012 | Saudi Arabia | Asian | Genetically-matched | TT/CT+CC | case control | MAF | 164 | 174 | N |
| Vithana (k) | 2012 | United Kingdom | Caucasian | Genetically-matched | TT/CT+CC | case control | MAF | 127 | 4701 | Y |
MAF: minor allele frequency, Nongpiur’s article included 4 independent studies; HWE: Hardy-Weinberg equilibrium.
Table 2.
Characteristics of the selected studies evaluating the association of PCMTD1-ST18 rs1015213 and AL/ACD
| Authors | year | country | population | allele | study design | outcomes | effect allele Num. | unaffected allele Num. | HWE Deviation |
|---|---|---|---|---|---|---|---|---|---|
| Shi | 2013 | China (Jiangsu) | Asian | CC/CT+TT | population-based survey | AL/ACD | 11/11 | 221/221 | Y |
| Day | 2013 | United Kingdom | Caucasian | GG/GA+AA | population-based survey | AL/ACD | 183/160 | 951/825 | N |
| Nongpiur (a) | 2013 | Singapore | Asian | A/G | population-based survey | AL/ACD | 19/18 | 1671/1595 | Not available |
| Nongpiur (b) | 2013 | Singapore | Asian | A/G | population-based survey | AL/ACD | 90/90 | 2117/2118 | Not available |
| Nongpiur (c) | 2013 | Singapore | Asian | A/G | population-based survey | AL/ACD | 308/304 | 2212/2184 | Not available |
| Nongpiur (d) | 2013 | China | Asian | A/G | population-based survey | AL/ACD | 20/20 | 806/577 | Not available |
AL: axial length; ACD: anterior chamber depth.
Quantitative synthesis
As shown in Table 1, a total of 24764subjects (4737 PACG patients and 20027 controls) were tested for genotype of rs101513, among them 19416 were Asian subjects (4378 PACG patients and 15038 controls) and 5348 were Caucasian subjects (359 PACG patients and 4989 controls). Low heterogeneity was detected among studies (for Asian subgroups P=0.80, I2=0%, for Caucasian subgroups P=0.78, I2=0%, for all groups, P=0.89, I2=0%), thus, only a fixed-effects model was used in this study.
The overall result showed that TT genotype of rs1015213 were significant higher (OR=1.51, 95% CI 1.28-1.78, P<0.01) in PACG group than the controls. If Vithana’s 11 studies were not included, the rest of data still show a low heterogeneity (P=0.71, I2=0%), and in a fix-effects model the rs1015213 (TT genotype) were also significantly associated with PACG (OR=1.57, 95% CI 1.10-2.25, P=0.01, forest plot omitted).
Stratified analyses were conducted based on Asian and Caucasian ethnicities, which have been reported to have differences in the prevalence of PACG [18]. The results showed that the association of rs1015213 (TT genotype) with PACG was significant in Asians populations (OR=1.51, 95% CI 1.27-1.79, P<0.01) but not in Caucasians populations (OR=1.54, 95% CI 0.94-2.54, P=0.09). In Asian subgroups, five studies (Duvesh and Vithana a, c, f, j) showed a significant association with rs1015213 (TT genotype) and PACG risk (all with 95% CI of OR above 1 and P<0.05). Actually all these studies contained more than 20 PACG patients with TT genotype, while all those studies which only contained less than 20 PACG patients with TT genotype didn’t showed significant association, more details are given in Figure 2.
Figure 2.
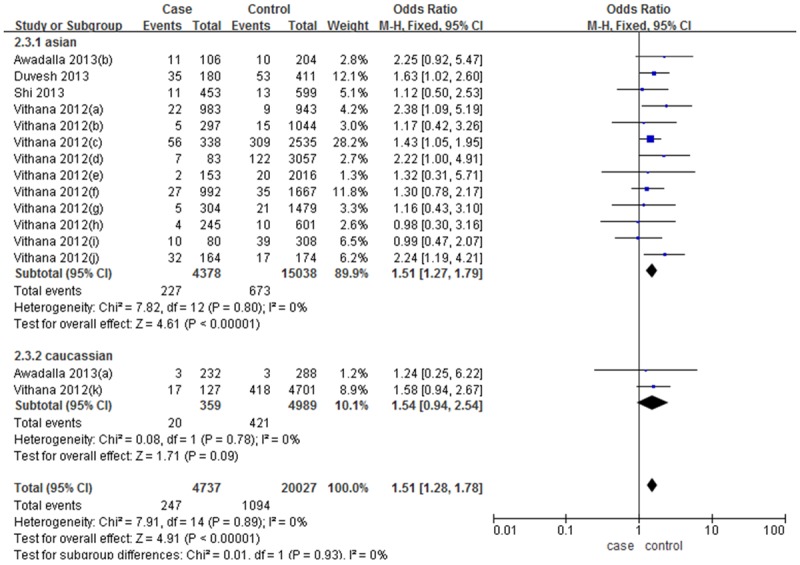
Forest plot (Fixed effects model) describing the association of PACG risk correlated with PCMTD1-ST18 rs1015213. The rs1015213-TT was associated with an increased risk of PACG (TT vs. CC+CT).
Sensitivity analysis of each study on the pooled OR was assessed by sequential omission of individual studies especially for the study with significant Hardy-Weinberg equilibrium deviation. When removing studies individually, the significance of the pooled OR remained almost the same. Publication bias was also analyzed; the funnel plot shows the points as evenly distributed and symmetric. After excluding all the populations in Vithana’s study, the funnel plot still shows the points as evenly distributed and symmetric (Figure 3A, 3B). Therefore, our effect estimates were reliable.
Figure 3.
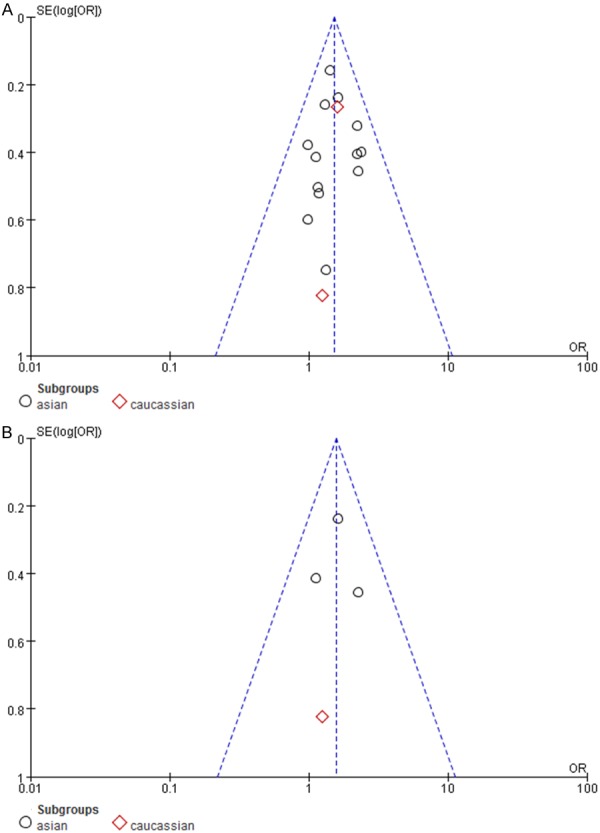
Funnel plots showed symmetric distribution (A for all 15 selected studies; B for four studies excluding Vithana’s studies). Log OR is plotted against the standard error of log OR for studies on the association of PACG risk correlated with PCMTD1-ST18 rs1015213. The dots represent specific studies for the indicated association.
There were 6 studies about the association of rs1015213 polymorphism to AL and ACD. All of them showed that 1015213 polymorphism is independent with AL (Shi, P=0.528; Day, P=0.74; Nongpiur, pooled P=0.067, respectively), more details are given in Tables 3, 4. While for ACD, one study showed that the presence of at least one A allele (AG or AA) for rs1015213 to be associated with a shallower ACD significantly (P=0.046) after adjusting for the effects of age and sex in both additive model and dominant model [10]. Four other studies reported by Nongpiur showed nominal significant association (in every single study P>0.05, but the pooled P=0.021) with rs1015213 (A allele) 16. But the other study showed no significant association (P=0.617) with rs1015213 (CT+TT genotype) 15. Unfortunately, these genetic polymorphism data were measured by different method and analyzed in different ways, so the data could not be used for doing the meta-analysis. These data showed that polymorphism of rs1015213 was not associated with AL, but the association between rs1015213 polymorphism and ACD is not clear and need more data.
Table 3.
Polymorphism of rs1015213 (genotype) and AL/ACD from selected studies. Values from Shi study were given in mean ± standard deviation and compared by t-test, Values from Day study were given in mean with 95% CI and compared by analysis of variance
| Authors | year | country | population | genotype | sample size | AL/mm | sample size | ACD/mm |
|---|---|---|---|---|---|---|---|---|
| Shi | 2013 | China (Jiangsu) | Asian | CC | 516 | 22.15±0.72 | 516 | 2.46±0.22 |
| CT+TT | 22 | 22.29±0.99 | 22 | 2.42±0.27 | ||||
| P | 0.528 | 0.617 | ||||||
| Day | 2013 | United Kingdom | Caucasian | GG | 951 | 23.55 (23.47-23.62) | 825 | 3.07 (3.04-3.10) |
| AG+AA | 183 | 23.51 (23.34-23.69) | 160 | 3.00 (2.94-3.06) | ||||
| P | 0.74 | 0.046 |
AL: axial length; ACD: anterior chamber depth.
Table 4.
Pooled measures for the association between polymorphism of rs1015213 (allele A) and AL/ACD from selected studies. The pooled value of the four studies for the simple size, change/allele, P value, P het and I2 are list in the last line
| Authors | year | study | country | population | AL | ACD | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||||
| sample size | MAF | change/allele | P | P het | I 2 | sample size | MAF | change/allele | P | P het | I 2 | |||||
| Nongpiur | 2013 | SEC | Singapore | Chinese | 1690 | 0.011 | -0.041 | 0.410 | 1613 | 0.011 | -0.19 | 0.520 | ||||
| Nongpiur | 2013 | SiMES | Singapore | Malay | 2207 | 0.04055 | -0.021 | 0.137 | 2208 | 0.04055 | -0.112 | 0.404 | ||||
| Nongpiur | 2013 | SINDI | Singapore | Indian | 2520 | 0.1222 | -0.033 | 0.301 | 2488 | 0.1222 | -0.048 | 0.089 | ||||
| Nongpiur | 2013 | BES | China (Beijng) | Chinese | 826 | 0.02427 | -0.068 | 0.622 | 591 | 0.02427 | -0.1 | 0.139 | ||||
| ALL | 7243 | -0.033 | 0.067 | 0.766 | 0 | 6900 | -0.007 | 0.021 | 0.870 | 0 | ||||||
SEC: Singapore Chinese Eye Study; SiMES: Singapore Malay Eye Study; SINDI; Singapore Indian Eye Study; BES: Beijing Eye Study; AL: axial length; ACD; anterior chamber depth; Phet: P value for heterogeneity between studies; I2: index for heterogeneity between studies.
Discussion
rs1015213 is located on chromosome 8 between PCMTD1 gene and ST18 gene, PCMTD1 encodes protein-L-isoaspartate O-methyltransferase domain-containing protein, there is very little known about the function of the protein but PCMTD1 can be expressed in the human foetal eye, and in human and mouse retina. ST18 is expressed in eye tissues including iris and trabecular meshwork which are involved in the pathogenesis of PACG. It encodes the suppression of tumorigenicity 18 protein which can downregulated in breast cancer cell lines [7] and may be a mediator in apoptosis and inflammation [19].
In the present study, 15 studies were included in a meta-analysis showed that the genetic polymorphisms of rs1015213 were associated with a risk of PACG at least in Asian ethnic groups (OR=1.51, 95% CI 1.51-2.79). But in the subgroup of Caucasian, the OR=1.55 (95% CI 0.94-2.54) suggested the risk of rs1015213 polymorphism to PACG was not statistically significant. The ethnic differences between them remain unclear. The possible reason might be the low MAF for rs1015213, in many of the sample collections the MAF was even lower than 1%, which can lead to more deviation in every single research. Meta-analysis might be a better way to lower the deviation and makes the result more stable because of the large sample size and multi central subject resources. In this meta-analysis, we found that all of the studies contained more than 20 PACG patients with TT genotype in rs105213 showed a significant association between rs101521 polymorphism and PACG, while all those studies contained less than 20 PACG patients with TT genotype in rs105213 didn’t showed significant association, but during meta-analysis the sample size became larger and the association of them became more significant. This means that the sample size is very important in epidemiological researches especially when MAF is low.
The number of subjects with rs1015213 genotype TT were 441 from a total sample size of 5348 in Caucasian groups and the mean MAF=0.082; while the number of subjects with rs1015213 genotype TT were 900 from a total sample size of 19416 in Asian groups and the mean MAF=0.046. We can see the MAF value was even lower in Asian group than in Caucasian groups. Recent epidemiological data shows incidence of narrow-angle glaucoma (NAG) in Chinese ethnicity (3.75%) was much higher than in White ethnicity (1.35%) including Caucasian [18]. PACG were much more common in Asian populations than the Caucasian population while the MAF of rs1015213 genotype was lower in Asian ethnicity than in Caucasian, so this result seems like that the MAF of rs1015213 genotype did not always proportional to the incidence of PACG in every ethnicity. One possible reason might be the low MAF for rs1015213 and the sample size in Caucasian groups were not big enough but the exact mechanism remains unclear. If rs1015213 polymorphism really leads to a higher risk of PACG only in Asian ethnicity but not in Caucasian ethnicity, this may implied that another genetic mechanism is involved to protect against with the destruction of rs1015213 polymorphism thought the mechanism is not known until now.
In addition, the statistic analyze did not support the association between rs1015213 and AL. The ocular biometric parameters such as a shallow ACD and AL were demonstrated as strong risk factors for PACG [20]. A Singapore study shows that eyes with ACD less than 2.80 mm were 42.5 times more likely to have angle closure than eyes with ACD above 3.00 mm [21]. Both of the six included studies showed that there is no statistic difference between different rs1015213 genotype groups in AL [10,15]. But for ACD, five studies supported the association while only one study not, This means rs1015213 polymorphism has a high potential risk of shallow ACD even though the evidence are not sufficient yet. It should be noted that, at least in one study [10] the author said his study had a lot of limitations, only once third of the participants’ ocular biometry data were available and lack of data for the TT genotype polymorphism. In spite of the limitations, these data still suggested a potential association of rs1015213 polymorphism and ACD.
Even if rs1015213 is associated with PGCA, this does not mean that the loci will change the ocular anatomical features of candidate patient, the risk loci can be independent with AL but work through other ways. PACG can result from one or a combination of anatomical and/or physiological changes of the anterior and posterior segment structures [16]. Thus some other ocular biometrics might be the phenotype of rs1015213 to get a higher risk of PACG, such as anterior chamber width, area and volume [16], and greater iris thickness, area, and curvature [22], and larger lens vault etc [23].
There are some limitations in this study. The first limitation is the origin of the articles may not be representative enough. There are 15 group of subjects included in this meta-analysis but 74% cases and 92% controls are origin from Vithana [7], it very easy to get big population bias and langue bias. Thus we analyzed the data twice, one included all of the studies as previously mentioned, the other meta-analysis only included 4 of the studies didn’t origin from Vithana. Fortunately these four studies still show a low heterogeneity (P=0.71, I2=0%), and the result didn’t changed as rs1015213 (TT genotype) were still significantly associated with PACG (OR=1.57, 95% CI 1.10-2.25, P=0.01). We also got an evenly distributed and symmetric funnel plot after excluding all the populations in Vithana’s study. Thus the bias from this limitation seemed acceptable for the meta-analysis. The second limitation was the low MAF of rs1015213, this lead to more deviation and need a larger sample size for data analysis, actually the sample size of some studies included in this meta-analysis is not large enough, thus the ORs and P values from every single study is usually not stable, maybe this is one of the reason why there is no significant association between rs1015213 polymorphism and PACG risk. Third, the data for analyze the ocular anatomical parameters such as AL and ACD are far away from enough, and the studies’ methodologies are different from each other, so the data cannot be used for doing meta-analysis. Some results of the six studies are not consistent with each other or even controversial; this implies an urgent requirement of more multi-central studies with standard design and big sample size to get a higher level evidence of the association between rs1015213 polymorphism and PACG.
This meta-analysis showed that rs1015213 (TT genotype) is associated with PACG in Asian populations, but the association in Caucasian population is not convinced and need more data. It seems like rs1015213 polymorphism associated with a shallow ACD but not a short AL; however the evidence is not sufficient yet.
Acknowledgements
This study was supported by grants from the Natural Science Foundation of China (81202726 (L. L.). The authors would like to thank Ahmed Bata, Feng Zhou, and Lou Lixia for searching articles and editing the paper. We thank the grant support from National Natural Science Foundation of China (81202726).
Disclosure of conflict of interest
None.
References
- 1.Jun W, Wei C. Progress in epidemiological studies of primary glaucoma in China. Int Eye Sci. 2012;12:667–670. [Google Scholar]
- 2.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster PJ, Johnson GJ. Glaucoma in China: how big is the problem? Br J Ophthalmol. 2001;85:1277–1282. doi: 10.1136/bjo.85.11.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowe RF. Primary angle-closure glaucoma. Inheritance and environment. Br J Ophthalmol. 1972;56:13–20. doi: 10.1136/bjo.56.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin X, Wang DJ, Qu LH, Hou BK, Gong Y, Xu WW. Haplotype Analysis of Association of the MYOC Gene with Primary Angle-Closure Glaucoma in a Han Chinese Population. Genet Test Mol Biomarkers. 2015;19:3–8. doi: 10.1089/gtmb.2014.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Awadalla MS, Thapa SS, Hewitt AW, Craig JE, Burdon KP. Association of eNOS polymorphisms with primary angle-closure glaucoma. Invest Ophthalmol Vis Sci. 2013;54:2108–2114. doi: 10.1167/iovs.12-11391. [DOI] [PubMed] [Google Scholar]
- 7.Vithana EN, Khor CC, Qiao C, Nongpiur ME, George R, Chen LJ, Do T, Abu-Amero K, Huang CK, Low S, Tajudin LS, Perera SA, Cheng CY, Xu L, Jia H, Ho CL, Sim KS, Wu RY, Tham CC, Chew PT, Su DH, Oen FT, Sarangapani S, Soumittra N, Osman EA, Wong HT, Tang G, Fan S, Meng H, Huong DT, Wang H, Feng B, Baskaran M, Shantha B, Ramprasad VL, Kumaramanickavel G, Iyengar SK, How AC, Lee KY, Sivakumaran TA, Yong VH, Ting SM, Li Y, Wang YX, Tay WT, Sim X, Lavanya R, Cornes BK, Zheng YF, Wong TT, Loon SC, Yong VK, Waseem N, Yaakub A, Chia KS, Allingham RR, Hauser MA, Lam DS, Hibberd ML, Bhattacharya SS, Zhang M, Teo YY, Tan DT, Jonas JB, Tai ES, Saw SM, Hon do N, Al-Obeidan SA, Liu J, Chau TN, Simmons CP, Bei JX, Zeng YX, Foster PJ, Vijaya L, Wong TY, Pang CP, Wang N, Aung T. Genome-wide association analyses identify three new susceptibility loci for primary angle closure glaucoma. Nat Genet. 2012;44:1142–1146. doi: 10.1038/ng.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Awadalla MS, Thapa SS, Hewitt AW, Burdon KP, Craig JE. Association of genetic variants with primary angle closure glaucoma in two different populations. PLoS One. 2013;8:e67903. doi: 10.1371/journal.pone.0067903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Chen X, Wang L, Hughes G, Qian S, Sun X. Extended association study of PLEKHA7 and COL11A1 with primary angle closure glaucoma in a Han Chinese population. Invest Ophthalmol Vis Sci. 2014;55:3797–3802. doi: 10.1167/iovs.14-14370. [DOI] [PubMed] [Google Scholar]
- 10.Day AC, Luben R, Khawaja AP, Low S, Hayat S, Dalzell N, Wareham NJ, Khaw KT, Foster PJ. Genotype-phenotype analysis of SNPs associated with primary angle closure glaucoma (rs1015213, rs3753841 and rs11024102) and ocular biometry in the EPIC-Norfolk Eye Study. Br J Ophthalmol. 2013;97:704–707. doi: 10.1136/bjophthalmol-2012-302969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua B, Hui L, Juan W, Guohui L, Yifei H. A common genetic variant as an effect modifier for primary angle closure glaucoma. Int J Clin Exp Med. 2015;8:883–889. [PMC free article] [PubMed] [Google Scholar]
- 12.Shuai P, Yu M, Li X, Zhou Y, Liu X, Liu Y, Zhang D, Gong B. Genetic associations in PLEKHA7 and COL11A1 with primary angle closure glaucoma: a meta-analysis. Clin Experiment Ophthalmol. 2015;43:523–30. doi: 10.1111/ceo.12516. [DOI] [PubMed] [Google Scholar]
- 13.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 14.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28:105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Shi H, Zhu R, Hu N, Shi J, Zhang J, Jiang L, Jiang H, Guan H. An Extensive Replication Study on Three New Susceptibility Loci of Primary Angle Closure Glaucoma in Han Chinese: Jiangsu Eye Study. J Ophthalmol. 2013;2013:641596. doi: 10.1155/2013/641596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nongpiur ME, Wei X, Xu L, Perera SA, Wu RY, Zheng Y, Li Y, Wang YX, Cheng CY, Jonas JB, Wong TY, Vithana EN, Aung T, Khor CC. Lack of Association Between Primary Angle-Closure Glaucoma Susceptibility Loci and the Ocular Biometric Parameters Anterior Chamber Depth and Axial Length. Invest Ophthalmol Vis Sci. 2013;54:5824–5828. doi: 10.1167/iovs.13-11901. [DOI] [PubMed] [Google Scholar]
- 17.Duvesh R, Verma A, Venkatesh R, Kavitha S, Ramulu PY, Wojciechowski R, Sundaresan P. Association study in a South Indian population supports rs1015213 as a risk factor for primary angle closure. Invest Ophthalmol Vis Sci. 2013;54:5624–5628. doi: 10.1167/iovs.13-12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein JD, Kim DS, Niziol LM, Talwar N, Nan B, Musch DC, Richards JE. Differences in rates of glaucoma among Asian Americans and other racial groups, and among various Asian ethnic groups. Ophthalmology. 2011;118:1031–1037. doi: 10.1016/j.ophtha.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Siqueira MF, Behl Y, Alikhani M, Graves DT. The transcription factor ST18 regulates proapoptotic and proinflammatory gene expression in fibroblasts. FASEB J. 2008;22:3956–3967. doi: 10.1096/fj.08-111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowe RF. Aetiology of the anatomical basis for primary angleclosure glaucoma. Biometrical comparisons between normal eyes and eyes with primary angle-closure glaucoma. Br J Ophthalmol. 1970;54:161–169. doi: 10.1136/bjo.54.3.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavanya R, Wong TY, Friedman DS, Aung HT, Alfred T, Gao H, Seah SK, Kashiwagi K, Foster PJ, Aung T. Determinants of angle closure in older Singaporeans. Arch Ophthalmol. 2008;126:686–691. doi: 10.1001/archopht.126.5.686. [DOI] [PubMed] [Google Scholar]
- 22.Wang B, Sakata LM, Friedman DS, Chan YH, He M, Lavanya R, Wong TY, Aung T. Quantitative iris parameters and association with narrow angles. Ophthalmology. 2010;117:11–17. doi: 10.1016/j.ophtha.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 23.Nongpiur ME, He M, Amerasinghe N, Friedman DS, Tay WT, Baskaran M, Smith SD, Wong TY, Aung T. Lens vault, thickness, and position in Chinese subjects with angle closure. Ophthalmology. 2011;118:474–479. doi: 10.1016/j.ophtha.2010.07.025. [DOI] [PubMed] [Google Scholar]


