Abstract
An artificial neuron network (ANN) model combining both the genetic risk factors and clinical factorsmay be effective in prediction of chemotherapy-induced adverse events. Purpose: To identify genetic factors and clinical factors associated with bone marrow suppression in cervical cancer patient, and to build a model for chemotherapy-induced neutropenia prediction. Methods: We performed a genome wide association study on a cohort to identify genetic determinants. Samples were genotyped using the Axiom CHB 1.0. The primary analyses focused on the scan of 657178 single-nucleotide polymorphisms (SNPs). Artificial neural network were used to integrating clinical factors and genetic factors to predict the occurrence of neutropenia. Results: 32 variants associated with neutropenia in the patients after chemotherapy were found (P<1 × 10-4). During internal validation and external validation, artificial neural network performed well in predicting neutropenia with considerable accuracy, which is 88.9% and 81.7% respectively. ROC analysis had acceptable areas under the curve of 0.897 for the internal validation sample and 0.782 for the external validation sample. Conclusion: Neutropenia may be associated with both genetic factors and clinical factors. Our study found that the artificial neural networks model based on the multiple risk factors jointly, can effectively predict the occurring of neutropenia, which provides some guidance before the starting of chemotherapy.
Keywords: Cervical cancer, genome-wide association study, artificial neuron network, platinum, single-nucleotide polymorphism
Introduction
Cervical cancer is the leading cause of cancer death among females in less developed country according to data in 2012 [1]. And it was also the third common female cancer worldwide, accounting for about 9% (529, 800) of total new cancer patients among women in 2008 [2]. Chemotherapy with platium plus other agents like taxanes or CPT-11 may be effective, but is poorly tolerated by some patients. Bone marrow suppression is one of the most common adverse effects of chemotherapy, leading to neutropenia, with risk of occurring of secondary sepsis. Identifying patients at greatest risk for these complications would be often clinically useful for selecting patients for chemotherapy. This is also useful for planning the frequency of monitor and clinical treatment with colony-stimulating factor. Patients treated with CPT-11 are at highest risk, and bone marrow suppression is also prevalent in patients with other platium-based treatment regimens. It is urgent to identify more accurate factors, including biomarkers and clinical factors.
For complex disorders, both genetics, environment and chance affect the pathogenetic processes [3]. A number of researchers suggest that genetic variants may be associated with chemotherapy-induced cytopenia [4-10]. The relevance of some variants to chemotherapy-induced neutropenia has been realized gradually. Secondly, polymorphism in the region of the UGT1A1 gene has recently been identified to be strongly associated with neutropenia [11-17]. The polymorphism is believed to regulate neurotoxicity to the anti-tumor agents. Other genetic variants were also reported by a series of studies [11,13,15]. Genetic biomarkers from human’s genome for predicting risk of chemotherapy induced mylosuppression may be particularly useful if treated as a pre-treatment test.
In this study, we have performed a genome-wide association study (GWAS) for determinants of chemotherapy-related myelosuppression in a large, well characterized cohort of cervical cancer patients treated with platium plus taxanes or platium plus CPT-11. We have focused primarily on chemotherapy-induced neutropenia.
Screening of patients at high risk, may enable preventative medical intervention in an economical way. Nowadays such a predictive model can be made available by using computer-based systems.
Artificial neural network (ANN) models, which are based on a series of multilayered interconnected equations, use non-linear statistical method to discover previously unknown relations between input variables and an output variable [18]. Researchers have revealed that ANN models are accurate and reliable in prediction of diverse clinical settings, including diagnosis and prognosis [19-23].
Our study aims to develop and validate a model through ANN method, which predicts WHO grade II-IV bone marrow depression in a group of cervical cancer patients presenting with neutropenia during chemotherapy.
Methods
Eligibility
Eligible patients were diagnosed with cervical cancer by pathological experts according to cervical biopsy by clinicians according to the International Federation of Gynecology and Obstetrics (FIGO). The exclusion criteria included preexistingsensory or motor neuropathy greater than WHO grade 1, a history of myocardial infarction and cardiac insufficiency ≥grade 3 (New York Heart Association scale). Patients previously treated for cervical cancer (i.e., surgery, chemotherapy orradiotherapy) and a past or current history of other neoplasmwere excluded. Patients with active infectious disease or other medically complicating condition were excluded. Women who were pregnant or lactating were also excluded from this study. This study was approved by each participating center’s Institutional Ethical Committee and was conducted according to the principles of the Declaration of Helsinki. Written informed consents were obtained from all subjects (ClinicalTrials.gov Identifier: NCT01628757).
Samples collection and DNA extraction
A case-control analysiswas performed, and all cervical cancer patients were unrelated ethnic Han Chinese. Ethylene diaminetetraacetic acid disodium salt (EDTA-2Na)-anticoagulated venous blood samples were collected from all participants.
Genomic DNA was extracted from peripheral blood by standard procedures using Flexi Gene DNA kits (Qiagen) and the QuickGene DNA whole-blood kit (Fujifilm). The extracted DNA was diluted to working concentrations of 50 ng/μL for genome-wide genotyping.
GWAS genotyping and quality control
Genome-wide genotyping was performed using the Axiom Genome-Wide CHB1.0 Array (Affymetrix). Quality-control filtering of the GWAS data required a dish quality control (DQC) value >0.82 for further data analysis. DQC is a metric developed by Affymetrix that takes both interchannel and intrachannel signal separation and spread into account and is the recommended quality-control metric for Axiom arrays. For sample filtering, arrays with generated genotypes for <98% of the loci were excluded (11 samples). PLINK’s identity by descent analysis was used to detect hidden relatedness. When pairs of individuals had PIHAT >0.25, the member of the pair with the lower call rate was excluded from the analysis (no sample was found); 57 cases and 218 controls were retained for further analysis. For SNP filtering (after sample filtering), SNPs with call rates <98% in the samples were removed. SNPs with MAF <5% or SNPs that deviated significantly (P≤1 × 10-5) from Hardy-Weinberg equilibrium in controls were also excluded. A total of 627,203 SNPs passed the quality criteria and were used in subsequent analyses. SNP with P≤1 × 10-4 was selected for follow-up study to construct the ANN model.
ANN: training and internal validation
ANN model was constructed using Matlab software (version 8.5). The model was trained with back propagation. Samples from Tongji hospital and ZhejiangUniversity were used asinternal group for training and internal validation. Data from three quarters of the first internal group were used to train the ANN. Data from the rest one quarter of internal group were used for internal validation of the ANN. During training, the input variables were entered as either categorical or continuous data into the ANN, while the output variables were entered as 0 or 1. Instructed by the pre-designed code, the programme was allowed to run and a prediction was made, then the output value was correlated with the actual outcome value. If the output was not correct, a process of back propagation readjusted weights within the hidden layer until the correct prediction result was achieved. This process was repeated thousands of times until the training completed. During the validation process, the actual outcome value was concealed from the networks, and output value was compared with the actual outcome value. During validation, all the input variables for training were used by the ANN to predict outcome.
ANN: external validation
Clinical data on a group of 60 patients from a cohort study (ClinicalTrials. gov Identifier: NCT01628757), were used for external validation of the model. Between 2008 and 2012, they were admitted to XiangfanHospital with chemotherapy treatment and the samples were genotyped by BeadChip. Compared with the internal validation group, the patients in the external validation group were from an independent hospital from central area of China. And the area has not so good economy and health care as internal group. Just like before, all the input variables for training and internal validation were entered in the external validation database.
Statistical analyses
The associations of single SNPs with cervical cancer were analyzed using PLINKv1.04 software. The genomic inflation factor (λ=1.013) in our analyses suggested that cases and controls matched well, indicating no populationstratification, and ourresults were based on the uncorrected P values. A quantile-quantile plot created by the R Programming Language was then used to evaluate the overall significance of the GWAS results (Figure 3). The per-allele ORs were calculated and presented for the minor allele of each SNP, unless stated otherwise. A genome-wide association analysis was carried out using an additive model in a logistic regression analysis with chemotherapy regimen as a covariate. Accuracy of the predictive model wascalculated by sum of correct prediction divided by totalpredictions. Sensitivity, specificity, positive predictivevalue, negative predictive valuewere also calculated. Also, receiver-operating characteristic (ROC) curves for the ANN were generated for the outcome variables in internal validation sample and external validation sample. The statistical methods used included the χ2 test and Fisher’s exact test forcategorical variables. All P-values were two-tailed, and values<0.05 were considered statistically significant. These statistical analyses werecarried out using the SPSS13.0 statistical software package.
Figure 3.
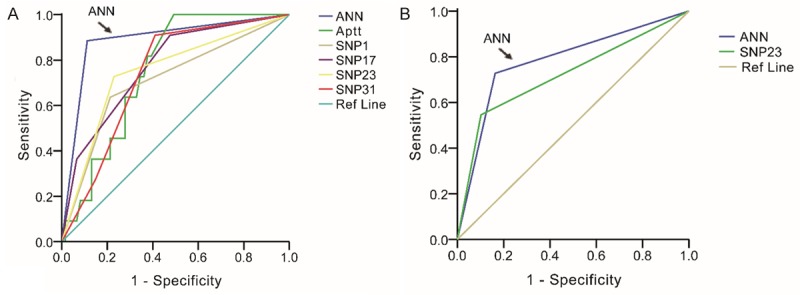
ROC for factors in internal validation and external validation groups. The largest area under the curve was covered by the ANN model in both the groups. The factors with low level of predicting were excluded, and only factors with area more than 0.7 were included in our analysis.
Results
The main characteristics for cases and controls are listed in Table 1. ALL patients were Han ancestry. Most of basic clinical variables were similar in both case and control data sets except chemotherapy regimens. Neutropenia was more frequent in CP arm than TP arm (P<0.001).
Table 1.
Patient Characteristics
| Characteristic | Control | Case | P-Value |
|---|---|---|---|
| All patients | 227 | 59 | |
| Age, years | |||
| Medium | 44 | 45 | |
| Q1-Q3 | 40-49 | 40-50 | |
| Weight, kg | |||
| Medium | 56 | 55 | |
| Q1-Q3 | 51-61 | 49-60 | |
| Height, cms | |||
| Medium | 158 | 159 | |
| Q1-Q3 | 155-161 | 154-161 | |
| Menarche, years | |||
| Medium | 14 | 14 | |
| Q1-Q3 | 13-16 | 13-15 | |
| Menstruation, days | |||
| Medium | 5 | 5 | |
| Q1-Q3 | 4-6 | 4-7 | |
| Menstrual cycle, days | |||
| Medium | 30 | 30 | |
| Q1-Q3 | 29-30 | 29-30 | |
| Gravidity | |||
| Medium | 3 | 3 | |
| Q1-Q3 | 2-5 | 3-5 | |
| Produce | |||
| Medium | 2 | 2 | |
| Q1-Q3 | 1-3 | 1-3 | |
| Abortion | |||
| Medium | 1 | 1 | |
| Q1-Q3 | 0-2 | 0-3 | |
| Basic Fg value | |||
| Medium | 3.43 | 3.47 | |
| Q1-Q3 | 3.06-3.98 | 3.05-4.04 | |
| Basic APTT value | |||
| Medium | 30.2 | 31.0 | |
| Q1-Q3 | 27.7-32.8 | 28.8-33.4 | |
| Basic PT value | |||
| Medium | 11.0 | 11.6 | |
| Q1-Q3 | 10.3-12.2 | 10.5-12.4 | |
| Basic WBC value | |||
| Medium | 6.27 | 5.0 | |
| Q1-Q3 | 5.17-7.70 | 4.10-6.21 | |
| Basic N value | |||
| Medium | 3.94 | 3.08 | |
| Q1-Q3 | 3.02-5.11 | 1.99-3.96 | |
| Basic Hb value | |||
| Medium | 111.0 | 105.0 | |
| Q1-Q3 | 98.8-125.0 | 96.0-119.0 | |
| Basic PLT value | |||
| Medium | 274 | 220 | |
| Q1-Q3 | 219-345 | 187-310 | |
| Chemo-regimens | |||
| TP | 158 | 25 | <0.001 |
| CP | 59 | 32 | |
| Missing | 10 | 2 | |
| Patients location | |||
| Urban | 83 | 22 | 0.87 |
| Rural | 123 | 31 | |
| Missing | 21 | 6 | |
| Pathology | |||
| Squamous carcinoma | 196 | 49 | 0.72 |
| Adenocarcinoma | 19 | 5 | |
| Adenosquamous carcinoma | 4 | 2 | |
| FIGO stage | |||
| IA1 | 5 | 0 | 0.10 |
| IA2 | 3 | 0 | |
| IB1 | 71 | 19 | |
| IB2 | 47 | 10 | |
| IIA | 37 | 17 | |
| IIB | 54 | 9 | |
| IIIA | 1 | 1 | |
| IIIB | 8 | 2 | |
| IV | 0 | 1 | |
| Missing | |||
| Surgery | |||
| Pre-surgery | 121 | 29 | 0.57 |
| Post-surgery | 106 | 30 | |
| Menstrual bleeding | |||
| Excessive | 13 | 6 | 0.49 |
| Moderate | 204 | 51 | |
| Inadequate | 4 | 1 | |
| Missing | |||
| Dysmenorrhea | |||
| Yes | 198 | 49 | 0.13 |
| No | 19 | 9 | |
| Missing | 10 | 1 | |
| Menopause | |||
| Yes | 48 | 16 | 0.44 |
| No | 137 | 35 | |
| Missing | 42 | 8 | |
| Smoking status | |||
| No | 193 | 51 | 1.00 |
| Yes | 2 | 0 | |
| Missing | 32 | 8 | |
| Drinking status | |||
| No | 192 | 49 | 0.28 |
| Yes | 3 | 2 | |
| Missing | 32 | 8 | |
| HIV | |||
| Negative | 170 | 51 | none |
| Positive | 0 | 0 | |
| Missing | 57 | 8 | |
| HBV | |||
| Negative | 154 | 49 | 0.25 |
| Positive | 15 | 2 | |
| Missing | 49 | 6 |
Genomic association analysis
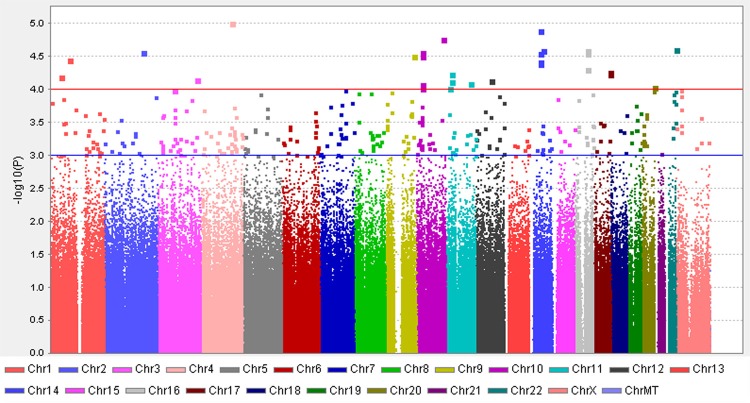
Assuming the additive mode, the top32 SNPs (P≤1 × 10-4) were chosen, whichwere most highly associated with neutropenia with adjustment of chemotherapy regimens. The selected SNPs were listed in Table 2. All genotype distributions followed the Hardy-Weinberg equilibrium law. The quantile-quantile plots showed some evidence for inflation due to population stratification (genomic inflation factor (λ = 1.013) (Figure 2).
Table 2.
Top 32 SNPs identified by genome-wide association study
| CHR | SNP | RS | BP | A1 | OR | L95 | U95 | P |
|---|---|---|---|---|---|---|---|---|
| 4 | SNP1 | NA | 136871784 | T | 3.178 | 1.907 | 5.295 | 9.06E-06 |
| 14 | SNP2 | NA | 47140657 | G | 4.307 | 2.241 | 8.277 | 1.18E-05 |
| 10 | SNP3 | rs1638410 | 118515516 | T | 4.454 | 2.24 | 8.857 | 2.05E-05 |
| 14 | SNP4 | rs9323332 | 58313237 | G | 2.919 | 1.776 | 4.799 | 2.40E-05 |
| 22 | SNP5 | rs1108364 | 46341876 | G | 2.686 | 1.697 | 4.25 | 2.46E-05 |
| 10 | SNP6 | rs11011962 | 20651003 | C | 2.867 | 1.757 | 4.678 | 2.50E-05 |
| 2 | SNP7 | NA | 170944892 | T | 4.986 | 2.359 | 10.54 | 2.59E-05 |
| 16 | SNP8 | rs4436775 | 54281857 | A | 2.66 | 1.686 | 4.198 | 2.63E-05 |
| 16 | SNP9 | rs4564560 | 54282265 | A | 2.618 | 1.672 | 4.102 | 2.63E-05 |
| 14 | SNP10 | NA | 47073011 | T | 6.163 | 2.636 | 14.41 | 2.71E-05 |
| 10 | SNP11 | rs10764221 | 20649311 | G | 2.859 | 1.748 | 4.675 | 2.86E-05 |
| 1 | SNP12 | rs4351663 | 83855776 | C | 6.458 | 2.676 | 15.58 | 3.32E-05 |
| 14 | SNP13 | rs10137341 | 46905759 | A | 3.639 | 1.972 | 6.714 | 3.58E-05 |
| 14 | SNP14 | NA | 47135262 | T | 4.028 | 2.077 | 7.812 | 3.73E-05 |
| 14 | SNP15 | NA | 47138095 | T | 4.028 | 2.077 | 7.812 | 3.73E-05 |
| 9 | SNP16 | rs72759216 | 125894343 | T | 3.07 | 1.791 | 5.263 | 4.53E-05 |
| 17 | SNP17 | NA | 70069186 | T | 2.668 | 1.661 | 4.284 | 4.92E-05 |
| 16 | SNP18 | rs11862589 | 54281443 | C | 2.53 | 1.614 | 3.966 | 5.17E-05 |
| 11 | SNP19 | rs340986 | 22463511 | T | 0.2363 | 0.1174 | 0.4755 | 5.26E-05 |
| 17 | SNP20 | rs55981110 | 70069449 | G | 2.594 | 1.634 | 4.119 | 5.35E-05 |
| 1 | SNP21 | rs34200648 | 44328065 | G | 3.372 | 1.863 | 6.103 | 5.94E-05 |
| 3 | SNP22 | NA | 177154093 | G | 3.142 | 1.791 | 5.513 | 6.57E-05 |
| 11 | SNP23 | rs1399096 | 22452671 | G | 0.2414 | 0.12 | 0.4858 | 6.79E-05 |
| 12 | SNP24 | rs11176925 | 66568062 | C | 2.981 | 1.741 | 5.102 | 6.84E-05 |
| 10 | SNP25 | rs11011954 | 20638640 | A | 2.89 | 1.708 | 4.893 | 7.73E-05 |
| 11 | SNP26 | rs1509728 | 108418533 | G | 2.494 | 1.583 | 3.93 | 8.11E-05 |
| 20 | SNP27 | rs7264385 | 57223196 | G | 2.94 | 1.717 | 5.037 | 8.59E-05 |
| 10 | SNP28 | rs987548 | 20639027 | G | 2.74 | 1.657 | 4.532 | 8.63E-05 |
| 11 | SNP29 | rs12799890 | 12490718 | G | 0.347 | 0.2043 | 0.5893 | 8.95E-05 |
| 23 | SNP30 | rs7051411 | 22358259 | A | 2.492 | 1.575 | 3.944 | 9.65E-05 |
| 7 | SNP31 | rs7796627 | 115873952 | T | 4.389 | 2.085 | 9.239 | 9.81E-05 |
| 3 | SNP32 | rs9822882 | 73736463 | A | 7.577 | 2.733 | 21.01 | 9.93E-05 |
A1, minor alleles. BP, Base pair position of the SNP. OR,odds ratio. OR and P werecalculated by an additive model in logistic regression analysis adjusted for chemo-regimens. MAF, minor allele frequency. L95, OR’s lower limitation for 95% CI. U95, OR’s upper limitation for 95% CI.
Figure 2.
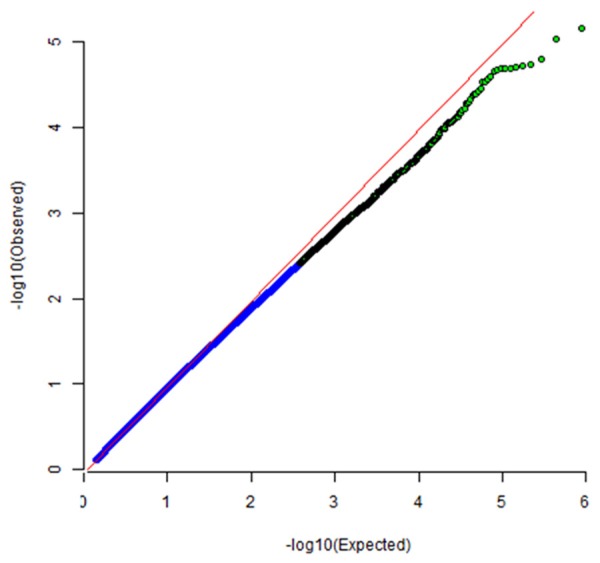
QQ plot for cases and controls study. The plot shows inflation factor for this genome-wide association study; and the effect of SNPs on the occurring of neutropenia.
Predictive factors (either clinical or genetic) for neutropenia
The details of the chemotherapy regimens and neutropenia cases in each group are shown in Table 2. Data from three quarters of the first internal group were used to train the ANN. Data from the rest one quarter of internal group were used for internal validation of the ANN. The two groups were similar in terms of chemotherapy regimens and clinical presentation (Table 2). All clinical and genetic variables shown in the Table 1 were used to construct the ANN model.
ANN model accuracy
The predictive accuracy of the ANN model was similar in the internal and external validation groups. The ANN model selected for analysis had an accuracy of 88.9% in predicting neutropenia (WHO grade 2-4) in the internal validation group, and 81.7% in theexternal validation group. Although the positive predictive value (PPV) of the ANN was not high in either validation group, the negative predictive value (NPV) was high in both the internal and external validation groups (98.2 and 93.2%, respectively) (Details was shown in Table 3).
Table 3.
Predictive performance of the ANN model in the internal and external validation group
| Model Validation | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|
| Internal Validation | 88.9 | 90.9 | 88.5 | 0.897 | 58.8 | 98.2 |
| External Validation | 81.7 | 72.7 | 83.7 | 0.782 | 50 | 93.2 |
ROC curve
The ROCcurves for clinical factors as well as each individual genomic marker and the ANN classifier illustrate the maximum area under the curve (AUC) for each factor. In the subset of evaluated cases in each validation cohort, the AUC for the ANN classifier (0.897 in training cohort, Figure 1A; 0.782 in validation cohort, Figure 1B) was greater than the AUC for all other factors considered in Table 1.
Figure 1.
The Manhattan plot shows a genome-wide view of the p values [log10 (P)] for association between SNPs tested and week 4 neutropenia in cervical cancer patients. 32 SNPs were discovered with P<10-4, with some SNPs clustered in some loci.
Discussion
As we know, this is the first study toinvestigate genetic determinants forchemotherapy-related cytopenia using a genome wide association approach in cervical cancer patients. In this study, we have identified a group of genetic variants causing neutropenia, SNPs with P value less than 10-4. In the drug of CPT-11 treated individuals, some genetic variants have been identified to associate with neutropenia,but no such studies for taxanes treated cervical cancer patients [14,15].
Screeningthe patients genetically predisposed to severe toxicity of classical cytotoxic agents is a critical issue. To analyze the role of genetic variants may shed light on the determinant factors of chemotherapy side effects. Understanding the genetic predisposition of cancer patients for developing severe toxicity has major clinical consequences. Our data indicate that platinum-based chemotherapy is relatively safe in patients with certain genetic background. On the contrary, severe and potentially life-threatening toxicity may occur and should be avoided in patients with suchbackground. This patient subset should probably reduce doses or, alternatively, with prepared Granulocyte Colony-Stimulating Factor (G-CSF), no matter where they are, in hospital or at home.
ANN-based modeling techniques were used in an attempt to predict the occurrence of moderate and serious adverse events. As predictive instruments for making a clinical decision, one of the important features is that only data that are readily available to the clinician at the time of occurrence of neutropenia are used. Our ANN predictive model combined data derived from genomic andclinical variantsby initial laboratory investigation, clinical history, and physical examination. We also emphasize that our ANN-based model is not meant to surrogate an experienced doctor. However, we advise that the ANN can be used as a decision aid for the clinical doctors. False positive rate andfalse negative rate were calculated to evaluate the model. The ANN model performed well in predicting the adverse events in both the internal and external validation groups.
This feature suggests that ANN may have a role in identifying patients who are at low risk of neutropenia and unlikely need therapeutic intervention during the treatment period. These patients could conceivably be discharged from the department with close outpatient follow-up, which may help save patients’ time and money. Such management strategy has obvious advantage on efficient utilization of health-care resources.
When our ANN model was tested in the independent, external cohort of patients, its performance was also impressive. Considering external cohort differs from those of the internal group, the clinical features between the two groups of patients were also different. Although the positive predictive value of the ANN was not high in either validation group, the negative predictive value was high in both the internal and external validation groups. This result was similar to the study made by doctors in University Hospitals of Cleveland [24]. Compared with conventional predictive models like multiple logistic regression model, ANN-based models are more universally applicable [20]. This kind of computer-based systems have generated exciting result for bettering care of patients [25].
Although the ANN software (usually Matlab) allowed researchers to identify the specific input parameters that improve predictive accuracy, these parameters should not be regarded as independent prognostic factors as perceived by a physician. Black-box approach was used in ANN-based models, while direct cause and effect relations between independent and dependent variables are usually unclear. Generally speaking, ANN-based models process input parameters in a non-linear style, and the network logic of calculation cannot be broken down into simple factors of clinical reasoning [26]. In the future, besides artificial neural network (ANN), other data-mining methods such as decision tree, logistic regression and support vector machines (SVMs) should be employed to analyze the massive data [27,28]. And pharmacogenetic studies also made contribution to identify biomarkers [29], which include genetic variants for evaluation of drug response [29-31]. Thus, the most suitable one can be applied. Prospective cohort studies are also needed to test the ability of ANN prediction models to improve management of chemotherapeutic patients in daily clinical practice.
Acknowledgements
We thank Professor Zengzhen Wang and Wenjun Sheng for the advice on statistical analysis. Grant support: This research was supported by thegrant from International S&T Cooperation Program of China (No.2013DFA31400), Program for New Century Excellent Talents in University (NO.NECT-12-0646), the Foundation of China (973 Program; No.2009CB521808) and by grants from the National Natural Science Foundation of China (No. 81300460; 8140211; 81370469; 81302264; 81201639; 81300453; 81072132; 81372781; 81071663; 81370469; 81230038; 81230052; 30973472; 81001151; 81071663; 30973205; 30973184; 81172464; 81101964) and National Major Science and Technology Project (No.2009ZX09103-739). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure of conflict of interest
None.
Abbreviations
- TP
Taxanesplus platinum
- CP
CPT-11plus platinum
- LACC
Locally advanced cervical cancer
- NACT
Neoadjuvant chemotherapy
- FIGO
International Federation of Gynecology and Obstetrics
- DFS
Disease-free survival
- OS
Overall survival
- pCR
Pathological completeresponse
- OPT
Optimal pathologic response
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Kere J. Genetics of complex disorders. Biochem Biophys Res Commun. 2010;396:143–146. doi: 10.1016/j.bbrc.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Huang RS, Ratain MJ. Pharmacogenetics and pharmacogenomics of anticancer agents. CA Cancer J Clin. 2009;59:42–55. doi: 10.3322/caac.20002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toffoli G, Cecchin E, Corona G, Russo A, Buonadonna A, D’Andrea M, Pasetto LM, Pessa S, Errante D, De Pangher V, Giusto M, Medici M, Gaion F, Sandri P, Galligioni E, Bonura S, Boccalon M, Biason P, Frustaci S. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J. Clin. Oncol. 2006;24:3061–3068. doi: 10.1200/JCO.2005.05.5400. [DOI] [PubMed] [Google Scholar]
- 6.Boige V, Mendiboure J, Pignon JP, Loriot MA, Castaing M, Barrois M, Malka D, Tregouet DA, Bouche O, Le Corre D, Miran I, Mulot C, Ducreux M, Beaune P, Laurent-Puig P. Pharmacogenetic assessment of toxicity and outcome in patients with metastatic colorectal cancer treated with LV5FU2, FOLFOX, and FOLFIRI: FFCD 2000-05. J. Clin. Oncol. 2010;28:2556–2564. doi: 10.1200/JCO.2009.25.2106. [DOI] [PubMed] [Google Scholar]
- 7.Cecchin E, Innocenti F, D’Andrea M, Corona G, De Mattia E, Biason P, Buonadonna A, Toffoli G. Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan. J. Clin. Oncol. 2009;27:2457–2465. doi: 10.1200/JCO.2008.19.0314. [DOI] [PubMed] [Google Scholar]
- 8.Yao S, Barlow WE, Albain KS, Choi JY, Zhao H, Livingston RB, Davis W, Rae JM, Yeh IT, Hutchins LF, Ravdin PM, Martino S, Lyss AP, Osborne CK, Abeloff M, Hortobagyi GN, Hayes DF, Ambrosone CB. Gene polymorphisms in cyclophosphamide metabolism pathway, treatment-related toxicity and disease-free survival in SWOG 8897 clinical trial for breast cancer. Clin Cancer Res. 2010;16:6169–6176. doi: 10.1158/1078-0432.CCR-10-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han B, Gao G, Wu W, Gao Z, Zhao X, Li L, Qiao R, Chen H, Wei Q, Wu J, Lu D. Association of ABCC2 polymorphisms with platinum-based chemotherapy response and severe toxicity in non-small cell lung cancer patients. Lung Cancer. 2011;72:238–243. doi: 10.1016/j.lungcan.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Joerger M, Burgers JA, Baas P, Doodeman VD, Smits PH, Jansen RS, Vainchtein LD, Rosing H, Huitema AD, Beijnen JH, Schellens JH. Gene polymorphisms, pharmacokinetics and hematological toxicity in advanced non-small-cell lung cancer patients receiving cisplatin/gemcitabine. Cancer Chemother Pharmacol. 2012;69:25–33. doi: 10.1007/s00280-011-1670-4. [DOI] [PubMed] [Google Scholar]
- 11.Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst. 2007;99:1290–1295. doi: 10.1093/jnci/djm115. [DOI] [PubMed] [Google Scholar]
- 12.Sugiyama E, Kaniwa N, Kim SR, Kikura-Hanajiri R, Hasegawa R, Maekawa K, Saito Y, Ozawa S, Sawada J, Kamatani N, Furuse J, Ishii H, Yoshida T, Ueno H, Okusaka T, Saijo N. Pharmacokinetics of gemcitabine in Japanese cancer patients: the impact of a cytidine deaminase polymorphism. J. Clin. Oncol. 2007;25:32–42. doi: 10.1200/JCO.2006.06.7405. [DOI] [PubMed] [Google Scholar]
- 13.McLeod HL, Sargent DJ, Marsh S, Green EM, King CR, Fuchs CS, Ramanathan RK, Williamson SK, Findlay BP, Thibodeau SN, Grothey A, Morton RF, Goldberg RM. Pharmacogenetic predictors of adverse events and response to chemotherapy in metastatic colorectal cancer: results from North American Gastrointestinal Intergroup Trial N9741. J. Clin. Oncol. 2010;28:3227–3233. doi: 10.1200/JCO.2009.21.7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Innocenti F, Kroetz DL, Schuetz E, Dolan ME, Ramirez J, Relling M, Chen P, Das S, Rosner GL, Ratain MJ. Comprehensive pharmacogenetic analysis of irinotecan neutropenia and pharmacokinetics. J. Clin. Oncol. 2009;27:2604–2614. doi: 10.1200/JCO.2008.20.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han JY, Lim HS, Shin ES, Yoo YK, Park YH, Lee JE, Jang IJ, Lee DH, Lee JS. Comprehensive analysis of UGT1A polymorphisms predictive for pharmacokinetics and treatment outcome in patients with non-small-cell lung cancer treated with irinotecan and cisplatin. J. Clin. Oncol. 2006;24:2237–2244. doi: 10.1200/JCO.2005.03.0239. [DOI] [PubMed] [Google Scholar]
- 16.Stewart CF, Panetta JC, O’Shaughnessy MA, Throm SL, Fraga CH, Owens T, Liu T, Billups C, Rodriguez-Galindo C, Gajjar A, Furman WL, McGregor LM. UGT1A1 promoter genotype correlates with SN-38 pharmacokinetics, but not severe toxicity in patients receiving low-dose irinotecan. J. Clin. Oncol. 2007;25:2594–2600. doi: 10.1200/JCO.2006.10.2301. [DOI] [PubMed] [Google Scholar]
- 17.Lamas MJ, Duran G, Balboa E, Bernardez B, Candamio S, Vidal Y, Mosquera A, Giraldez JM, Lopez R, Carracedo A, Barros F. The value of genetic polymorphisms to predict toxicity in metastatic colorectal patients with irinotecan-based regimens. Cancer Chemother Pharmacol. 2012;69:1591–1599. doi: 10.1007/s00280-012-1866-2. [DOI] [PubMed] [Google Scholar]
- 18.Baxt WG. Application of artificial neural networks to clinical medicine. Lancet. 1995;346:1135–1138. doi: 10.1016/s0140-6736(95)91804-3. [DOI] [PubMed] [Google Scholar]
- 19.Bottaci L, Drew PJ, Hartley JE, Hadfield MB, Farouk R, Lee PW, Macintyre IM, Duthie GS, Monson JR. Artificial neural networks applied to outcome prediction for colorectal cancer patients in separate institutions. Lancet. 1997;350:469–472. doi: 10.1016/S0140-6736(96)11196-X. [DOI] [PubMed] [Google Scholar]
- 20.Cross SS, Harrison RF, Kennedy RL. Introduction to neural networks. Lancet. 1995;346:1075–1079. doi: 10.1016/s0140-6736(95)91746-2. [DOI] [PubMed] [Google Scholar]
- 21.Lapuerta P, Rajan S, Bonacini M. Neural networks as predictors of outcomes in alcoholic patients with severe liver disease. Hepatology. 1997;25:302–306. doi: 10.1053/jhep.1997.v25.pm0009021938. [DOI] [PubMed] [Google Scholar]
- 22.Djavan B, Remzi M, Zlotta A, Seitz C, Snow P, Marberger M. Novel artificial neural network for early detection of prostate cancer. J. Clin. Oncol. 2002;20:921–929. doi: 10.1200/JCO.2002.20.4.921. [DOI] [PubMed] [Google Scholar]
- 23.Mian S, Ugurel S, Parkinson E, Schlenzka I, Dryden I, Lancashire L, Ball G, Creaser C, Rees R, Schadendorf D. Serum proteomic fingerprinting discriminates between clinical stages and predicts disease progression in melanoma patients. J. Clin. Oncol. 2005;23:5088–5093. doi: 10.1200/JCO.2005.03.164. [DOI] [PubMed] [Google Scholar]
- 24.Das A, Ben-Menachem T, Cooper GS, Chak A, Sivak MV Jr, Gonet JA, Wong RC. Prediction of outcome in acute lower-gastrointestinal haemorrhage based on an artificial neural network: internal and external validation of a predictive model. Lancet. 2003;362:1261–1266. doi: 10.1016/S0140-6736(03)14568-0. [DOI] [PubMed] [Google Scholar]
- 25.Johnston ME, Langton KB, Haynes RB, Mathieu A. Effects of computer-based clinical decision support systems on clinician performance and patient outcome. A critical appraisal of research. Ann Intern Med. 1994;120:135–142. doi: 10.7326/0003-4819-120-2-199401150-00007. [DOI] [PubMed] [Google Scholar]
- 26.Baxt WG. Use of an artificial neural network for the diagnosis of myocardial infarction. Ann Intern Med. 1991;115:843–848. doi: 10.7326/0003-4819-115-11-843. [DOI] [PubMed] [Google Scholar]
- 27.Liu W, Xie H. Prediction of regulation relationship between protein interactions in signaling networks. Biochem Biophys Res Commun. 2013;440:388–392. doi: 10.1016/j.bbrc.2013.09.093. [DOI] [PubMed] [Google Scholar]
- 28.Luk JM, Lam BY, Lee NP, Ho DW, Sham PC, Chen L, Peng J, Leng X, Day PJ, Fan ST. Artificial neural networks and decision tree model analysis of liver cancer proteomes. Biochem Biophys Res Commun. 2007;361:68–73. doi: 10.1016/j.bbrc.2007.06.172. [DOI] [PubMed] [Google Scholar]
- 29.Chai H, Pan J, Zhang X, Shen X, Li H, Zhang K, Yang C, Sheng H, Gao H. ERCC1 C118T associates with response to FOLFOX4 chemotherapy in colorectal cancer patients in Han Chinese. Int J Clin Exp Med. 2012;5:186–194. [PMC free article] [PubMed] [Google Scholar]
- 30.Gu AQ, Wang WM, Chen WY, Shi CL, Lu JH, Han JQ. XRCC1 genetic polymorphisms and sensitivity to platinum-based drugs in non-small cell lung cancer: an update meta-analysis based on 4708 subjects. Int J Clin Exp Med. 2015;8:145–154. [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez-Chavez A, Thomas A, Rajan A, Raffeld M, Morrow B, Kelly R, Carter CA, Guha U, Killian K, Lau CC, Abdullaev Z, Xi L, Pack S, Meltzer PS, Corless CL, Sandler A, Beadling C, Warrick A, Liewehr DJ, Steinberg SM, Berman A, Doyle A, Szabo E, Wang Y, Giaccone G. Molecular Profiling and Targeted Therapy for Advanced Thoracic Malignancies: A Biomarker-Derived, Multiarm, Multihistology Phase II Basket Trial. J. Clin. Oncol. 2015;33:1000–1007. doi: 10.1200/JCO.2014.58.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]