Abstract
Glioma is the most common and aggressive brain tumor with poor clinical outcome. Identification and development of new biomarkers could be beneficial for diagnosis and prognosis of glioma patients. Recent studies have showed evidences that dysregulation of microRNAs (miRNAs) is involved in glioma tumorigenesis. Therefore, we attempted to identify specific miRNAs as prognostic and predictive markers for glioma. We statistically compared expression profile of 365 miRNAs between WHO grade IV and grade III gliomas, by qRT-PCR. MiR-105 was identified as a remarkably decreased miRNA in grade IV gliomas compared with grade III gliomas (P=0.012, fold change =0.04). We subsequently examined its expression levels in an independent series of gliomas, and statistically analyzed the associations between miR-105 expression and clinicopathological characteristics and survivals of these glioma patients. MiR-105 showed remarkably decreased expression in gliomas as compared to non-neoplastic brains. And grade IV gliomas had significantly lower miR-105 expression compared with grade III and II gliomas (both P<0.001). Additionally, low miR-105 expression was statistically associated with advanced tumor grade, advanced patient’s age and low pre-operative Karnofsky performance score (all P<0.001). Furthermore, patients with low miR-105 expression had significantly poorer survival by Kaplan-Meier method (P<0.001). Multivariate analysis indicated miR-105 as an independent prognostic indicator for glioma patients (P=0.018, risk ratio =4.2). Our results suggested that low expression of miR-105 may correlate with unfavorable clinical outcome and be involved in tumorigenesis and aggressive progression of glioma. And miR-105 may be a novel biomarker in prognostic prediction for glioma.
Keywords: microRNA, miR-105, glioma, glioblastoma, down-regulation, prognosis
Introduction
MicroRNAs (miRNAs) are small, noncoding regulatory RNA molecules of about 18-25 nucleotides, first discovered in early 1990s in C. elegans [1]. They are considered to play important roles in a variety of biological processes including cell proliferation, apoptosis, migration and differentiation, through post-translationally regulating expression of their target genes [2,3]. In recent years, a growing number of evidences have suggested that miRNAs are involved in human tumorigenesis by functioning as oncogenes or tumor suppressors [4,5]. Identification of specific miRNAs in cancer cells is thought to have substantial value for diagnostic and prognostic determinations as well as for eventual therapeutic interventions [6,7].
Gliomas are the most frequent and malignant primary brain tumors in human adults. The World Health Organization (WHO) classifies human gliomas into pilocytic astrocytoma (PA, WHO grade I), diffuse astrocytoma (DA, WHO grade II), anaplastic astrocytoma (AA, WHO grade III), and glioblastoma (GBM, WHO grade IV) in the order of increasing malignancy [8]. Despite recent advances in surgery, radiotherapy, and chemotherapy, the prognosis for patients with this tumor remains poor. The grade IV glioma, also known as glioblastoma multiforme (GBM), is the most common and aggressive form of glioma, with a median survival of only 12-15 months as compared to 2-5 years for patients with grade III gliomas and 6-8 years for low grade (I and II) gliomas [8]. Therefore, it is necessary to explore new diagnostic and prognostic biomarkers and effective therapeutics that may be beneficial for improving the clinical management of glioma.
Recent studies have indicated that expression profiles of miRNAs are associated with patients’ survival and are able to function as prognostic and predictive indicators in glioma, based on public database entries [9-13] or independent tissue cohorts [14-17]. Most of these identified miRNAs have also been proved to be involved in the tumorigenesis and function as oncogenes or tumor suppressors in human gliomas, such as miR-21 [18,19], miR-155 [20-22], miR-196 [23,24], miR-221/222 [25] and miR-326 [26,27]. Thus, the identification of the miRNA expression signature for gliomas, in particular glioblastoma, is of great significance not only for predicting clinical outcomes, but also for understanding the molecular mechanisms of tumorigenesis and developing novel therapeutics of these malignancies.
In the attempt to identify molecular determinants associated with clinical outcome aggressive behavior in gliomas, we statistically compared the expression levels of 365 miRNAs between 4 grade IV and 4 grade III gliomas with the worst and a relative favorable clinical outcome, respectively. We identified 23 and 11 miRNAs that showed remarkably up- and down-regulation in grade IV gliomas versus grade III gliomas, respectively. Several identified miRNAs, such as miR-21, miR-155 and miR-326, have been demonstrated to be involved in tumorigenesis and have prognostic implications of human glioma [18-22,26,27]. We then focused on a significantly down-regulated miRNA in grade IV gliomas, miR-105, whose clinical significance in glioma remains unclear, for further analysis. Expression levels of miR-105 were subsequently examined in an independent series of 76 gliomas including 50 grade IV, 13 grade III and 13 grade II tumors, as well as 10 normal brain tissues, by qRT-PCR method. MiR-105 showed remarkably decreased expression in gliomas when compared with control brain tissues and grade IV gliomas had significantly lower miR-105 expression than grade III and II tumors. In addition, low miR-105 expression was statistically associated with advanced tumor grade, advanced patient’s age and low pre-operative Karnofsky performance score. Furthermore, patients with low miR-105 expression had significantly poorer survival and multivariate analysis indicated miR-105 as an independent prognostic indicator for glioma patients.
Materials and methods
Glioma specimens and patients
Glioma specimens were obtained from patients during surgery at First Affilated Hospital of China Medical University. A portion of the tumor tissue was saved and made into paraffin sections for histopathologic diagnosis in strict accordance with World Health Organization (WHO) criteria by two established neuropathologists, with differences resolved by careful discussion. And the remaining tissue was snap-frozen in liquid nitrogen then stored at -80°C for RNA extraction and other biological molecular experiments. Before the RNA extraction from frozen tissues, the adjacent tumor tissues were subjected to frozen sections and reviewed by a pathologist to ensure that a minimum of 80% tumor cells were included in the sample. To profile the global miRNAs expression of malignant glioma, 8 high-pathological gliomas including 4 GBMs and 4 AAs were selected for the global miRNA expression screening by TanMan real-time quantitative PCR array. Subsequently, expression levels of miR-105 were examined on an independent series of 76 gliomas including 50 GBMs, 13 AAs and 13 DAs, as well as 10 non-neoplastic brain tissues for calibration purpose, by conventional miRNA real-time PCR assay. These non-neoplastic brain tissues used as controls were obtained by collecting donations with consents from individuals who died in traffic accidents and were confirmed to be free of any prior pathological lesions. For glioma patients, none of them had received chemotherapy or radiotherapy prior to surgery, and all patients were well followed up. Overall survival time was calculated from the date of the initial surgical operation to death. Patients, who died of diseases not directly related to their gliomas or due to unexpected events, were excluded from this study. The present study was approved by the Ethics Committee of China Medical University.
RNA extraction, reverse transcription and real-time PCR quantification for miRNA detection
Total RNA was extracted from frozen tissues of glioma and non-neoplastic brain using a mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacture’s instruction. RNA concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), and RNA quality was measured using a denaturing 15% polyacrylamide gel.
To compare the global miRNAs expression levels between GBMs and AAs, PCR array assays were applied by using TaqMan Human MiRNA Arrays v1.0 (Applied Biosystems, Foster City, CA, USA). In brief, 365 human miRNAs and enddogenous controls RNU6B were reverse transcribed using 8 predefined reverse transcription primer pools containing up to 48 reverse transcription primers each. The reverse transcription products were subsequently loaded onto the TaqMan Array to perform real-time PCR amplification using Applied Biosystems 7900HT Fast Real-Time PCR System. Relative quantification of miRNA expression was calculated with the 2-ΔΔCt method.
To examine the expression levels of miR-105 in glioma tissues and cell lines, cDNA synthesis and subsequent quantitative real-time PCR were porformed using a TaqMan MiRNA Reverse Transcription Kit (Applied Biosystems) and individual TaqMan miRNA assay (Applied Biosystems), and Applied Biosystems 7500HT Fast Real-Time PCR System (Applied Biosystems), as previously described [28]. RNU6B were used as endogenous controls, non-neoplastic brain tissues and human astrocyte were used for calibrations. Relative quantification of miR-105 expression was calculated with the 2-ΔΔCt method.
Statistical analysis
All computations were carried out using the software of SPSS version 19.0 for Windows (SPSS Inc., Chicago, IL, USA). Data were expressed as mean ± standard deviation (SD). Student’s t-test was used to compare the expression levels of miRNAs between different subtypes of glioma, as well as between gliomas and non-neoplastic brains. The χ2 test was used to analyze the relationship between miRNA expression and the clinicopathological characteristics. A life table was calculated according to the Kaplan-Meier method. Risk ratios for the time-to-event endpoint were estimated using the multivariate Cox regression analysis in a forward stepwise method to evaluate the effect of multiple independent prognostic factors on overall survival outcome. Differences were considered statistically significant when P was less than 0.05.
Results
Comparison of 365 miRNA expression between WHO grade IV and III gliomas
To identify specific miRNAs that significantly correlate with tumor grade and prognosis of glioma, we examined the expression levels of 365 human miRNAs in 8 high-grade gliomas including 4 GBMs (WHO grade IV) and 4 AAs (WHO grade III), by TaqMan microRNA PCR array assays. Subsequently, we statistically compared expression levels of these miRNAs between GBMs and AAs, by student’s t-test. In total, 43 miRNAs showed significantly different expression between GBMs and AAs (P<0.05). Among these, 34 were identified as candidate miRNAs (Table 1) that showed remarkably increased (23 miRNAs; fold change >3) or decreased (11 miRNAs; fold change <1/3) expression in GBMs when compared with AAs (Figure 1). Furthermore, we focused on miR-105, which was significantly down-regulated (P=0.012, fold change =0.04, Figure 1) in GBMs as compared with AAs, for further analyses in another panel of gliomas.
Table 1.
miRNAs showed remarkably differential expression between GBMs and AAs
| Down-regulated microRNA | P value (Student’s t test) | Fold change (GBM vs. AA) |
|
| ||
| hsa-miR-504 | 0.0009 | 0.11 |
| hsa-miR-184 | 0.0052 | 0.14 |
| hsa-miR-326 | 0.0055 | 0.25 |
| hsa-miR-601 | 0.007 | 0.19 |
| hsa-miR-105 | 0.0115 | 0.04 |
| hsa-miR-128b | 0.0125 | 0.21 |
| hsa-miR-517c | 0.0144 | 0.14 |
| hsa-miR-383 | 0.0153 | 0.25 |
| hsa-miR-101 | 0.0167 | 0.32 |
| hsa-miR-575 | 0.0278 | 0.18 |
| hsa-miR-367 | 0.0436 | 0.03 |
|
| ||
| Up-regulated microRNA | P value (Student’s t test) | Fold change (GBM vs. AA) |
|
| ||
| hsa-miR-155 | <0.0001 | 10.35 |
| hsa-miR-15b | 0.0023 | 6.26 |
| hsa-miR-335 | 0.0033 | 3.09 |
| hsa-miR-196a | 0.0035 | 289.86 |
| hsa-miR-551b | 0.0039 | 4.53 |
| hsa-miR-34a | 0.006 | 3.85 |
| hsa-miR-148a | 0.0076 | 3.83 |
| hsa-miR-378 | 0.0113 | 3.55 |
| hsa-miR-130b | 0.0117 | 4.51 |
| hsa-miR-21 | 0.0169 | 5.72 |
| hsa-miR-135b | 0.0172 | 9.06 |
| hsa-miR-296 | 0.0206 | 3.15 |
| hsa-miR-199a | 0.0228 | 3.58 |
| hsa-miR-429 | 0.0279 | 6.01 |
| hsa-miR-214 | 0.032 | 5.19 |
| hsa-miR-93 | 0.0322 | 3.08 |
| hsa-miR-432* | 0.0373 | 5.4 |
| hsa-miR-363 | 0.0378 | 24.85 |
| hsa-miR-382 | 0.0396 | 3.05 |
| hsa-miR-449b | 0.0414 | 4.71 |
| hsa-miR-126 | 0.0423 | 3.08 |
| hsa-miR-196b | 0.0449 | 10.52 |
| hsa-miR-615 | 0.0473 | 61.37 |
Abbreviations: AA, anaplastic astrocytoma; GBM, glioblastoma.
Figure 1.
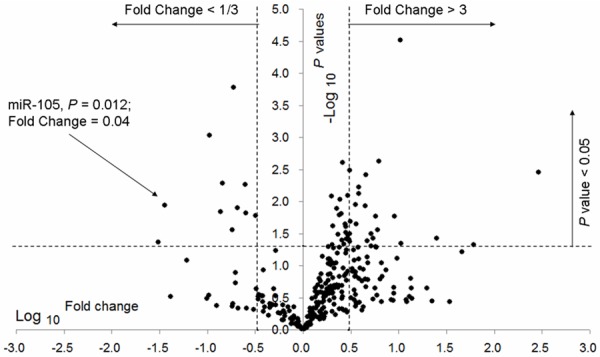
A volcano plot showing the fold change in the expression 365 miRNAs between 4 GBMs and 4 AAs, detected by miRNA qPCR array analysis. The volcano plot shows the distribution of these 365 miRNAs according to their P values. The horizontal dotted line, left and right vertical dotted lines indicate P value =0.05, fold change <1/3 and >3, respectively. A set of 34 miRNAs were identified to be remarkably increased (fold change >3 and P<0.05) or decreased (fold change <1/3 and P<0.05) in GBMs versus AAs. MiR-105 had significantly lower expression in GBMs as compared to AAs (left arrow; P=0.012 and fold change =0.04).
Down-regulation of miR-105 in glioma tissues
To further evaluate the dysregulation of miR-105 in gliomas, we examined its expression levels in an independent panel of 76 gliomas including 50 GBMs (grade IV), 13 AAs (grade III) and 13 DAs (grade II), as well as 10 non-neoplastic brain tissues for control purpose, by qRT-PCR. As shown in Figure 2, expression of miR-105 were significantly decreased in glioma tissues compared with non-neoplastic brain tissues (fold change =0.47; P≤0.001, Student’s t-test). GBMs showed remarkably lower miR-105 expression relative to brain tissues (fold change =0.23; P≤0.001). However, neither AAs nor DAs had significantly differential expression of miR-105 than normal brain tissues. In addition, miR-105 expression in GBMs was statistically lower than that in both AAs and DAs (both P≤0.001). No significant difference was observed between AAs and DAs.
Figure 2.
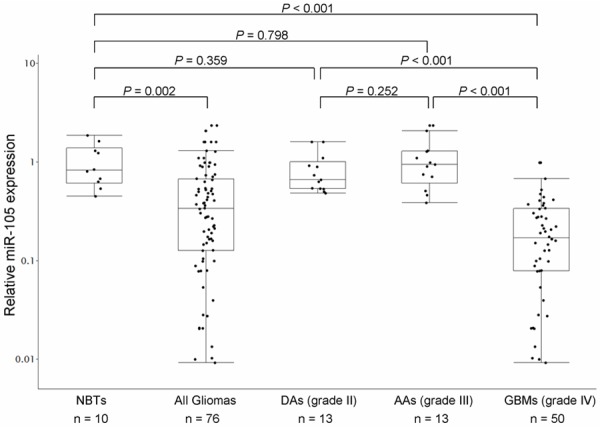
MiR-105 expression in 76 glioma tissues (50 GBMs, 13 AAs and 13 DAs) and 10 non-neoplastic brain tissues, detected by quantitative reverse transcriptive real-time polymerase chain reaction (qRT-PCR) analysis. Dots indicate the relative quantification (RQ) values of miRNA expression level, normalized by RNU6B. miR-105 expression was found to be remarkably decreased in glioma tissues compared to normal brain tissues (P=0.002). GBMs showed significantly lower expression of miR-105 than brain tissues (P≤0.001). Statistically significantly lower expression of miR-105 was observed in GBMs versus AAs (P≤0.001), as well as in GBMs versus DAs (P≤0.001).
Correlation of low miR-105 expression with clinicopathological features of glioma
Subsequently, we statistically evaluated the associations of miR-105 expression with several clinicopathological features including tumor grade, patients’ age at diagnosis, gender and pre-operative Karnofsky performance scale (KPS) of these 76 glioma patients by Χ2 test as summarized in Table 2. We assigned gliomas to high-miR-105 group and low-miR-105 group that were tumors with miR-105 expression above and under the mean value of miR-105 expression in all of the 76 gliomas, respectively (n=29 and 47 for high-miR-105 and low-miR-105 groups, respectively). As shown in Table 2, low miR-105 expression was significantly associated with advanced tumor grade, advanced patient’s age and low pre-operative Karnofsky performance score (all P≤0.001, χ2 test). However, no statistically significant correlation was observed between miR-105 expression and patient’s gender (Table 2).
Table 2.
Correlation of miR-105 expression with clinicopathological characteristics of gliomas
| Clinicopathological features | No. of cases | miR-105 expression | P | |
|---|---|---|---|---|
|
| ||||
| High (n, %) | Low (n, %) | |||
| WHO grade | ≤0.001 | |||
| II | 13 | 13 (100.0) | 0 (0.0) | |
| III | 13 | 11 (84.6) | 2 (15.4) | |
| IV | 50 | 5 (10.0) | 45 (90.0) | |
| Age | ≤0.001 | |||
| >50 | 48 | 9 (18.8) | 39 (81.2) | |
| ≤50 | 28 | 20 (71.4) | 8 (28.6) | |
| Gender | 0.866 | |||
| Male | 41 | 16 (39.0) | 25 (61.0) | |
| Female | 35 | 13 (37.1) | 22 (62.9) | |
| KPS | ≤0.001 | |||
| <90 | 42 | 8 (19.0) | 34 (81.0) | |
| ≥90 | 34 | 21 (61.8) | 13 (38.2) | |
Abbreviations: KPS, Karnofsky performance scale.
Prognostic implication of miR-105 down-regulation in glioma patients
Furthermore, we evaluate the potential prognostic performance of miR-105 expression in glioma patients, by performing Kaplan-Meier analysis. We observed that miR-105 expression displayed significant correlations with glioma patients’ overall survival. As shown in Figure 3A, patients in low-miR-105 group (n=47) had significantly shorter overall survival compared to that in high-miR-105 group (n=29) (mean overall survival times for patients in low- and high-miR-105 groups were 426 and 1618 days, respectively, P≤0.001, logrank test). In addition, we performed univariate and multivariate analysis using the Cox propotional harzard regression model to determine whether miR-105 expression and other clinical parameters are independent factors for prognostic prediction in glioma patients. Our assessment revealed that both low-miR-105 (P = 0.012; risk ratio 4.2, multivaratie Cox regression analysis) and high pathological grade (P≤0.001; risk ratio 8.0, multivaratie Cox regression analysis) were independent predictors of poor prognosis in glioma patients (Table 3). Moreover, the prognostic value of miR-105 expression was also analyzed in patients with high-grade glioma (grade III and IV) and we found that low-miR-105 expression was significantly associated with poor overall survival of patients with these aggressive tumors (P≤0.001, Figure 3B).
Figure 3.
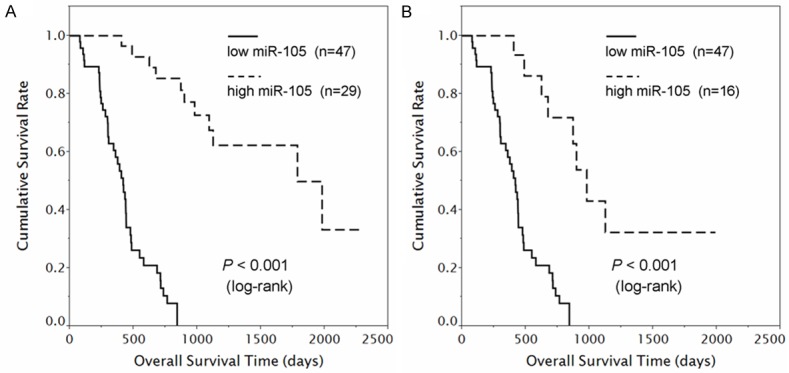
Prognostic performance of miR-105 expression in gliomas. A. Among the 76 glioma patients (50 GBMs, 13 AAs and 13 DAs), those with low miR-105 expression (left, solid line, n=47) had significantly shorter survival periods than did patients with high miR-105 expression (right, dotted line, n=29; P≤0.001, log-rank test). B. Among the 63 high-grade glioma patients (50 GBMs and 13 AAs), those with low miR-105 expression (left, solid line, n=47) had significantly shorter survival periods than did patients with high miR-105 expression (right, dotted line, n=16; P≤0.001).
Table 3.
Univariate and multivariate Cox regression analysis for overall survival in glioma patients
| Univariate analysis | Multivariate analysis | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Variate | No. of case (%) | Mean OS | 95% CI | P (log-rank) | Variate | RR | 95% CI | P |
| WHO grade | ≤0.001 | WHO grade | ≤0.001 | |||||
| II | 13 (17.1%) | 1947 | 1688-2205 | IV vs. III vs. II | 8.0 | 2.6-25.0 | ||
| III | 13 (17.1%) | 1330 | 936-1724 | |||||
| IV | 50 (65.8%) | 444 | 378-509 | |||||
| Age | ≤0.001 | Age | 0.055 | |||||
| >50 | 48 (63.2%) | 547 | 409-685 | >50 vs. ≤50 | 0.4 | 0.2-1.0 | ||
| ≤50 | 28 (36.8%) | 1378 | 1070-1686 | |||||
| Gender | 0.888 | Gender | 0.121 | |||||
| Male | 41 (53.9%) | 953 | 699-1208 | Male vs. Female | 1.6 | 0.9-3.0 | ||
| Female | 35 (46.1%) | 808 | 599-1017 | |||||
| KPS | ≤0.001 | KPS | 0.292 | |||||
| <90 | 42 (55.3%) | 505 | 405-605 | <90 vs. ≥90 | 1.5 | 0.7-3.0 | ||
| ≥90 | 34 (44.7%) | 1352 | 1057-1648 | |||||
| Surgical resection | 0.182 | Surgery | 0.232 | |||||
| GTR | 38 (50.0%) | 1018 | 753-1284 | PR vs. GTR | 1.4 | 0.8-2.6 | ||
| PR | 38 (50.0%) | 771 | 556-985 | |||||
| miR-105 expression | ≤0.001 | miR-105 expression | 0.018 | |||||
| Low | 47 (61.8%) | 426 | 362-490 | Low vs. High | 4.2 | 1.3-13.6 | ||
| High | 29 (31.2%) | 1618 | 1339-1896 | |||||
Abbreviations: KPS, Karnofsky performance scale; GTR, gross total resection; PR, partial resection; OS, overall survival; RR, risk ratio.
Discussion
Prognostic predictions and treatment options for patients with glioma mainly depend on the histological grade of the tumor due to the fact that the biological behaviors and clinical outcomes are distinctly different between glioma histological subtypes [8]. However, as the currently used histology-based grading is subjective, it is necessary to develop more accurate methods of glioma classification. Recent integrated genomic analyses based on global gene expression profiles have allowed molecular subclassification of malignant glioma and also provided biomarkers for use in diagnosis and clinical management [29-31]. However, these findings and related signatures are yet to be translated to utility in clinical settings, suggesting that these studies require further characterization and validation. Recent studies have indicated that expression-based clustering using miRNA profile can yield more accurate histological and prognostic cancer classification than clustering based on mRNA expression profile [32,33]. Several groups have identified candidate miRNAs that are associated with patients’ survival and involved in tumorigenesis of human glioma, by microarray-based high-throughput miRNA expression profiling [9-17]. However, these studies mentioned above were mainly performed on GBM cohorts.
We in the present study statistically compared the 365-miRNA expression profile of 4 WHO grade IV gliomas (GBMs) with 4 WHO grade III tumors (AAs), by qRT-PCR array. In total, 34 remarkably altered miRNAs including 23 up- and 11 down-regulated ones between the two glioma groups, were identified (Table 1). Among these 34 identified candidate miRNAs, several have been demonstrate to have prognostic significance and/or be involved in tumorigenesis and aggressive progression of glioma, by functioning as oncogenes or tumor suppressors. For example, as one of the most frequently dysregulated miRNAs in glioma, miR-21 has been revealed to act as an antiapoptotic factor that targets a network of p53, transforming growth factor (TGF)-β, and mitochondrial apoptosis tumor suppressive genes in glioblastoma cells [18,19], a recent study by Hermansen et al. indicated that miR-21 overexpression have unfavorable prognostic value in glioma patients [15]; miR-155, which showed the most significantly differential expression among the up-regulated candidate miRNAs (P<0.0001, fold change =10.35, Table 1) in our present study, has been previously indicated to promote the cell proliferation and contribute to progression of glioma [20,22], in addition, its overexpression predicts poor prognosis in glioma patients [21]; miR-196a showed a hundreds-fold increased expression level in GBMs vs. AAs in our data (P=0.004, fold change =289.86, Table 1). Previous studies have showed the overexpression of miR-196a in glioblastoma cell lines and tissues and indicated the prognostic significance of this miRNA in glioma patients [14,23]. A recent study showed evidence that miR-196a exerted its oncogenic effect in glioblastoma by inhibition of IκBα [24]; the tumor-suppressive miRNA, miR-326, has been revealed to inhibit glioma cell survival by directly regulating expression of its target genes, Notch-1 and PKM2 [26,27], furthermore, low miR-326 expression was reported to be associated with unfavorable outcome of glioma patients [17]. These miRNAs might have significant values for diagnostic and prognostic determinations as well as for eventual therapeutic interventions. Therefore, they should be included into the molecular-based glioma classification system as key miRNAs and synthetically analyzed in the future studies. Taken together, expression profiles of miRNA are useful for predicting glioma patient survival, and have the potential to identify efficacious therapeutic targets.
Among the 34 candidate miRNAs that showed remarkably increased or decreased expression level in our present study, miR-105 was significantly down-regulated in GBMs compared with AAs (P=0.012, fold change =0.04, Table 1). However, there are few studies about function and clinical significance in human glioma of this miRNA. Barbano et al. in 2014 compared miRNA expression profile between low-grade and high-grade gliomas and identified 22 miRNAs distinguishing these two types of the tumor, including miR-105, by miRNA-microarray assay. They subsequently validated expression patterns of these candidate miRNAs in an independent panel of glioma tissues by qRT-PCR, and indicated the significant down-regulation of miR-105 in high-grade gliomas compared with low-grade tumors, as well as normal brain tissues [13]. A simultaneously published study by Yan et al developed a five-miRNA signature including miR-105 as a protective miRNA that could identify patients with a high risk of unfavorable outcome in anaplastic gliomas regardless of histology type [34]. Being consistent with both of these two reports, our results demonstrated that: i), miR-105 was remarkably down-regulated in glioma tissues compared with normal brains and had significantly lower expression in GBMs relative to both AAs and DAs; ii), down-regulation of miR-105 was associated with aggressive clinicopathological features including advanced tumor grade, advanced patient’s age and low pre-operative Karnofsky performance score of glioma; iii), miR-105 expression significantly correlated with overall survival and was an independent prognostic indicator of glioma patients. Taken together, results of these studies suggest that miR-105 may function as a tumor suppressor and its down-regulation may contribute to tumorigenesis and aggressive progression of human glioma.
Our present study focused on the expression level and clinical significance of miR-105 in glioma. However, the dysregulation and involvement in tumorigenesis of miR-105 have been indicated in other kinds of human cancer, by recent investigations. Lee et al. in 2008 initially reported significantly decreased expression levels of miR-105 in multiple types of cancer cell lines including HS766T, MiaPaca2 and Panc1 (pancreatic carcinomas), HCT-116 (colorectal carcinoma), HeLa (cervical carcinoma), and HL-60 (promyelocytic leukemia) [35]. Interestingly, they found that the mature form of miR-105 is undetectable in certain cancer cell lines compared with abundant precursor miR-105 molecules present in the nucleus of these same cells [35]. Such phenomenon could be attributed to the fact that some cancers posses the loss-of-function mutations in the gene of expotin-5, an important miRNA biogenesis protein mediating pre-miRNA nuclear transport into the cytoplasm [36]. In addition, two recent studies showed evidence that miR-105 function as a potential tumor suppressor [37,38]. Honeywell et al. found that miR-105 is decreased in prostate cell lines compared with normal prostate epithelial cells and inhibits tumor growth through suppressing CDK6 levels [37]. Shen et al. demonstrated that miR-105 suppresses cell proliferation and inhibits PIK3/AKT signaling pathway in human hepatocellular carcinoma [38]. On the contrary, several other groups have reported the overexpression and oncogenic role of miR-105 in cancer cells. Using high-throughput sequencing, Hamfjord et al. in 2012 identified miR-105 as one of the up-regulated miRNAs in colorectal cancers compared with the paired adjacent normal tissues [39]. Similarly, up-regulation of miR-105 was also indicated in gastric cancer by Liu et al. in 2014 [40]. As a cancer-secreted miRNA, miR-105 was recently reported to be increased in circulation at the pre-metastatic stage and significantly correlate with occurrence of metastatic in breast cancer patients, by Zhou et al. [41]. They further demonstrated that cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis by directly targeting the tight junction protein ZO-1 [41]. From above discussion, miR-105 is differently expressed in tumors originate from diverse organic tissues and could function as both oncogenic and tumor suppressive miRNA. And the detailed biological mechanism of miR-105 dysregulation in tumorigenesis and aggressive progression of human cancers including glioma deserves further study.
In conclusion, we in the current study statistically compared miRNA expression profile of WHO grade IV gliomas with WHO grade III tumors and identified 34 dysregulated miRNAs, 23 up-regulated and 11 down-regulated. We further provided a novel prognostic indicator, miR-105, which serves as a biomarker for patient with poor overall survival in gliomas. Our results suggested that miRNA profiling are useful for predicting glioma patient survival, and have the substantial value for identifying novel therapeutic targets.
Acknowledgements
This study was supported by Natural Science Foundation of China (Grant No: 81302190) and the Scientific Research Foundation for the Returned Overseas Chinese Scholars of China (Grant No: 2013-1792).
Disclosure of conflict of interest
None.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2005;12:580–7. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Lowery AJ, Miller N, McNeill NE, Kerin MJ. MicroRNAs as prognostic indicators and therapeutic targets: potential effect on breast cancer management. Clin Cancer Res. 2008;14:360–5. doi: 10.1158/1078-0432.CCR-07-0992. [DOI] [PubMed] [Google Scholar]
- 7.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumors of central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivasan S, Patric IR, Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One. 2011;6:e17438. doi: 10.1371/journal.pone.0017438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim TM, Huang W, Park R, Park PJ, Johnson MD. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res. 2011;71:3387–99. doi: 10.1158/0008-5472.CAN-10-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu S, Lin S, Hu D, Feng Y, Tan Y, Peng Y. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J Transl Med. 2013;11:10. doi: 10.1186/1479-5876-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, Zhang J, Yan W, You G, Bao Z, Li S, Kang C, Jiang C, You Y, Zhang Y, Chen CC, Song SW, Jiang T. Whole-genome microRNA expression profiling identifies a 5-microRNA signature as a prognostic biomarker in Chinese patients with primary glioblastoma multiforme. Cancer. 2013;119:814–24. doi: 10.1002/cncr.27826. [DOI] [PubMed] [Google Scholar]
- 13.Barbano R, Palumbo O, Pasculli B, Galasso M, Volinia S, D’Angelo V, Icolaro N, Coco M, Dimitri L, Graziano P, Copetti M, Valori VM, Maiello E, Carella M, Fazio VM, Parrella P. A miRNA signature for defining aggressive phenotype and prognosis in gliomas. PLoS One. 2014;9:e108950. doi: 10.1371/journal.pone.0108950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan Y, Mizoguchi M, Yoshimoto K, Hata N, Shono T, Suzuki SO, Araki Y, Kuga D, Nakamizo A, Amano T, Ma X, Hayashi K, Sasaki T. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin Cancer Res. 2010;16:4289–97. doi: 10.1158/1078-0432.CCR-10-0207. [DOI] [PubMed] [Google Scholar]
- 15.Zhi F, Chen X, Wang S, Xia X, Shi Y, Guan W, Shao N, Qu H, Yang C, Zhang Y, Wang R, Zen K, Zhang CY, Zhang J, Yang Y. The use of has-miR-21, has-miR-181b and has-miR-106a as prognositic indicator of astrocytoma. Eur J Cancer. 2010;46:1640–9. doi: 10.1016/j.ejca.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Hermansen S, Dahlrot RH, Nielsen BS, Hansen S, Kristensen BW. MiR-21 expression in the tumour cell compartment holds unfavorable prognostic value in gliomas. J Neurooncol. 2013;111:71–81. doi: 10.1007/s11060-012-0992-3. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Lu S, Geng S, Ma S, Liang Z, Jiao B. Expression and clinical significance of microRNA-326 in human glioma miR-326 expression in glioma. Med Oncol. 2013;30:373. doi: 10.1007/s12032-012-0373-y. [DOI] [PubMed] [Google Scholar]
- 18.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptitic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 19.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–72. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Wang W, Gao Z, Peng X, Chen X, Chen W, Xu W, Xu H, Lin MC, Jiang S. MicroRNA-155 promotes glioma cell proliferation via the regulation of MXI1. PLoS One. 2013;8:e83055. doi: 10.1371/journal.pone.0083055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun J, Shi H, Lai N, Liao K, Zhang S, Lu X. Overexpression of microRNA-155 predicts poor prognosis in glioma patients. Med Oncol. 2014;31:911. doi: 10.1007/s12032-014-0911-x. [DOI] [PubMed] [Google Scholar]
- 22.Yan Z, Che S, Wang J, Jiao Y, Wang C, Meng Q. miR-155 contributes to the progression of glioma by enhancing Wnt/-catenin pathway. Tumour Biol. 2015;36:5323–31. doi: 10.1007/s13277-015-3193-9. [DOI] [PubMed] [Google Scholar]
- 23.Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–68. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 24.Yang G, Han D, Chen X, Zhang D, Wang L, Shi C, Zhang W, Li C, Chen X, Liu H, Zhang D, Kang J, Peng F, Liu Z, Qi J, Gao X, Ai J, Shi C, Zhao S. MiR-196a exerts its oncogenic effect in glioblastoma multiforme by inhibition of IκBα both in vitro and in vivo. Neuro Oncol. 2014;16:652–61. doi: 10.1093/neuonc/not307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–8. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 26.Kefas B, Comeau L, Floyd DH, Seleverstov O, Godlewski J, Schmittgen T, Jiang J, diPierro CG, Li Y, Chiocca EA, Lee J, Fine H, Abounader R, Lawler S, Purow B. The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J Neurosci. 2009;29:15161–8. doi: 10.1523/JNEUROSCI.4966-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kefas B, Comeau L, Erdle N, Montgomery E, Amos S, Purow B. Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro Oncol. 2010;12:1102–12. doi: 10.1093/neuonc/noq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time PCR of microRNA by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy SP, Britto R, Vinnakota K, Aparna H, Sreepathi HK, Thota B, Kumari A, Shilpa BM, Vrinda M, Umesh S, Samuel C, Shetty M, Tandon A, Pandey P, Hegde S, Hegde AS, Balasubramaniam A, Chandramouli BA, Santosh V, Kondaiah P, Somasundaram K, Rao MR. Novel glioblastoma markers with diagnostic and prognostic value identified through transcriptome analysis. Clin Cancer Res. 2008;14:2978–87. doi: 10.1158/1078-0432.CCR-07-4821. [DOI] [PubMed] [Google Scholar]
- 30.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayers DN Cancer Genome Atlas Research Network. Intergrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houshmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K Cancer Genome Atlas Research Network. Identification of a CpG island methlator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classfy human cancer. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 33.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, Lebanony D, Goren Y, Silberschein E, Targan N, Ben-Ari A, Gilad S, Sion-Vardy N, Tobar A, Feinmesser M, Kharenko O, Nativ O, Nass D, Perelman M, Yosepovich A, Shalmon B, Polak-Charcon S, Fridman E, Avniel A, Bentwich I, Bentwich Z, Cohen D, Chajut A, Barshack I. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–9. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 34.Yan W, Li R, Liu Y, Yang P, Wang Z, Zhang C, Bao Z, Zhang W, You Y, Jiang T. MicroRNA expression patterns in the malignant progression of gliomas and a 5-microRNA signature for prognosis. Oncotarget. 2014;5:12908–15. doi: 10.18632/oncotarget.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines and tumors. RNA. 2008;1:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya G, Yamamoto H, Schwartz S Jr, Esteller M. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–15. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Honeywell DR, Cabrita MA, Zhao H, Dimitroulakos J, Addison CL. miR-105 inhibits prostate tumor growth by suppressing CDK6 levels. PLoS One. 2013;8:e70515. doi: 10.1371/journal.pone.0070515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen G, Rong X, Zhao J, Yang X, Li H, Jiang H, Zhou Q, Ji T, Huang S, Zhang J, Jia H. MicroRNA-105 suppresses cell proliferation and inhibits PIK3/AKT signaling in human hepatocellular carcinoma. Carcinogenesis. 2014;12:2748–55. doi: 10.1093/carcin/bgu208. [DOI] [PubMed] [Google Scholar]
- 39.Hamfjord J, Stangeland AM, Hughes T, Skrede ML, Tveit KM, Ikdahl T, Kure EH. Differential expression of miRNAs in colorectal cancer: comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PLoS One. 2012;7:e34150. doi: 10.1371/journal.pone.0034150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu D, Hu X, Zhou H, Shi G, Wu J. Identification of aberrantly expressed miRNAs in gastric cancer. Gastroenterol Res Pract. 2014;2014:473817. doi: 10.1155/2014/473817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O’Connor ST, Chin AR, Yen Y, Wang Y, Marcusson EG, Chu P, Wu J, Wu X, Li AX, Li Z, Gao H, Ren X, Boldin MP, Lin PC, Wang SE. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–15. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


