Abstract
This study aimed to investigate the effect of a selective cyclooxygenase-2 (COX-2) inhibitor (celecoxib) on the expression of arachidonate-associated inflammatory genes in cultured human normal chondrocytes. Normal chondrocytes were obtained from the cartilage of three different amputated patients without osteoarthritis (OA). Affymetrix Human microarray was used to assess the alterations in gene expression in three groups of cells: untreated cells (negative control group), cells treated with interleukin-1β (IL-1β) (positive control group), and cells treated with IL-1β and celecoxib. The patterns of up-regulation and down-regulation of gene expression were further validated by real-time PCR. A total of 1091 up-regulated genes and 1252 down-regulated genes were identified in the positive control group compared with the negative control group. Among them, PTGS2, ADAMTS5, PTGER2, mPTGES and PTGER4 are known to be involved in chondrocyte inflammation, while VEGFA, BCL2, TRAF1, CYR61, BMP6, DAPK1, DUSP7, IL1RN, MMP13 and TNFSF10 were reported being associated with cytokine and chemokine signaling. 189 up-regulated genes and 177 down-regulated genes were identified in the positive control group compared with intervention group. PTGS1, PTGS2, ADAMTS5, PTGER2, mPTGES and PTGER4 were among the genes down-regulated upon the treatment with celecoxib. Our results demonstrated that the OA chondrocytes are the site of active eicosanoid production. IL-1β can activate inflammation in chondrocytes and trigger the production of various proteins involved in cyclooxygenase pathway. The expression of genes corresponding to these proteins can be down-regulated by celecoxib. The findings indicate that the therapy with prostaglandin E2 (PGE2)-blocking agents may decrease the PGE2 production not only by direct inhibition of COX-2 activity, but also by down-regulating the expression of genes encoding for COX-2, microsomal prostaglandin-endoperoxide synthase 1 (mPGES-1) and prostaglandin E receptors 4 (EP4) in the articular chondrocytes.
Keywords: Celecoxib, COX-2, PGE2, articular chondrocytes, osteoarthritis
Introduction
Osteoarthritis (OA) is associated with the loss of balance between degeneration and reparation of cartilage [1,2]. These processes are targeted by active cytokines and inflammatory mediators. Previous studies reported that prostaglandin E2 (PGE2) and its oxidative product can be produced by pathological chondrocytes. These compounds play an important role in the degeneration of cartilage and thus the development of OA [1,3-7].
Cyclooxygenases-1 (COX-1) and COX-2 are enzymes responsible for the sequential oxidation of arachidonic acid (AA) or eicosapentaenoic acid (EPA) during the production of PGE2. While both COX-1 and COX-2 are active in the blood vessels, stomach and the kidneys, the increase of PGE2 levels is mainly mediated by COX-2 in the scenarios of inflammation and growth [8].
Celecoxib, a COX-2 selective inhibitor, has been found useful in relieving pain, improving articular function and augmenting the life quality of OA patients [9]. Although celecoxib has been proven to inhibit COX-2 function and PGE2 production, the molecular mechanisms and associated changes in gene expression in the down-stream pathways remains poorly studied.
In this study, we performed the genomic microarray analysis to investigate the changes in the expression of genes associated with COX-2 and arachidonate pathway upon treatment of chondrocytes with celecoxib. The findings might help to gain insight into the pathogenesis of OA in general, and the mechanisms involved in the action of celecoxib on the OA chondrocytes in particular.
Material and methods
Chondrocytes isolation and growth
Cartilages were obtained from three non-arthritis accident victims after knee amputation, following the guidelines of the Institution Review Board (IRB) of Peking Union Medical College Hospital. The cartilage slices were cut into 3 mm discs and treated by 0.2% collagenase at 37°C with shaking for 8 h. The supernatant was then filtered, centrifuged for 5 min, and cultured in the flask containing DMEM with 10% FBS. The cultural media were changed every 2 or 3 days. When the chondrocytes reached 80% confluence, they were transferred to culture plates.
Treatment of chondrocytes and preparation of total RNA
The chondrocytes were divided into three groups and transferred to the respective culture plates. The three groups consisted of negative control group (no treatment), positive control group (interleukin-1β (IL-1β) stimulation), and intervention group (Celecoxib + IL-1β stimulation). After serum-starvation for 12 h in the media without FBS, the media of intervention group was changed to DMEM containing 10 µM celecoxib. After additional 12 h, IL-1β in the final concentration of 5 ng/mL was added into the media of positive and intervention groups. After incubation for 24 h, the chondrocytes in three groups were harvested.
Total RNA extraction and microarray assay
RNA was separated from the cells using Trizol (Invitrogen) according to the manufacturer’s protocol. RNA was extracted with water-saturated phenol, followed by phenol or chloroform extraction and isopropanol precipitation. The RNA pellets were then dissolved in water and precipitated with alcohol in the presence of acetic acid. The total RNA from the cartilage chondrocytes samples were used for labeling as described: RT-cDNA labeling reactions were performed using protocol and materials from R&D Systems (USA) and 33P-labelled dATP from NEN Life Sciences Products (USA). Pooled total RNA samples (2 µg each, n=3) were used in one set of labeling reactions with cytokine specific primers (R&D Systems). Three other 2 µg samples of pooled total RNA were used in another set of labeling reactions with 16- to 18-mer oligo-dT primers (Gibco-BRL, Rockville, Maryland, USA). Labeled cDNAs obtained from both reaction sets were pooled together as recommended by the manufacturer and hybridized to the R&D Systems Cytokine Expression Array blots, using the manufacturer’s hybridization protocol. The array blots were then exposed to PhosphorImager plates (Molecular Dynamics). After exposure period, the plates were read on a Molecular Dynamics Storm apparatus. Quantitation of the signals was performed using ImageQuaNT software and the data was normalized to β-actin and (Glyceraldehyde-3-phosphate dehydrogenase) GAPDH as reference genes.
Microarray data analysis
Raw data of gene expression files from Affymetrix GeneChips were imported into GeneSpring, version 7.2 software (Silicon Genetics, Redwood City, CA). GC-robust multichip analysis preprocessing was performed. Data sets from negative control group were assigned to the “normal” treatment group, and thus defined baseline expression for each probe. The remaining data sets (samples from positive control and intervention groups) were assigned to “diseased” group and treatment group, and used in subsequent analysis. All data were interpreted using the log-ratio setting. Post hoc Bonferroni multiple comparison testing was performed to identify statistically significant changes in expression between negative control groups and positive control groups, and between positive control groups and intervention groups, with P values less than 0.05 considered significant. GeneSpring 7.2 software was used to create gene lists based on fold change.
Real-time polymerase chain reaction (RT-PCR)
To quantitatively determine relative gene expression in cartilage chondrocytes groups, realtime PCR was carried out. The reactions were prepared with the TaqMan One-Step Master Mix kit (Applied Biosystems, Foster City, CA). TaqMan GAPDH (forward primer 5’-GAAGGTGAAGGTCGGAGTC-3’, reverse primer 5’-GAAGATGTGATGGGATTTC-3’, probe JOE-CAAGCTTCCCGTTCTCAGCC-TAMRA) control reagents were used for the internal control. The target primer/probe sets for all tested genes were purchased as TaqMan gene expression assays (Applied Biosystems). An ABI Prism 7900 HT Real-Time PCR system (PerkinElmer, Emeryville, CA) was used to detect amplification for over 40 cycles in these experiments. Three groups of RNA samples from negative control group, positive control group, and intervention group were assayed. All relative expression values were calculated using the ΔCt method [10], normalized to GAPDH expression, and expressed in arbitrary units relative to the expression values in the negative control group (set at 1). Tukey’s post hoc comparison was performed to compare the means for all three groups. Expression values are shown as the mean ± SEM (standard error of mean). P values less than 0.05 were considered significant. Analyses were carried out using GraphPad Prism, version 4 (GraphPad Software, San Diego, CA).
Reagents
Human gene 1.0 microarray was purchased from Affymetrix (USA). Reagents for RT-PCR were obtained from ABI (USA). Recombinant human IL-1β was purchased from R&D Systems (Minneapolis, MN). Celecoxib was purchased from Pharmacia (Skokie, IL).
Results
Chondrocytes culture and total RNA quality examination
Chondrocytes were isolated and grown to 80% confluence (Figure 1A). The RNA quality examination showed that OD value of each group was in the range of 2.0~2.1. Agarose gel electrophoresis showed the presence of 28S and 18S rRNA bands, the width of 28S rRNA band was the double of 18S rRNA (Figure 1B).
Figure 1.
Chondrocytes culture and total RNA quality examination. A. Chondrocytes grown to 80% confluence; B. Results of RNA quality examination.
Gene microarray
Comparison between negative control group and positive control group
Gene expression profiling of negative control and positive control groups using Affymetrix microarray showed the presence of various mRNA representing proteins associated with eicosanoids pathway, Mitogen-activated protein kinase (MAPK) signal pathway, and cartilage degeneration enzymes (Table 1). A total of 1091 up-regulated genes and 1252 down-regulated genes were identified in the positive control group compared with the negative control group. Clustering analysis showed significant differently expressed genes (Figure 2A). Up-regulated genes in the IL-1β group were associated with COX-2/PGES pathway and included PTGES, PTGS2, PTGER4, IL-1B and IL1RN. The expression of PTGS1 was down-regulated. Our results showed that IL-1β triggers the inflammatory response in the chondrocytes which could be involved in the cartilage degeneration.
Table 1.
Genes in positive control group with the expression up-regulated and down-regulated compared to negative control group
| Genebank encode number | Expression ratio positive control group/negative control group | Gene name (abbreviation) | Gene function |
|---|---|---|---|
| NM_000963 | 19.62 | Prostaglandin-endoperoxide synthase 2 (PTGS2) | Protease |
| NM_000576 | 11.24 | Interleukin 1, beta (1L-1B) | Mediator of inflammation |
| NM_004878 | 10.05 | Tumor necrosis factor (ligand) superfamily, member 18 (TNFSF18) | Cytokine |
| NM_001561 | 9.81 | Tumor necrosis factor (ligand) superfamily, member 18 (TNFRSF9) | Cytokine |
| NM_007038 | 9.58 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 (ADAMTS5) | Protease |
| NM_004878 | 8.13 | Prostaglandin E synthase (PTGES) | Protease |
| NM_002478 | 6.28 | Mitogen activated kinase-like protein (MAPK) | Signal transduction |
| NM_000577 | 4.54 | Interleukin 1 receptor antagonist (IL-1RN) | Signal transduction |
| NM_002427 | 4.02 | Matrix metallopeptidase 13 (MMP13) | Protease |
| NM_000958 | 2.15 | Prostaglandin E receptor 4 (PTGER4) | Signal transduction |
| NM_008005 | 0.73 | Fibroblast growth factor 18 (FGF18) | Signal transduction |
| NM_000089 | 0.62 | Collagen, type I, alpha 2 (COL1A2) | Structural protein |
| NM_000088 | 0.61 | Collagen, type I, alpha 1 (COL1A1) | Structural protein |
| NM_001271166 | 0.38 | Prostaglandin-endoperoxide synthase 1 (PTGS1) | Protease |
| NM_001554 | 0.13 | Cysteine-rich angiogenic inducer, 61 (CYR61) | Apoptosis |
Figure 2.
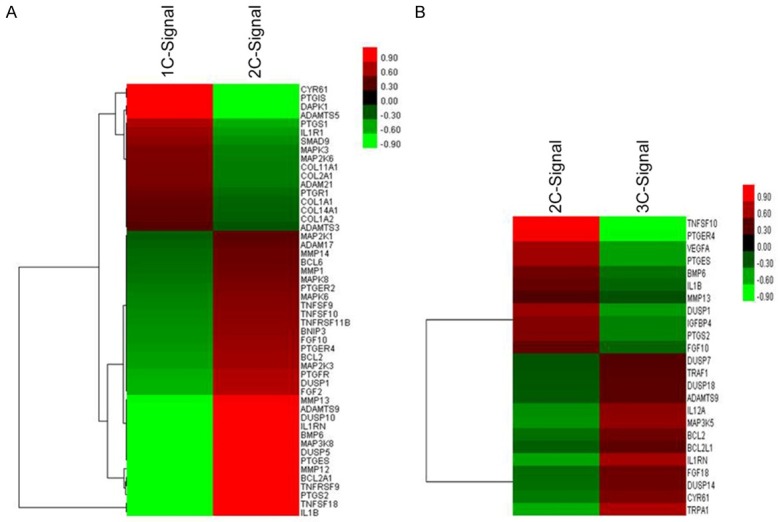
Cluster analysis of differently expressed genes. The change of colors from green to red represents the increasing expression of genes. Sample names are listed in the horizontal axis (1c, 2c, and 3c represent negative control group cells, positive control group cells, and intervention group cells, respectively). Right vertical axis shows significantly up- and down-regulated genes.
Comparison between positive control group and intervention group
In comparison with the positive control group, the genes with changed levels of expression in the intervention group were associated with eicosanoids pathway, apoptosis signal pathway, cartilage degeneration, structural proteins, and signal transduction (Table 2). A total of 189 up-regulated genes and 177 down-regulated genes were identified in the positive control group compared with the intervention group. Clustering analysis showed significant differently expressed genes (Figure 2B). Upon the treatment with celecoxib, the expression of genes associated with COX-2/PGES pathway was significantly down-regulated. These genes included PTGES, PTGS2, PTGER4 and IL1RN. The genes PTGS1 and IL-1RN were up-regulated. These results showed that celecoxib could inhibit the inflammatory response of chondrocytes and thus may play role in the prevention of the cartilage degeneration.
Table 2.
Genes in intervention group with the expression up-regulated and down-regulated compared to positive control group
| Genebank encode number | Expression ratio intervention group/positive control group | Gene name (abbreviation) | Gene function |
|---|---|---|---|
| NM_001271166 | 2.75 | Prostaglandin-endoperoxide synthase 1 (PTGS1) | Protease |
| NM_000633 | 2.60 | B-cell CLL/lymphoma 2 (BCL2) | Apoptosis |
| NM_000577 | 2.20 | Interleukin 1 receptor antagonist (IL1RN) | Cytokine |
| NM_001190945 | 1.80 | TNF receptor-associated factor 1 (TRAF1) | Signal transduction |
| NM_001554 | 1.80 | Cysteine-rich angiogenic inducer, 61 (CYR61) | Apoptosis |
| NM_004938 | 1.60 | Death-associated protein kinase 1 (DAPK1) | Apoptosis |
| NM_001947 | 1.40 | Dual specificity phosphatase 7 (DUSP7) | Apoptosis |
| NM_001718 | 0.58 | Bone morphogenetic protein 6 (BMP6) | Cytokine |
| NM_002427 | 0.53 | Matrix metallopeptidase 13 (MMP13) | Protease |
| NM_000963 | 0.51 | Prostaglandin-endoperoxide synthase2 (PTGS2) | Protease |
| NM_001552 | 0.51 | Insulin-like growth factor binding protein 4 (IGFBP4) | Signal transduction |
| NM_004878 | 0.46 | Prostaglandin E synthase (PTGES) | Protease |
| NM_001171623 | 0.46 | Vascular endothelial growth factor A (VEGFA) | Cytokine |
| NM_000958 | 0.29 | Prostaglandin E receptor 4 (PTGER4) | Signal transduction |
| NM_001190942 | 0.27 | Tumor necrosis factor (ligand) superfamily, member 10 (TNFSF10) | Apoptosis |
RT-PCR
Compared with the negative control group, the up-regulated mRNAs in positive control group included COX-2, mPGES-1, Prostaglandin E receptors 2 and 4 (EP2 and EP4), whereas the COX-1 and mPGES-2 were down-regulated. Compared with the positive control group, down-regulated mRNAs included COX-2, mPGES-1 and EP4 while COX-1 and mPGE-2 were up-regulated. EP2 expression was up-regulated, but not to the statistically significant level. With the exception of EP2, the results of RT-PCR were consistent with the results of gene microarray analysis, thus proving the reliability of our system (Figure 3).
Figure 3.
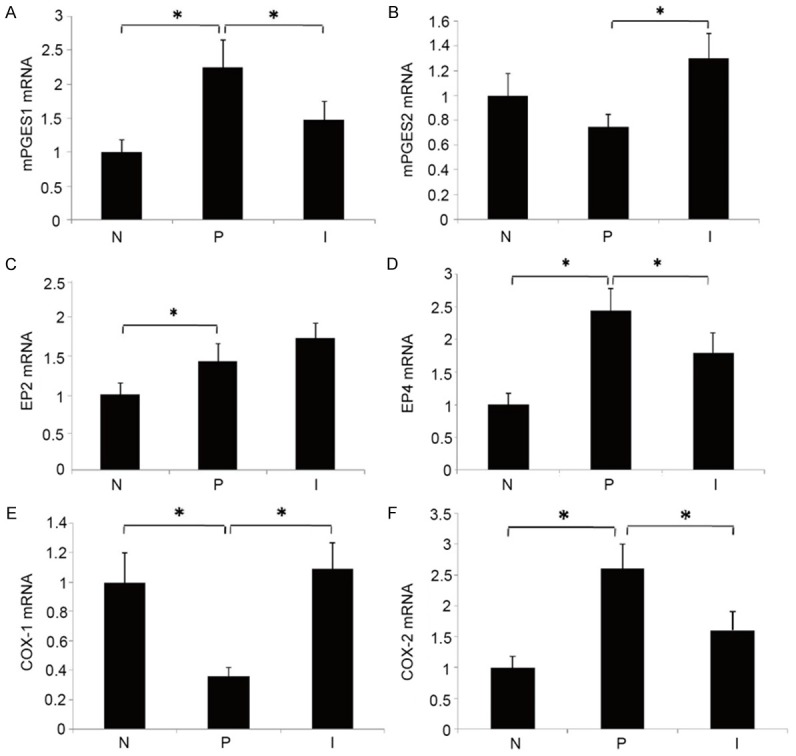
Results of RT-PCR. N: Negative control group; P: Positive control group; I: Intervention group; *P<0.05.
Discussion
Osteoarthritis is the most common degenerative joint disease in the world [11]. Prostaglandin E2 functions as one of the major catabolic mediators of the joint cartilage degradation in vitro and is abundantly produced in the OA cartilage [7,12]. Due to this association with the disease, prostaglandin E2 is considered a possible target for therapeutic treatment of osteoarthritis. Two isoforms of COX are known: COX-1 (encoded by PTGS1) and COX-2 (encoded by PTGS2). There are also 3 isoforms of prostaglandin-endoperoxide synthases (PGES): microsomal PGES-1 (mPGES-1) (encoded by PTGES), mPGES-2 (encoded by PTGES2), and cytosolic PGES (cPGES) (encoded by PTGES3). COX-1, mPGES-2 and cPGES are expressed constitutively in various tissues and function to maintain the homeostasis. COX-2 and mPGES-1, on the other hand, can be induced by various stimuli during catabolic and inflammatory processes.
A human genetic study identified the prostaglandin-endoperoxide synthase 2 gene variant as being involved in the risk of knee OA, suggesting the importance of the COX-2 signaling in the pathogenesis of this disease [13]. Previous study of the genome-wide changes in gene expression also demonstrated the changes in regulation in a number of genes known to be involved in human OA. These genes include MMP13, ADAMTS5, PTGS2 and others [14].
Although non-steroidal anti-inflammatory drugs (NSAIDs) that inactivate COX enzymes are commonly used as symptom-modifying treatments to suppress pain and inflammation in OA, it remains unclear if these drugs can be used as disease-modifying treatment due to their chondroprotective effects [15]. Traditional NSAIDs and selective COX-2 inhibitors are commonly employed to treat the OA. The effects of NSAIDs are derived mainly from their abilities to inhibit the COX activity, thereby decreasing the PGE2 synthesis. In vitro and in vivo studies have demonstrated that the selective COX-2 inhibitor celecoxib, in addition to mediating pain and inflammation in OA, has chondroprotective effect in the chondrocytes cultures and in cartilages [9,16,17]. Nakamura and co-authors [18] found that celecoxib increased IL-1-induced production of RANTES and MIP-1α and decreased IL-1-induced NO production by chondrocytes. Celecoxib was found to repress apoptosis in chondrocytes [19]. A recent article of Takenouchi et al. [20] reported significant changes in the protein expression profile of chondrocytes upon treatment with celecoxib. Kim and co-workers [21] analyzed the changes in gene expression using complementary DNA microarray in a stable COX-2 knockdown A549 lung cancer cell line and a mock line following the treatment with celecoxib. They found that celecoxib could regulate the RAS homolog gene family B in a COX-2 dependent manner in the irradiated cells. Study showed that the exposure to celecoxib normalized the release of PGE2 and diminished the CTS-induced COX-2, MMP-1, MMP-3, MMP-9 and ADAMTS-5 gene expressions [22]. Although celecoxib is commonly employed to treat the OA, its effect on the gene expression in chondrocytes and cartilage remains poorly understood.
In this work, we demonstrated that stimulation of chondrocytes by IL-1β could trigger inflammatory response. Considerable number of genes associated with cartilage degeneration and joint inflammation, including genes for matrix metalloproteinases, ADAMTS and genes involved in apoptosis and PGE2 pathways become activated. In the intervention group, the treatment with celecoxib significantly amended the inflammatory response in chondrocytes by relieving the inflammation reactions and decreasing the expression of genes associated with cartilage degeneration. Among those genes, the expression of genes involved in the PGE2 pathway, such as PTGS2 (encoding for COX-2) and PTGES (encoding for mPGES-1), was significantly influenced by celecoxib. COX-2 and mPGES-1 are key enzymes of the PGE2 pathway in the OA [7]. Previously study showed that celecoxib could inhibit the COX-2 expression and synthesis in the OA synovial membrane [23]. Our results are also in agreement with the previously published data on the inhibition of COX-2 expression in leukemic cells, lung carcinoma cells and in synoviocytes [24,25]. The current study demonstrated that celecoxob could suppress the expression of PTGS and PTGES, thus inhibiting the COX-2 activity and decreasing the PGE2 production. Celecoxib might relieve the inflammation of cartilage and produce a protective effect in OA through this mechanism.
Four different prostaglandin receptors (EP1-4) had been identified to date [26]. The influence of celecoxib on the EPs expression in human OA chondrocytes is poorly studied [27]. Our study showed that IL-1β stimulation could increase both EP2 and EP4 gene expression. In our experiments, celecoxib was able to significantly decrease the IL-1β-induced expression of EP4 receptor, but exert no effect on the expression of EP2 receptor. PGE2 induces cartilage degeneration mainly through EP4 in the joint inflammation and in the extracellular matrix degradation in different experimental models of arthritis [28].
The reduction of the EP4 expression observed in the celecoxib-treated chondrocytes would help to control the inflammatory response and prevent the progression of the cartilage damage [29]. The targeting of EP4 was previously suggested as a potential strategy for OA disease modification [30].
In summary, our study demonstrated that OA chondrocytes are the sites of active eicosanoid production. IL-1β can activate chondrocytes inflammation and produce a broad spectrum of products derived from the cyclooxygenase pathways. The current experiment showed that celecoxib decreased PGE2 production not only by direct inhibition of COX-2 activity, but also via down-regulating the expression of the genes encoding for key enzymes in the PGE2 biosynthesis pathway.
Disclosure of conflict of interest
None.
References
- 1.Roach HI, Aigner T, Soder S, Haag J. Pathobiology of osteoarthritis: pathomechanisms and potential therapeutic targets. Curr Drug Targets. 2007;8:271–282. doi: 10.2174/138945007779940160. [DOI] [PubMed] [Google Scholar]
- 2.Swales C, Athanasou NA. The pathobiology of osteoarthritis. Orthopaedics and Trauma. 2010;24:399–404. [Google Scholar]
- 3.Abramson SB, Attur M, Yazici Y. Prospects for disease modification in osteoarthritis. Nat Clin Pract Rheumatol. 2006;2:304–312. doi: 10.1038/ncprheum0193. [DOI] [PubMed] [Google Scholar]
- 4.Martel-Pelletier J, Pelletier JP, Fahmi H. Cyclooxygenase-2 and prostaglandins in articular tissues. Semin Arthritis Rheum. 2003;33:155–167. doi: 10.1016/s0049-0172(03)00134-3. [DOI] [PubMed] [Google Scholar]
- 5.Sandell LJ. Modern molecular analysis of a traditional disease: progression in osteoarthritis. Arthritis Rheum. 2007;56:2474–2477. doi: 10.1002/art.22760. [DOI] [PubMed] [Google Scholar]
- 6.Aigner T, Dudhia J. Genomics of osteoarthrosis. Curr Opin Rheumatol. 2003;15:634–640. doi: 10.1097/00002281-200309000-00019. [DOI] [PubMed] [Google Scholar]
- 7.Murakami M, Kudo I. Recent advances in molecular biology and physiology of the prostaglandin E2-biosynthetic pathway. Prog Lipid Res. 2004;43:3–35. doi: 10.1016/s0163-7827(03)00037-7. [DOI] [PubMed] [Google Scholar]
- 8.Ricciotti E, FitzGerald GA. Prostaglandins and Inflammation. Arterioscler Thromb Vasc Biol. 2011;31:986–1000. doi: 10.1161/ATVBAHA.110.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zweers MC, de Boer TN, van Roon J, Bijlsma JW, Lafeber FP, Mastbergen SC. Celecoxib: considerations regarding its potential disease-modifying properties in osteoarthritis. Arthritis Res Ther. 2011;13:239. doi: 10.1186/ar3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 22DDCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Dillon CF, Rasch EK, Gu Q, Hirsch R. Prevalence of knee osteoarthritis in the united States: Arthritis Data from the Third National health and nutrition examination survey 1991-94. J Rheumatol. 2006;33:2271–2279. [PubMed] [Google Scholar]
- 12.Hardy MM, Seibert K, Manning PT, Currie MG, Woemer BM, Edwards D. Cyclooxygenase 2-dependent prostaglandin E2 modulates cartilage proteoglycan degradation in human osteoarthritis explants. Arthritis Rheum. 2002;46:1789–1803. doi: 10.1002/art.10356. [DOI] [PubMed] [Google Scholar]
- 13.Valdes AM, Loughlin J, Timms KM, van Meurs JJ, Southam L, Wilson SG. Genome-wide association scan identifies a prostaglandin-endoperoxide synthase 2 variant involved in risk of knee osteoarthritis. Am J Hum Genet. 2008;82:1231–1240. doi: 10.1016/j.ajhg.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Appleton CTG, Pitelka V, Henry J. Global Analyses of Gene Expression in Early Experimental Osteoarthritis. Arthritis Rheum. 2007;56:1854–1868. doi: 10.1002/art.22711. [DOI] [PubMed] [Google Scholar]
- 15.Ding C. Do NSAIDS affect the progression of osteoarthritis? Inflammation. 2002;26:139–142. doi: 10.1023/a:1015504632021. [DOI] [PubMed] [Google Scholar]
- 16.Mastbergen SC, Jansen NW, Bijlsma JW, Lafeber FP. Differential direct effects of cyclooxygenase-1/2 inhibition on proteoglycan turnover of human osteoarthritis cartilage: an in vitro study. Arthritis Res Ther. 2006;8:R2. doi: 10.1186/ar1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Boer TN, Huisman AM, Polak AA, Niehoff AG, van Rinsim AC, Saris D. The chondroprotective effect of selective COX-2 inhibition in osteoarthritis: ex vivo evaluation of human cartilage tissue after in vivo treatment. Osteoarthritis Cartilage. 2009;17:482–488. doi: 10.1016/j.joca.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura H, Masuko K, Yudoh K, Kato T, Nishioka K. Effects of celecoxib on human chondrocytes-enhanced production of chemokines. Clin Exp Rheumatol. 2007;25:11–16. [PubMed] [Google Scholar]
- 19.Ou Y, Tan C, An H, Jiang D, Quan Z, Tang K, Luo X. Selective COX-2 inhibitor ameliorates osteoarthritis by repressing apoptosis of chondrocyte. Med Sci Monit. 2012;18:BR247–252. doi: 10.12659/MSM.882901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takenouchi K, Arito M, Sato T, Takahashi K, Kurokawa MS, Yudoh K, Takai S, Kato T, Nakamura H. Proteomic Analysis of Celecoxib on Chondrocytes from Patients with Osteoarthritis. Modern Research in Inflammation. 2014;3:90–98. [Google Scholar]
- 21.Kim YM, Shin YK, Jun HJ, Rha SY. Systematic analyses of genes associated with radiosensitizing effect by celecoxib, a specific cyclooxygenase-2 inhibitor. J Radiat Res. 2011;52:752–765. doi: 10.1269/jrr.10146. [DOI] [PubMed] [Google Scholar]
- 22.Su SC, Tanimoto K, Tanne Y, Kunimatsu R, Hirose N. Celecoxib exerts protective effects on extracellular matrix metabolism of mandibular chondrocytes under excessive mechanical stress. Osteoarthritis Cartilage. 2014;22:845–851. doi: 10.1016/j.joca.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Alvarez-Soria MA, Herrero-Beaumont G. Long-term NSAID treatment directly decreases COX-2 and mPGES-1 production in the articular cartilage of patients with osteoarthritis. Osteoarthritis Cartilage. 2008;16:1484–1493. doi: 10.1016/j.joca.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 24.Shishodia S, Koul D, Affarwal BB. Cyclooxygenase (COX)-2 inhibitor celecoxib abrogates TNF-induced NF-kappa B activation through inhibition of activation of kappa B alpha kinase and Ake in human non-small cell lung carcinoma: correlation with suppression of COX-2 synthesis. J Immunol. 2004;173:2011–2022. doi: 10.4049/jimmunol.173.3.2011. [DOI] [PubMed] [Google Scholar]
- 25.Takada Y, Bhardwai A, Potdar P. Nonsteroidal anti-inflammatory agents differ in their ability to suppress NF-kappa B activation, inhibition of expression of cyclooxygenase-2 and cyclin D1 and abrogation of tumor cell proliferation. Oncogene. 2004;9:9247–9258. doi: 10.1038/sj.onc.1208169. [DOI] [PubMed] [Google Scholar]
- 26.Sugimoto Y, Narumiya S. Prostaglandin E receptors. J Biol Chem. 2007;282:11613–11617. doi: 10.1074/jbc.R600038200. [DOI] [PubMed] [Google Scholar]
- 27.Brochhausen C, Neuland P, Kirkpatrick CJ. Cyclooxygenases and prostaglandin E2 receptors in growth plate chondrocytes in vitro and in situ prostaglandin E2 dependent proliferation of growth plate chondrocytes. Arthritis Res Ther. 2006;8:R78. doi: 10.1186/ar1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honda T, Segi-Nishida E, Miyachi Y, Narumiy S. Prostacyclin-IP signaling and prostaglandin E2-EP2/EP4 signal both mediate joint inflammation in mouse collagen-induced arthritis. J Exp Med. 2006;203:325–335. doi: 10.1084/jem.20051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe Y, Namba A, Honda K. IL-1beta stimulates the expression of prostaglandin receptor EP4 in human chondrocytes by increasing production of prostaglandin E2. Rheumatol Int. 2008;28:727–736. doi: 10.1080/03008200802588451. [DOI] [PubMed] [Google Scholar]
- 30.Attur M, Al-Mussawir HE, Patel J, Kitay A, Dave M, Palmer G, Pillinger MH, Abramson SB. Prostaglandin E2 exerts catabolic effects in osteoarthritis cartilage: evidence for signaling via the EP4 receptor. J Immunol. 2008;181:5082–5088. doi: 10.4049/jimmunol.181.7.5082. [DOI] [PubMed] [Google Scholar]


