Abstract
Ventilator-associated pneumonia (VAP) results in considerable morbidity and mortality in neonatal intensive care units. VAP is associated with polymicrobial biofilms that form on endotracheal tubes (ETTs). We aimed to evaluate the diversity and the bacterial community in biofilms on ETTs extubated from mechanically ventilated newborns. ETTs (N = 29) and aerobic sputum cultures were obtained from 20 mechanically ventilated newborns. Denaturing gradient gel electrophoresis (DGGE) was used to characterize the bacterial species in the biofilms on the ETTs. Species-specific PCR was used to detect common oropharyngeal Streptococcus species and known ETT-associated pathogens. DGGE profiling of ETT biofilms showed multiple banding patterns indicating a diverse bacterial community. The dominant bacterial species were Klebsiella spp. (29/29), Streptococcus spp. (27/29), and Pseudomonas spp. (24/29). The most frequently occurring Streptococcus species was Streptococcus mitis (N = 18). Oropharyngeal bacteria were present in 25 of 29 ETT specimens. Streptococcus spp. often co-existed with K. pneumoniae and/or P. aeruginosa. In contrast, only one bacterial species was isolated from each sputum culture, K. pneumoniae or Acinetobacter baumannii. Our results demonstrated that Klebsiella spp., Streptococcus spp., and Pseudomonas spp. were the most frequent microbes on the surface of neonatal ETTs. The co-existence of oral commensals and pathogenic bacteria on the same tubes may play a crucial role for biofilm formation.
Keywords: Mechanical ventilation, biofilm, denaturing gradient gel electrophoresis (DGGE), neonate, Streptococcus spp
Introduction
Ventilator-associated pneumonia (VAP) is one of the most common iatrogenic infections in neonatal intensive care units (NICU). VAP is associated with significantly prolonged hospitalization, increased medical costs, and a rising rate of morbidity. The VAP-related mortality rate is reported to be 32.2% [1]. Apart from factors such as birth weight, parenteral alimentation, and mechanical ventilation [2], endotracheal intubation is often considered an independent risk factor for VAP [3]. Scanning electron microscopy (SEM) studies have found that bacteria can colonize endotracheal tubes (ETTs) within 24 hours of implantation. The ETT provides an ideal biological niche for bacterial adhesion, and biofilms can form on both the inner luminal and outer surface [4]. In fact, it has been suggested that the term “endotracheal tube-associated pneumonia” is more accurate than “ventilator-associated pneumonia” [5].
Under most circumstances, biofilms contain multiple species of bacteria [6]. Multi-species biofilm formation is a dynamic process leading to a three-dimensional structure. As compared with planktonic bacteria, bacteria embedded in biofilms can adapt to the environment more easily by secreting polysaccharides, lipids, extracellular DNA, and proteins. Biofilm formation is also an effective immune evasion mechanism. The matrix of the biofilm prevents antimicrobial agents from penetrating into the interior of the biofilm, which contributes to the antibiotic-resistance of bacteria.
Current research focused on ETT-associated biofilms encompasses biofilm development, architecture, and interactions [6,7]. Historically, VAP was diagnosed using conventional culture-based methods to assess the composition of the bacterial community. However, conventional methods are insufficient as up to 99% of bacteria in the natural environment cannot be cultured [8]. Indeed, over 90% of aspirates from ETTs do not yield positive culture results in the early stages. In contrast, culture-independent methods provide a more comprehensive profile of bacterial diversity and can be used to assess complex bacterial community dynamics. Denaturing gradient gel electrophoresis (DGGE) has been successfully used to investigate the composition of the bacterial community on ETTs that were removed from mechanically ventilated adults [9]. However, there are limited studies using the 16S rRNA PCR-DGGE method to characterize the bacterial community on neonatal ETTs. Understanding the microbiota of ETTs from mechanically ventilated neonates is crucial for effective antimicrobial therapy.
The aim of this study was to characterize the diversity of the bacterial community in biofilms on ETTs extubated from mechanically ventilated newborns using the 16S rRNA PCR-DGGE method. In addition, species-specific PCR was performed to determine whether oropharyngeal Streptococci species and other known potentially pathogenic bacteria were colonized on the ETT.
Materials and methods
Sample collection
The protocol was approved by the Ethics Committee of the Chongqing Medical University, and written informed consent was obtained from all parents or guardians. Mechanically ventilated newborns were recruited from the NICU of the Children’s Hospital of Chongqing Medical University in China between October 2011 and March 2012. Demographic and clinical characteristics of the patients were recorded. Sputum specimens were collected before extubation as previously described [10]. Specimens were subjected to routine aerobic culture immediately after suctioning according to standard microbiological methods recommended by the American Society for Microbiology (ASM). The resulting colonies were Gram stained and identified using a MicroScan WalkAway-40 automated bacterial identification and susceptibility test instrument.
The distal part of the ETT is generally the most common area for biofilm formation [4]. Therefore, a 2-cm long cross-sectional segment was taken from the distal part of each ETT and cut longitudinally using a sterile scalpel for SEM and molecular analysis. All extubated ETTs were deposited in sterile tubes and transported to the laboratory within 1 hour. All the samples were stored at -80°C until further processing.
Scanning electron microscopy (SEM)
SEM was used to investigate the formation of biofilms on the interior luminal surface of the distal part of the ETT. The SEM samples were processed using standard techniques. Briefly, the ETT specimens were fixed for at least three hours with 2.5% glutaraldehyde and washed twice in sterile phosphate buffered saline (PBS). Dehydration was performed in graded concentrations of ethanol-30, 50, 70, 90, and 100%. Each sample was gold coated using a gold sputter and viewed with an S-3000N Hitachi SEM (Hitachi High-Technologies, Japan).
DNA extraction
The planktonic microorganisms on both outer and inner surfaces of the ETT were removed by washing with sterile PBS. Microorganisms within the biofilms were detached from the surface of the ETT by sonication and vortex. The bacterial suspension was placed in a 2-mL sterile tube, and the DNA was extracted using the TIANamp Bacteria DNA Kit (Tiangen, China) according to the manufacturer’s instructions. DNA extracts were stored at -20°C until further analysis.
16S rRNA PCR and DGGE
The variable V3 region of 16S rRNA was amplified using a universal bacterial primer set (357f, 5’-GC-clamp-CCTACGGGAGGCAGCAG-3’ and 518r, 5’-ATTACCGCGGCTGCTGG-3’). A 40bp GC-clamp (CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCAGGGG) was attached to the 5’ end of the 357f primer for analysis of the PCR products by DGGE. Primers were produced by Sangon (Shanghai, China). Each 50 µl PCR reaction mixture contained: 25 µl Premix Taq (TaKaRa), 1 µl of primer, 18 µl ddH2O, and 5 µl template DNA. PCR was performed using a GeneAmp PCR system 9700 (ABI, Foster City, CA) under the following conditions: initial denaturation at 94°C for 2 min; 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s; followed by extension at 72°C for 5 min. Prior to DGGE analysis, the PCR products were examined by electrophoresis in a 2% agarose gel and visualized under UV light (BenchTop 3UV, USA).
DGGE analysis of the PCR amplicons was performed using the DCode universal mutation System (Bio-Rad, CA, USA) based on a protocol described by Muyzer et al. [11]. Twenty microliters of PCR product from the amplified DNA of each sample were added to an 8% (w/v) polyacrylamide gel. Electrophoresis was carried out using a denaturing gradient ranging from 35-65% to separate the amplicons. The denaturant (100%) contained 7 M urea and 40% formamide. Electrophoresis was performed for 30 min at 45 V followed by 16 h at 85 V in 1xTAE buffer at a constant temperature of 60°C. After electrophoresis, gels were stained using 20 µl SYBR Green I (Bio Teke) in 200 mL 1×TAE buffer for 30 min in the dark. Stained gels were imaged using the GelDoc system (Bio-Rad), and gel images were analyzed using Quantity One software, v 4.4 (Bio-Rad).
Cloning and sequencing
After imaging, 16 of the 16S rRNA bands were identified for excision and further characterization. The bands were excised with a sterile razor blade and placed in sterile tubes, and 50 µl of ddH2O were added to each tube. DNA was released into the water from the gel at 4°C during an overnight incubation. The eluate, containing the template DNA, was amplified for the second time using the primer 357f and the primer 518r without the GC clamp under the conditions described above. The PCR product was purified with the TaKaRa Mini BEST Plasmid Purification kit (TaKaRa, China). Additional cloning was performed with the pMD 18-T Vector System (TaKaRa, China) according to the manufacturer’s instructions. The PCR product was cloned into the pMD18-T vector and transformed into Escherichia coli competent DH5a cells (Tiangen, China). The transformed cells were loaded onto Luria-Bertani agar plates mixed with ampicillin (100 mg/ml) and incubated overnight at 37°C. A single colony was picked and sequenced with an ABI 3730xl sequencer (Applied Biosystems, Carlsbad, CA, USA) by Sangon (Shanghai, China) according to the manufacturer’s instructions. The BLAST program (http://blast.ncbi.nlm.nih.gov/blast) was used to find the most similar sequences in the NCBI Genbank database. To be considered a match, sequences had to have at least 97% sequence identity with the typical genus phenotypes in the public database.
Species-specific PCR processing
Species-specific PCR was undertaken to detect the common oropharyngeal Streptococcus group microorganisms in biofilms on ETTs, and the ETT-associated pathogens P. aeruginosa and K. pneumoniae. The assays were performed using primers and conditions that have been described previously [12-17]. The seven sets of species-specific PCR primers used in this study are shown in Table 1.
Table 1.
PCR primers for species-specific PCR
| Bacteria species | Primer sequence (5’-3’) | Product size (bp) |
|---|---|---|
| P. aeruginosa | F-GGGGGATCTTCGGACCTCA | 956 |
| R-TCCTTAGAGTGCCCACCCG | ||
| K. pneumoniae | F-CGACCTGATTGCATTCGCCAC | 521 |
| R-CTTCGCGCCGCGTGATACGCG | ||
| S. pneumoniae | F-ACGCAACTGACGAGTGTGAC | 353 |
| R-GATCGCGACACCGAACTAAT | ||
| S.mutans | F-TCGCGAAAAAGATAAACAAACA | 479 |
| R-GCCCCTTCACAGTTGGTTAG | ||
| S. salivarius | F-GTGTTGCCACATCTTCACTCGCTTCGG | 544 |
| R-CGTTGATGTGCTTGAAAGGGCACCATT | ||
| S. oralis | F-TCCCGGTCAGCAAACTCCAGCC | 374 |
| R-GCAACCTTTGGATTTGCAAC | ||
| S. mitis | F-TGAAATCGAGGTTGGCCTAC | 259 |
| R-CGTTTAGGAAAATCTC(G/T)CCCTT |
Results
Neonatal demographic and clinical characteristics
A total of 29 extubated ETTs were obtained from 20 newborns during the study period. The clinical information of the neonates is presented in Table 2. The median gestational age was 29 weeks (29±7 weeks), and the postnatal age ranged from one to 10 days. The duration of intubation prior to ETT collection ranged from one to seven days (mean = 3.1 d). Patients 14-20 underwent continuous ventilation; therefore, they harbored more than one ETT. The underlying diseases included severe pneumonia, neonatal respiratory distress syndrome (NRDS), premature delivery, and respiratory failure.
Table 2.
Clinical data and the results of DGGE, species-specific PCR and sputum culture
| Patient number | ETT number | Intubated Period (d) | Disease | Species-specific PCR | Sputum culture | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| S. mitis | S. oralis | S. mutans | S. salivas | S. pneumoniae | K. pneumoniae | P. aeruginosa | |||||
| 1 | 1 | 4 | Premature, Respiratory failure | + | + | + | Normal flora | ||||
| 2 | 2 | 5 | NRDS, Respiratory failure | + | + | Normal flora | |||||
| 3 | 3 | 1 | NRDS, Respiratory failure | + | Normal flora | ||||||
| 4 | 4 | 2 | Pneumonia, Respiratory failure | + | Normal flora | ||||||
| 5 | 5 | 4 | NRDS, Respiratory failure | + | + | + | Normal flora | ||||
| 6 | 6 | 5 | NRDS, Respiratory failure | + | Normal flora | ||||||
| 7 | 7 | 4 | Premature, Respiratory failure | + | + | + | + | Normal flora | |||
| 8 | 8 | 7 | NRDS, Respiratory failure | + | + | + | Klebsiella pneumoniae | ||||
| 9 | 9 | 3 | Pneumonia, | + | + | Klebsiella pneumoniae | |||||
| 10 | 10 | 3 | NRDS, Respiratory failure | + | + | + | Klebsiella pneumoniae | ||||
| 11 | 11 | 2 | Pneumonia, Respiratory failure | + | + | + | Klebsiella pneumoniae | ||||
| 12 | 12 | 4 | Pneumonia | + | + | + | Klebsiella pneumoniae | ||||
| 13 | 13 | 2 | NRDS, Respiratory failure | Klebsiella pneumoniae | |||||||
| 14a | 14 | 1 | Pneumonia, Respiratory failure | + | + | + | + | Klebsiella pneumoniae | |||
| 14b | 15 | 6 | + | + | + | Klebsiella pneumoniae | |||||
| 15a | 16 | 6 | NRDS | + | + | Klebsiella pneumoniae | |||||
| 15b | 17 | 1 | + | + | Klebsiella pneumoniae | ||||||
| 15c | 18 | 2 | + | + | Klebsiella pneumoniae | ||||||
| 16a | 19 | 1 | Pneumonia | + | + | No bacterial growth | |||||
| 16b | 20 | 5 | + | No bacterial growth | |||||||
| 17a | 21 | 3 | NRDS | + | + | Normal flora | |||||
| 17b | 22 | 4 | + | + | + | Normal flora | |||||
| 18a | 23 | 3 | NRDS, Pneumonia | + | + | + | Klebsiella pneumoniae | ||||
| 18b | 24 | 3 | + | + | + | Klebsiella pneumoniae | |||||
| 18c | 25 | 2 | + | + | Klebsiella pneumoniae | ||||||
| 19a | 26 | 2 | Pneumonia, Respiratory failure | + | + | + | Acinetobacterbaumannii | ||||
| 19b | 27 | 1 | + | + | + | Acinetobacterbaumannii | |||||
| 20a | 28 | 3 | Pneumonia | + | -- | ||||||
| 20b | 29 | 2 | + | + | -- | ||||||
Note: Normal Flora-defined as no detectable outgrowth of common VAP associated pathogens from the sputum culture. The sputum culture for patient 20 was excluded due to contamination so no data is reported. NRDS, Neonatal respiratory distress syndrome.
Microbial morphology
A representative SEM micrograph (Figure 1) shows biofilm growth on the surface of the interior lumen of an ETT following extubation. Dense amorphous material covers most of the surface, and aggregates of bacteria adhere to the matrix. Cocci species account for the majority of the bacteria, and bacilli species are sparse.
Figure 1.
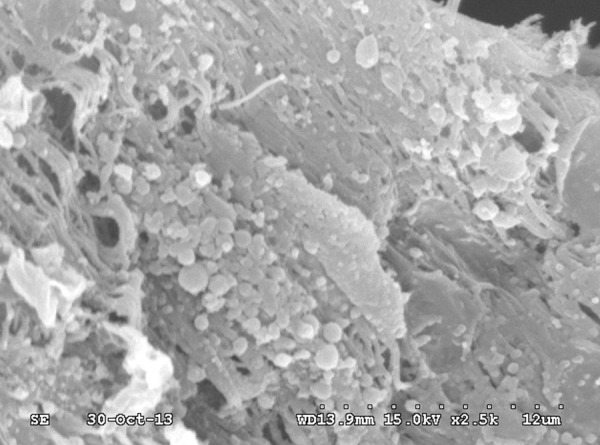
Representative SEM micrograph of an ETT associated biofilm. A dense amorphous material covers most of the surface. There is visible aggregation of bacteria adhered to the matrix.
Bacterial cultures of the sputum specimens
Sputum specimens were obtained from 19 of the 20 neonates. One specimen was discarded due to contamination. The microbiological profile of the sputum cultures is summarized in Table 2. K. pneumoniae was the most prevalent bacterium, which was found in 14 of 27 (52%) cultures. These results are consistent with previously published studies [18]. Acinetobacter baumannii was found in sputum cultures from two neonates (7%). Bacterial growth was observed in nine of the 27 (33%) specimens, but there was no indication of common pathogens associated with VAP. The sputum cultures from two of the neonates did not show any bacterial growth. No bacteria growth was detected from a short-term (less than 10 min intubation time) ETT used as a control (data not shown).
The diversity and sequencing results of DGGE
The DGGE profiles of the16S rRNA from the ETT associated biofilms are shown in Figure 2. The number of distinct DGGE bands from each ETT ranged from three to eight (mean = 5.6) (Table 2). There was a high diversity of DGGE profiles in each patient. To identify the bacterial species in the ETT associated biofilms, the 16 most prevalent bands were excised for further characterization (Table 3). All ETT samples showed polymicrobial colonization (Table 4). The three most common bacteria were Klebsiella spp. (identified in 100% of ETT specimens; 29/29), Streptococcus spp. (93%; 27/29), and Pseudomonas spp. (83%; 24/29). The high prevalence of Klebsiella reflects either the higher sensitivity of DGGE as compared to sputum culture or contamination from the extubation process, as Klebsiella spp. were also detected using DGGE on the short-term (less than 10 min intubation time) ETT used as a negative control (data not shown).The remaining sequences belonged to the genera Bacillus (19/29) and genera Enterobacter (6/29). Microorganisms that are unculturable were found in 16 of 29 (55%) specimens.
Figure 2.
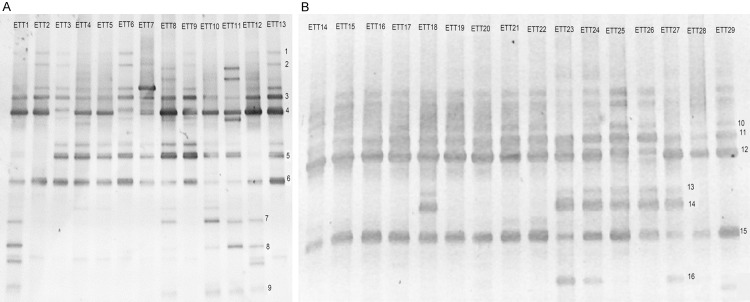
DGGE profiles of amplified 16S rRNA from ETT associated biofilms from 29 ventilated neonates. A. ETT specimens 1-13. B. ETT specimens 14-29. The Arabic numerals 1-16 indicate the DGGE bands that were excised for sequencing.
Table 3.
Sequencing results of the DGGE bands
| Band number | NCBI BLAST result | Accession number | Identity (%) |
|---|---|---|---|
| Band 1 | Unculturable bacterium | JQ462576.1 | 100% |
| Band 2 | Pseudomonas spp. | JX949982.1 | 100% |
| Band 3 | Streptococcus spp. | JX548436.1 | 100% |
| Band 4 | Streptococcus spp. | JX548381.1 | 98% |
| Band 5 | Bacillus spp. | JN316004.1 | 100% |
| Band 6 | Klebsiella spp. | JX457349.1 | 100% |
| Band 7 | Klebsiella spp. | JX310744.1 | 99% |
| Band 8 | Unculturable bacterium | HQ776847.1 | 100% |
| Band 9 | Klebsiella spp. | KC632210.1 | 100% |
| Band 10 | Pseudomonas spp. | GQ375136.1 | 100% |
| Band 11 | Streptococcus spp. | JX861486.1 | 100% |
| Band 12 | Bacillus spp. | JX490096.1 | 100% |
| Band 13 | Klebsiella spp. | JX457349.1 | 100% |
| Band 14 | Enterobacter spp. | HM074378.1 | 100% |
| Band 15 | Klebsiella spp. | JX282908.1 | 100% |
| Band 16 | Klebsiella spp. | JX489160.1 | 100% |
Table 4.
Distribution of Sequencing results of DGGE bands from neonatal ETTs
| Patient ID | ETT number | DGGE bands | Results of sequencing | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Klebsiella spp. | Streptococcus spp. | Pseudomonas spp. | Bacillus spp. | Enterobacter spp. | Unculturable bacteria | |||
| 1 | 1 | 9 | ● | ● | ● | ● | ||
| 2 | 2 | 6 | ● | ● | ● | ● | ||
| 3 | 3 | 7 | ● | ● | ● | ● | ● | |
| 4 | 4 | 5 | ● | ● | ● | |||
| 5 | 5 | 5 | ● | ● | ● | |||
| 6 | 6 | 7 | ● | ● | ● | ● | ||
| 7 | 7 | 6 | ● | ● | ● | ● | ||
| 8 | 8 | 8 | ● | ● | ● | ● | ● | |
| 9 | 9 | 7 | ● | ● | ● | ● | ● | |
| 10 | 10 | 6 | ● | ● | ● | ● | ||
| 11 | 11 | 7 | ● | ● | ● | ● | ● | |
| 12 | 12 | 7 | ● | ● | ● | |||
| 13 | 13 | 7 | ● | ● | ● | ● | ● | |
| 14a | 14 | 4 | ● | ● | ● | ● | ||
| 14b | 15 | 6 | ● | ● | ● | ● | ||
| 15a | 16 | 7 | ● | ● | ● | ● | ● | ● |
| 15b | 17 | 4 | ● | ● | ● | |||
| 15c | 18 | 3 | ● | ● | ● | |||
| 16a | 19 | 7 | ● | ● | ● | ● | ● | ● |
| 16b | 20 | 7 | ● | ● | ● | ● | ● | ● |
| 17a | 21 | 3 | ● | ● | ● | |||
| 17b | 22 | 3 | ● | ● | ● | |||
| 18a | 23 | 6 | ● | ● | ● | ● | ● | |
| 18b | 24 | 4 | ● | ● | ● | ● | ||
| 18c | 25 | 4 | ● | ● | ● | ● | ||
| 19a | 26 | 5 | ● | ● | ● | ● | ● | |
| 19b | 27 | 5 | ● | ● | ● | ● | ● | |
| 20a | 28 | 4 | ● | ● | ● | |||
| 20b | 29 | 3 | ● | ● | ● | |||
Interestingly, there was longitudinal consistency over time in individuals with more than one ETT analysis (patient 14-20). In these cases, an organism was present not only in the first specimen but also the second or third. This was especially apparent for Klebsiella spp., Streptococcus spp., and Pseudomonas spp (Table 4).
Detection of microorganisms by species-specific PCR
The results of species-specific PCR from 20 neonates (29 ETT specimens) are shown in Table 2. The three most common species identified were: P. aeruginosa (found in 86% of samples tested), S. mitis (66%), and K. pneumoniae (51%). Two bacterial species, S. oralis and S. pneumoniae were found only in four neonates and three neonates, respectively. The least common bacteria identified were S. salivas, found in one neonate (patient 18), and S. mutans, which was not detected. Oropharyngeal bacteria were found in 25 of 29 samples. In 10 of 20 neonates, the polymicrobial communities on the ETT specimens included at least one Streptococcus species, P. aeruginosa and K. pneumoniae.
Discussion
The profiles of ETT-associated microbial communities from mechanically ventilated neonates are not well understood and have been based primarily on culture techniques, which fail to detect the majority of bacteria. Molecular methods such as the 16S rRNA PCR-DGGE method can be used to characterize polymicrobial communities. Using these molecular methods, we found a high microbial diversity in the airway sample of neonates. A change in the prevalence of detectable organisms may be an indicator of VAP [10]. In the current study, we use molecular methods to demonstrate the high microbial diversity and population characteristics of polymicrobial communities on the surface of ETTs extubated from neonates. Bacterial species detected in this study included Klebsiella spp., Streptococcus spp., Pseudomonas spp., and Bacillus spp.; these bacteria, isolated from biofilms, tended to aggregate together on the same tubes.
Although molecular detection techniques can provide valuable information, it is important to utilize both conventional culture techniques and molecular techniques to fully characterize the neonatal microbiota. In the current study, we found a high degree of convergence between sputum cultures and DGGE, for example, K. pneumoniae was the most prevalent isolate by both methods. K. pneumoniae has been considered a major pathogen in nosocomial infections. In particular, extended spectrum β lactamases (ESBL) producing strains are causes for concern as they are associated with high rates of morbidity and mortality among infants in the NICU. The bacteria detected in sputum cultures and DGGE were not always consistent. Sequences corresponding to A. baumannii were not found using the DGGE method, but it is one of the most prevalent isolates in sputum samples of VAP neonates in our hospital [2].
In the current study, both DGGE and species-specific PCR methods demonstrated a remarkable diversity of the bacterial community in the biofilms on the ETTs. Oropharyngeal bacteria, alone or in combination with other bacteria, were recovered from most of the specimens. Interestingly, Klebsiella spp., Streptococcus spp. and Pseudomonas spp. seemed to persist on the tubes during the intubation period, which may be an important factor for pathogenesis. While conventional culture techniques can only detect few or even none of the bacterial species in biofilms [9], molecular methods may promote an improved understanding of the microbiota and allow us to better diagnose and treat ETT-associated infections. As automated systems become available, high throughput screening will facilitate the process.
Biofilm formation is a dynamic process; microorganisms in biofilms on extubated ETTs can reflect disease status. Cairns et al. suggested that normal oral microflora may act as the initial colonizers on ETTs and could promote subsequent biofilm development [9]. Co-aggregation of different bacterial species in biofilms was first described by Gibbons [19] in dental plaque, and is now known to occur in a variety of environments [20-22]. The mechanisms underlying co-aggregation of oral commensals and other bacteria are still unclear. One possibility is that quorum sensing (QS), a method of autoinducers (AIs)-mediated cross-talk between individual bacteria embedded in biofilms, could lead to polymicrobial biofilm formation. Autoinducer-2 (AI-2) is a well-studied QS signaling molecule produced by many Gram-positive and Gram-negative bacteria species [23,24]. AI-2 can facilitate interspecies communication in microbial environments [25]. It has also been shown to promote co-aggregation by increasing the expression of adhesion molecules of pathogenic bacteria [26]. In this context, we speculate that the AI-2 producers K. pneumoniae and Streptococcus spp. may facilitate biofilm formation incorporating P. aeruginosa.
Our study is associated with several limitations. First, DGGE data are descriptive rather than quantitative. Second, we had a limited sample size both in the number of patients and ETT specimens. Further study using a larger sample of ETT biofilms is necessary to elucidate whether there is a correlation between the microbial community on the ETT and different diseases.
In conclusion, we demonstrated that Klebsiella spp., Streptococcus spp. and Pseudomonas spp. were the most frequent microbes on the surface of neonatal ETTs, and they prefer to co-exist in biofilms. These data provide a more in-depth knowledge of the ETT associated microbiota, which may have clinical significance for the management of mechanically ventilated neonates. We speculate that the AI-2 mediated signaling pathway may play a role in biofilm formation; however, additional studies are required to support this speculation.
Acknowledgements
We are grateful to Yu He, Zhengli Wang and Ying Dong for their valuable advice to the linguistic revision of the manuscript. This study was supported by the National Natural Science Foundation of China (No. 81370744), the fund from Ministry of Education (No. X3387), National Science and Technology Support Project (2012BAI04B05), the Scientific Research Foundation of Chongqing Municipal Health Bureau (NO. 2013-2-051) and the Scientific Research Foundation of The science and Technology Commission of Yuzhong District of Chongqing (No. 20140103).
Disclosure of conflict of interest
None.
References
- 1.Garland JS. Strategies to prevent ventilator-associated pneumonia in neonates. Clin Perinatol. 2010;37:629–43. doi: 10.1016/j.clp.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Deng C, Li X, Zou Y, Wang J, Wang J, Namba F, Hiroyuki Y, Yu J, Yamauchi Y, Guo C. Risk factors and pathogen profile of ventilator-associated pneumonia in a neonatal intensive care unit in China. Pediatr Int. 2011;53:332–7. doi: 10.1111/j.1442-200X.2011.03382.x. [DOI] [PubMed] [Google Scholar]
- 3.Gibbs K, Holzman IR. Endotracheal tube: friend or foe? bacteria, the endotracheal tube, and the impact of colonization and infection. Semin Perinatol. 2012;36:454–61. doi: 10.1053/j.semperi.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Zur K. Electron microscopic analysis of biofilm on endotracheal tubes removed from intubated neonates. Otolaryngol Head Neck Surg. 2004;130:407–14. doi: 10.1016/j.otohns.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Pneumatikos IA, Dragoumanis CK, Bouros DE. Ventilator-associated pneumonia or endotracheal tube-associated pneumonia? An approach to the pathogenesis and preventive strategies emphasizing the importance of endotracheal tube. Anesthesiology. 2009;110:673–80. doi: 10.1097/ALN.0b013e31819868e0. [DOI] [PubMed] [Google Scholar]
- 6.Yang L, Liu Y, Wu H, Hóiby N, Molin S, Song ZJ. Current understanding of multi-species biofilms. Int J Oral Sci. 2011;3:74–81. doi: 10.4248/IJOS11027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gil-Perotin S, Ramirez P, Marti V, Sahuquillo JM, Gonzalez E, Calleja I, Menendez R, Bonastre J. Implications of endotracheal tube biofilm in ventilator-associated pneumonia response: a state of concept. Crit Care. 2012;16:R93. doi: 10.1186/cc11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaeberlein T, Lewis K, Epstein SS. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–9. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 9.Cairns S, Thomas JG, Hooper SJ, Wise MP, Frost PJ, Wilson MJ, Lewis MA, Williams DW. Molecular analysis of microbial communities in endotracheal tube biofilms. PLoS One. 2011;6:e14759. doi: 10.1371/journal.pone.0014759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu W, Yu J, Ai Q, Liu D, Song C, Li L. Increased constituent ratios of Klebsiella sp. , Acinetobacter sp., and Streptococcus sp. and a decrease in microflora diversity may be indicators of ventilator-associated pneumonia: a prospective study in the respiratory tracts of neonates. PLoS One. 2014;9:e87504. doi: 10.1371/journal.pone.0087504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoshino T, Kawaguchi M, Shimizu N, Hoshino N, Ooshima T, Fujiwara T. PCR detection and identification of oral streptococci in saliva samples using gtf genes. Diagn Microbiol Infect Dis. 2004;48:195–9. doi: 10.1016/j.diagmicrobio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Kotlovsky T, Shalginov R, Austin L, Sprecher H. Rapid detection of bla (KPC)-positive Klebsiella pneumoniae in a clinical setting. Eur J Clin Microbiol Infect Dis. 2009;28:309–11. doi: 10.1007/s10096-008-0615-2. [DOI] [PubMed] [Google Scholar]
- 14.Park HK, Lee SJ, Yoon JW, Shin JW, Shin HS, Kook JK, Myung SC, Kim W. Identification of the cpsA gene as a specific marker for the discrimination of Streptococcus pneumoniae from viridans group streptococci. J Med Microbiol. 2010;59:1146–52. doi: 10.1099/jmm.0.017798-0. [DOI] [PubMed] [Google Scholar]
- 15.Garnier F, Gerbaud G, Courvalin P, Galimand M. Identification of clinically relevant viridans group streptococci to the species level by PCR. J Clin Microbiol. 1997;35:2337–41. doi: 10.1128/jcm.35.9.2337-2341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spilker T, Coenye T, Vandamme P, LiPuma JJ. PCR-based assay for differentiation of Pseudo- monas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42:2074–9. doi: 10.1128/JCM.42.5.2074-2079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Saxena D, Caufield PW, Ge Y, Wang M, Li Y. Development of species-specific primers for detection of Streptococcus mutans in mixed bacterial samples. FEMS Microbiol Lett. 2007;272:154–62. doi: 10.1111/j.1574-6968.2007.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Apisarnthanarak A, Holzmann-Pazgal G, Hamvas A. Ventilator-associated pneumonia in extremely preterm neonates in a neonatal intensive care unit: characteristics, risk factors, and outcomes. Pediatrics. 2003;112:1283–9. doi: 10.1542/peds.112.6.1283. [DOI] [PubMed] [Google Scholar]
- 19.Gibbons RJ, Nygaard M. Interbacterial aggregation of plaque bacteria. Arch Oral Biol. 1970;15:1397–400. doi: 10.1016/0003-9969(70)90031-2. [DOI] [PubMed] [Google Scholar]
- 20.Vornhagen J, Tevens M, McCormick DW, Dowd SE, Eisenberg JN, Boles BR, Rickard AH. Coaggregation occurs amongst bacteria within and between biofilms in domestic showerheads. Biofouling. 2013;29:53–68. doi: 10.1080/08927014.2012.744395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ledder RG, Timperley AS, Friswell MK, Macfarlane S, McBain AJ. Coaggregation between and among human intestinal and oral bacteria. FEMS Microbiol Ecol. 2008;66:630–6. doi: 10.1111/j.1574-6941.2008.00525.x. [DOI] [PubMed] [Google Scholar]
- 22.Simoes LC, Simoes M, Vieira MJ. Intergeneric coaggregation among drinking water bacteria: evidence of a role for Acinetobacter calcoaceticus as a bridging bacterium. Appl Environ Microbiol. 2008;74:1259–63. doi: 10.1128/AEM.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNab R, Ford SK, El-Sabaeny A, Barbieri B, Cook GS, Lamont RJ. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol. 2003;185:274–84. doi: 10.1128/JB.185.1.274-284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan K, Dammel C, Stein J, Rabin H, Surette MG. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol. 2003;50:1477–91. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 25.Hardie KR, Heurlier K. Establishing bacterial communities by ‘word of mouth’: LuxS and autoinducer 2 in biofilm development. Nat Rev Microbiol. 2008;6:635–43. doi: 10.1038/nrmicro1916. [DOI] [PubMed] [Google Scholar]
- 26.Jang YJ, Choi YJ, Lee SH, Jun HK, Choi BK. Autoinducer 2 of Fusobacterium nucleatum as a target molecule to inhibit biofilm formation of periodontopathogens. Arch Oral Biol. 2013;58:17–27. doi: 10.1016/j.archoralbio.2012.04.016. [DOI] [PubMed] [Google Scholar]


