Abstract
The association between alcohol dehydrogenase 1C (ADH1C) gene polymorphism and alcoholic liver cirrhosis (ALC) has been analyzed in several studies, but results have been conflicting. In this study, a meta-analysis was performed to assess the associations between the ADH1C polymorphism and risk of ALC. Relevant studies were identified using PubMed, Web of Science, CNKI and Wanfang databases up to January 10, 2015. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess the strength of the association using the fixed or random effect model. A total of 16 case-control studies, including 1375 cases and 1802 controls, were included. Overall, no significant association between the ADH1C polymorphism and ALC risk was found (dominant model: OR=0.87, 95% CI: 0.62-1.23; recessive model: OR=1.30, 95% CI: 0.84-1.99; *1/*2 vs. *1/*1: OR=0.87, 95% CI: 0.63-1.21; *2/*2 vs. *1/*1: OR=1.10, 95% CI: 0.71-1.70). In the subgroup analysis by ethnicity, we observed a significant association in Asian descent (*1/*2 vs. *1/*1: OR=1.63, 95% CI: 1.07-2.49), while a decreased risk was found among Caucasians (dominant model: OR=0.81, 95% CI: 0.66-0.99; *1/*2 vs. *1/*1: OR=0.76, 95% CI: 0.61-0.95). This meta-analysis demonstrated that the ADH1C polymorphism might increase the risk of ALC in Asians, while it may be a protective factor for ALC among Caucasians.
Keywords: ADH1C, polymorphism, alcoholic liver cirrhosis, meta-analysis
Introduction
Alcoholic liver disease (ALD) refers to a wide spectrum of liver abnormalities, ranging from fatty liver to acute alcoholic hepatitis, and alcoholic liver cirrhosis (ALC). The most severe of these, ALC, causes an estimated 373, 000 deaths per year [1]. The burden of ALD is highest in the developed world, where it may account for as much as 9.2% of all disability-adjusted life years [2]. For example, the annual costs of hospitalization for ALD in the United States are estimated to be between $600 million and $1.8 billion [3]. It has been demonstrated that a clear correlation exists between cumulative alcohol intake and ALD; however, only a small portion of the alcohol abusers develop signs of liver disease, which suggests some of the genetic variations are involved in the etiology of ALD [4].
Alcohol dehydrogenase (ADH) is a dimeric protein consisting of two 40-kDa enzyme subunits. There are at least five different classes of human ADH isoenzymes based on differences at the molecular level [5]. The class I ADH enzymes, encoded by ADH1A, ADH1B, and ADH1C (previously known as ADH1, ADH2, and ADH3, respectively) are mainly involved in the oxidation of ethanol. Polymorphic variants exist among the class I ADH genes, specifically ADH1B and ADH1C, and are known to produce enzymes with distinct kinetic properties [6]. As for ADH1C, the polymorphic sites are Arg272Gln (rs1693482) and Ile350Val (rs698) [7], the 272Arg and 350Ile carriers have the ADH1C*1 allele, whereas 272Gln and 350 Val carriers have the ADH1C*2 allele. Individuals with ADH1C*1 allele have an ethanol oxidizing capacity 2.5-times higher when compared to ADH1C*2 allele [8] and therefore produces more acetaldehyde. Thus, it has been hypothesized that individuals with homozygosity for the allele ADH1C*1 carry an increased risk for alcohol-induced organ damage, such as liver cirrhosis, than patients with heterozygosity or those homozygous for ADH1C*2 [9].
To date, many studies have investigated the association between the ADH1C polymorphism and the risk of alcoholic liver cirrhosis [10-28]. However, the results remain controversial. In this study, we conduct a meta-analysis to evaluate the association between the polymorphism and alcoholic liver cirrhosis risk.
Materials and methods
Search strategy
Relevant articles published before January 10, 2015 were identified through a search of PubMed, Web of Science, CNKI, Wanfang and VIP databases using the following terms: “alcohol dehydrogenase 3 or ADH3 or alcohol dehydrogenase 1C or ADH1C” and “genetic polymorphism or polymorphisms or variant” and “alcoholic liver disease or ALD or alcoholic liver cirrhosis or ALC or cirrhosis”. The search was restricted to humans without language restrictions. Additional studies were identified by a hand search of references of original or review articles on this topic.
Inclusion criteria and exclusion criteria
Studies included in this meta-analysis have to meet the following criteria: (1) studies that evaluated the association between the ADH1C polymorphism and alcoholic liver cirrhosis, (2) in a case-control study design, (3) had detailed genotype frequency of cases and controls or could be calculated from the article text. Studies were excluded when they were: (1) case-only study, case reports, and review articles, (2) based on incomplete data, (3) duplicate of previous publication.
Data extraction
For each study, the following data were extracted independently by two investigators: the first author’s name, year of publication, country of origin, ethnicity, genotyping methods, number of cases and controls, and Hardy-Weinberg equilibrium (HWE) in controls (P value). The results were compared, and disagreements were discussed among all authors and resolved with consensus.
Statistical analysis
The strength of the association between the ADH1C polymorphism and alcoholic liver cirrhosis risk was estimated by odds ratio (ORs) and 95% confidence interval (CIs). Four different ORs were calculated: dominant model (*1/*2+*2/*2 vs. *1/*1), recessive model (*2/*2 vs. *1/*2+*1/*1), heterozygote comparison (*1/*2 vs. *1/*1), and homozygote comparison (*2/*2 vs. *1/*1). Genotype frequencies of the controls were tested for the HWE using the χ2 test. Heterogeneity among studies was assessed by χ2-based Q test as well as the I 2 statistic [29]. When a significant Q test (P>0.1) or I 2<50% indicated homogeneity across studies, the fixed effects model was used [30], or else the random effects model was used [31]. Then, we performed stratification analyses on ethnicity. Sensitivity analysis was performed by removing one individual study each time to evaluate the stability of the results. Begg’s funnel plot and Egger’s regression test were used to investigate potential publication bias [32,33]. P<0.05 was considered statistically significant. All statistical analyses were performed using the Cochrane Collaboration RevMan 5.2 and STATA package version 12.0 (Stata Corporation, College Station, Texas).
Results
Study characteristics
Initially, the searched keywords identified 52 articles. According to the inclusion criteria, 19 studies [10-28] with full-text were included in this meta-analysis and 33 studies were excluded. Because one study [28] did not present detailed genotyping information, we excluded it. We also excluded two studies [26,27] because they included the overlapped data with those included in the analysis [14]. Therefore, as shown in Table 1, there were 16 case-control studies with 1375 cases and 1802 controls concerning ADH1C polymorphism. Of the 16 eligible studies, two ethnicities were addressed: six studies [10-13,19-21] were conducted on Asian populations and ten studies [10,14-18,22-25] on Caucasian populations. The distribution of genotypes in the controls was consistent with the HWE for all selected studies, except for three studies [17,22,25].
Table 1.
Characteristics of studies included in the meta-analysis
| Author | Year | Country | Ethnicity | Genotyping methods | Genotype (case/control) | HWE | |||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| Total | *1/*1 | *1/*2 | *2/*2 | ||||||
| Borras [10] | 2000 | Mixed | Caucasian | PCR-RFLP | 180/224 | 62/66 | 82/117 | 36/41 | 0.387 |
| Chao [11] | 1994 | China | Asian | PCR-RFLP | 27/47 | 17/42 | 8/5 | 2/0 | 0.700 |
| Chao [12] | 1997 | China | Asian | PCR-RFLP | 75/100 | 57/88 | 16/11 | 2/1 | 0.342 |
| Chao [13] | 2000 | China | Asian | PCR-RFLP | 116/105 | 88/91 | 25/13 | 3/1 | 0.495 |
| Cichoz-Lach [14] | 2007 | Poland | Caucasian | PCR-RFLP | 57/54 | 26/10 | 19/24 | 12/20 | 0.559 |
| Couzigou [15] | 1990 | France | Caucasian | PCR | 46/39 | 13/14 | 26/17 | 7/8 | 0.504 |
| Day [16] | 1991 | England | Caucasian | PCR | 59/79 | 26/25 | 22/37 | 11/17 | 0.634 |
| Frenzer [17] | 2002 | Australia | Caucasian | PCR-RFLP | 57/200 | 13/56 | 21/120 | 23/24 | 0.001 |
| Homann [18] | 2006 | German | Caucasian | PCR-RFLP | 217/174 | 43/38 | 122/92 | 52/44 | 0.439 |
| Khan [19] | 2010 | India | Asian | PCR-RFLP | 175/255 | 99/84 | 76/171& | NA | |
| Kim [20] | 2004 | Korea | Asian | PCR-RFLP | 22/100 | 20/76 | 2/21 | 0/3 | 0.313 |
| Lee [21] | 2001 | Korea | Asian | PCR-RFLP | 56/64 | 50/57 | 5/7 | 1/0 | 0.644 |
| Monzoni [22] | 2001 | Italy | Caucasian | PCR | 15/92 | 6/31 | 4/53 | 5/8 | 0.028 |
| Poupon [23] | 1992 | France | Caucasian | Starch-Gel Electrophoresis | 23/42 | 12/12 | 8/23 | 3/7 | 0.472 |
| Sun [24] | 2005 | German | Caucasian | PCR-RFLP | 151/163 | 30/33 | 92/89 | 29/41 | 0.227 |
| Vidal [25] | 2004 | Spain | Caucasian | PCR-RFLP | 99/64 | 35/15 | 45/42 | 19/7 | 0.008 |
HWE: Hardy-Weinberg equilibrium;
Numbers of *1/*2+*2/*2;
PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; NA: not available.
Quantitative data synthesis
Overall, no significant association between the ADH1C polymorphism and ALC risk was found (dominant model: OR=0.87, 95% CI: 0.62-1.23; recessive model: OR=1.30, 95% CI: 0.84-1.99; *1/*2 vs. *1/*1: OR=0.87, 95% CI: 0.63-1.21; *2/*2 vs. *1/*1: OR=1.10, 95% CI: 0.71-1.70) (Table 2; Figure 1).
Table 2.
Summary of OR of the ADH1C polymorphism and alcoholic cirrhosis risk
| Variables | Na | Dominant model | Recessive model | *1/*2 vs. *1/*1 | *2/*2 vs. *1/*1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||
| OR (95% CI) | P b | I2 | OR (95% CI) | P b | I2 | OR (95% CI) | P b | I2 | OR (95% CI) | P b | I2 | ||
| Total | 16 | 0.87 (0.62, 1.23) | <0.00001 | 72 | 1.30 (0.84, 1.99) | 0.0005 | 63 | 0.87 (0.63, 1.21) | 0.002 | 58 | 1.10 (0.71, 1.70) | 0.009 | 52 |
| Ethnicity | |||||||||||||
| Asian | 6 | 1.19 (0.47, 3.04) | <0.00001 | 87 | 2.70 (0.90, 8.13) | 0.81 | 0 | 1.63 (1.07, 2.49) | 0.10 | 48 | 2.84 (0.96, 8.39) | 0.72 | 0 |
| Caucasian | 10 | 0.81 (0.66, 0.99) | 0.07 | 44 | 1.19 (0.75, 1.91) | <0.0001 | 74 | 0.76 (0.61, 0.95) | 0.07 | 43 | 0.97 (0.61, 1.56) | 0.004 | 63 |
Number of comparisons.
Test for heterogeneity.
Figure 1.

Forest plots for the association of ADH1C polymorphism and ALC risk (dominant model).
In the subgroup analysis by ethnicity, we observed a significant association in Asian descent (*1/*2 vs. *1/*1: OR=1.63, 95% CI: 1.07-2.49), while a decreased risk was found among Caucasians (dominant model: OR=0.81, 95% CI: 0.66-0.99; *1/*2 vs. *1/*1: OR=0.76, 95% CI: 0.61-0.95) (Table 2; Figure 2).
Figure 2.
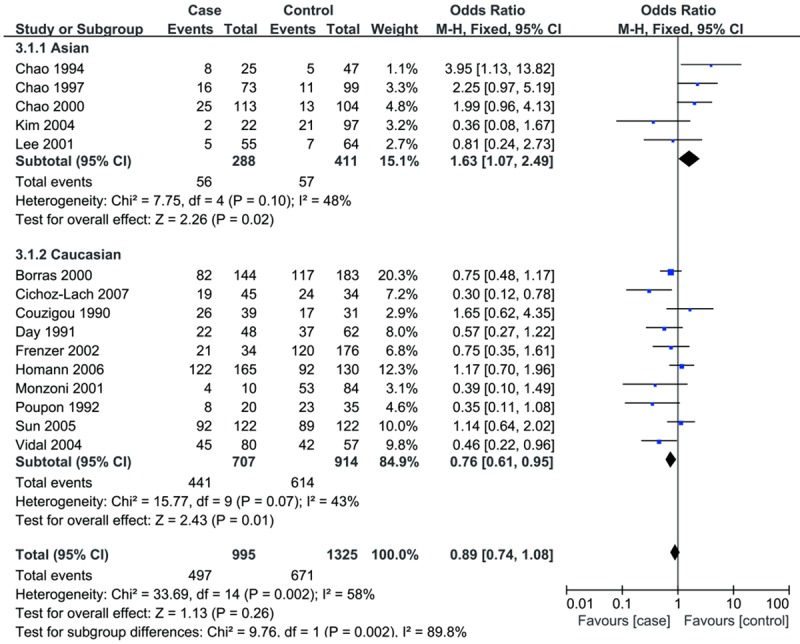
Forest plots for subgroup analysis by ethnicity for the association of ADH1C polymorphism and ALC risk (*1/*2 vs. *1/*1).
Sensitivity analyses were conducted to determine whether modification of the inclusion criteria of the meta-analysis affected the final results. We examined the influence of these studies on the pooled OR by repeating the meta-analysis while excluding one study at a time. The estimated pooled ORs change quite little, indicating that our results were statistically robust.
Test of heterogeneity
There was significant heterogeneity for overall comparisons (dominant model: P<0.00001, I2=72%; recessive model: P=0.0005, I2=63%; *1/*2 vs. *1/*1: P=0.002, I2=58%; *2/*2 vs. *1/*1: P=0.009, I2=52%). In the subgroup analysis by ethnicity, the heterogeneity was partially decreased or removed in Asian population. However, significant heterogeneity remain exist in the Caucasian population under recessive and homozygote comparison models.
Publication bias
The Begg’s funnel plot and Egger’s test was used to address potential publication bias in the available literature. The shape of funnel plots did not reveal any evidence of funnel plot asymmetry (data not shown). Egger’s test also showed that there was no statistical significance for the evaluation of publication bias (dominant model: P=0.395; recessive model: P=0.325; *1/*2 vs. *1/*1: P=0.645; *2/*2 vs. *1/*1: P=0.403).
Discussion
Pharmacokinetic difference in ethanol metabolism is a plausible determinant of susceptibility to the many diseases associated with alcohol. ADH is a zinc-containing cytosolic enzyme that oxidizes short-chain alcohols to aldehydes, mainly formaldehyde and acetaldehyde. Epidemiological studies have examined the association between the ADH1C*1 allele and risk of ALC with conflicting results. Some investigations demonstrated that the ADH1C*1/*1 genotype exhibited significant association with alcohol liver cirrhosis [14,19]; on the contrary, Frenzer et al [17] suggest the ADH1C*2*2 is associated with alcoholic cirrhosis. Additionally, the association of ADH1C variants and ALC risk was not validated by others [15,20,21,23,25]. In order to resolve this conflict, we performed a meta-analysis to derive a more precise estimation of the association.
In this study, 16 case-control studies included 1375 cases and 1802 controls were included. We found that the ADH1C*2 allele is not associated with ALC. That is, the ADH1C genotype distribution between ALC and control group was no significant difference. Interestingly, when the analysis was stratified by ethnicity, we observed a significant association in Asian descent, while a decreased risk was found among Caucasians. There are some possible explanations for the discrepant results. First, significant linkage disequilibrium has been detected between the ADH1B and ADH1C polymorphisms as well as the two variants in ADH1C [34]. These functional variants result in the production of enzymes with different kinetic properties [7] and subsequently the generation of different quantities of acetaldehyde, which might affect the risk of ALC and that could differ between Caucasians and Asians. Another reason may be found in the population genetics of alcohol metabolizing enzyme variants. In addition, the prevalence of the variant ADH1C allele is high in Caucasian population (40-50%) and lower in Asians (5%) [9], which may also contribute to the results.
Two significant issues should be addressed in this study, that is, heterogeneity and publication bias, which may influence the results of meta-analysis. We don’t detect a significant publication bias in this meta-analysis, suggesting the reliability of our results. With regard to heterogeneity, in this meta-analysis, heterogeneity was found in overall comparison under all four genetic models, when stratified by ethnicity, the heterogeneity was partly decreased or removed in Asian populations. However, heterogeneity still existed among Caucasian population. The results above suggest that the ethnic background might be the source of heterogeneity. Then sensitivity analyses were conducted by successively excluding one study, the estimated pooled odd ratio changed quite little, strengthening the results from this meta-analysis.
This meta-analysis has limitations that must be acknowledged. First, because of incomplete raw data or publication limitations, some relevant studies could not be included in our analysis. Second, moderate to higher heterogeneity existed for the analyses especially for the subgroup of Caucasian. Third, our results were based on unadjusted estimates, which may cause serious confounding bias. In addition, all of the studies were conducted in Asian and Caucasians, which may generate selective bias. More studies focused on Africans are needed.
In summary, this meta-analysis suggests that the ADH1C polymorphism might increase the risk of ALC in Asians, while it may be a protective factor for ALC among Caucasians. However, due to the limitations mentioned above, more researches with larger sample size are still required to provide a more reliable and representative statistical analysis precisely.
Disclosure of conflict of interest
None.
References
- 1.Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, Patra J. Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. Lancet. 2009;373:2223–2233. doi: 10.1016/S0140-6736(09)60746-7. [DOI] [PubMed] [Google Scholar]
- 2.O’Shea RS, Dasarathy S, McCullough AJ. Alcoholic liver disease. Hepatology. 2010;51:307–328. doi: 10.1002/hep.23258. [DOI] [PubMed] [Google Scholar]
- 3.Kim WR, Brown RS Jr, Terrault NA, EI-Serag H. Burden of liver disease in the United States: summary of a workshop. Hepatology. 2002;36:227–242. doi: 10.1053/jhep.2002.34734. [DOI] [PubMed] [Google Scholar]
- 4.Stickel F, Hampe J. Genetic determinants of alcoholic liver disease. Gut. 2012;61:150–159. doi: 10.1136/gutjnl-2011-301239. [DOI] [PubMed] [Google Scholar]
- 5.Duester G, Farres J, Felder MR, Holmes RS, Hoog JO, Pares X, Plabb BV, Yin SJ, Jornvall H. Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem Pharmacol. 1999;58:389–395. doi: 10.1016/s0006-2952(99)00065-9. [DOI] [PubMed] [Google Scholar]
- 6.Bosron WF, Lumeng L, Li TK. Genetic polymorphism of enzymes of alcohol metabolism and susceptibility to alcoholic liver disease. Mol Aspects Med. 1988;10:147–158. doi: 10.1016/0098-2997(88)90019-2. [DOI] [PubMed] [Google Scholar]
- 7.Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- 8.Höög JO, Hedén LO, Larsson K, Jörnvall H, von Bahr-Lindström H. The gamma 1 and gamma 2 subunits of human liver alcohol dehydrogenase. cDNA structures, two amino acid replacements, and compatibility with changes in the enzymatic properties. Eur J Biochem. 1986;159:215–218. doi: 10.1111/j.1432-1033.1986.tb09855.x. [DOI] [PubMed] [Google Scholar]
- 9.Bosron WF, Ehrig T, Li TK. Genetic factors in alcohol metabolism and alcoholism. Semin Liver Dis. 1993;13:126–135. doi: 10.1055/s-2007-1007344. [DOI] [PubMed] [Google Scholar]
- 10.Borràs E, Coutelle C, Rosell A, Fernández-Muixi F, Broch M, Crosas B, Hjelmqvist L, Lorenzo A, Gutiérrez C, Santos M, Szczepanek M, Heilig M, Quattrocchi P, Farrés J, Vidal F, Richart C, Mach T, Bogdal J, Jörnvall H, Seitz HK, Couzigou P, Parés X. Genetic polymorphism of alcohol dehydrogenase in europeans: the ADH2*2 allele decreases the risk for alcoholism and is associated with ADH3*1. Hepatology. 2000;31:984–989. doi: 10.1053/he.2000.5978. [DOI] [PubMed] [Google Scholar]
- 11.Chao YC, Liou SR, Chung YY, Tang HS, Hsu CT, Li TK, Yin SJ. Polymorphism of alcohol and aldehyde dehydrogenase genes and alcoholic cirrhosis in Chinese patients. Hepatology. 1994;19:360–366. [PubMed] [Google Scholar]
- 12.Chao YC, Young TH, Tang HS, Hsu CT. Alcoholism and alcoholic organ damage and genetic polymorphisms of alcohol metabolizing enzymes in Chinese patients. Hepatology. 1997;25:112–117. doi: 10.1002/hep.510250121. [DOI] [PubMed] [Google Scholar]
- 13.Chao YC, Wang LS, Hsieh TY, Chu CW, Chang FY, Chu HC. Chinese alcoholic patients with esophageal cancer are genetically different from alcoholics with acute pancreatitis and liver cirrhosis. Am J Gastroenterol. 2000;95:2958–2964. doi: 10.1111/j.1572-0241.2000.02328.x. [DOI] [PubMed] [Google Scholar]
- 14.Cichoz-Lach H, Partycka J, Nesina I, Celinski K, Slomka M, Wojcierowski J. Alcohol dehydrogenase and aldehyde dehydrogenase gene polymorphism in alcohol liver cirrhosis and alcohol chronic pancreatitis among Polish individuals. Scand J Gastroenterol. 2007;42:493–498. doi: 10.1080/00365520600965723. [DOI] [PubMed] [Google Scholar]
- 15.Couzigou P, Fleury B, Groppi A, Cassaigne A, Begueret J, Iron A. Genotyping study of alcohol dehydrogenase class I polymorphism in French patients with alcoholic cirrhosis. The French Group for Research on Alcohol and Liver. Alcohol Alcohol. 1990;25:623–626. doi: 10.1093/oxfordjournals.alcalc.a045058. [DOI] [PubMed] [Google Scholar]
- 16.Day CP, Bashir R, James OF, Bassendine MF, Crabb DW, Thomasson HR, Li TK, Edenberg HJ. Investigation of the role of polymorphisms at the alcohol and aldehyde dehydrogenase loci in genetic predisposition to alcohol-related end-organ damage. Hepatology. 1991;14:798–801. doi: 10.1002/hep.1840140509. [DOI] [PubMed] [Google Scholar]
- 17.Frenzer A, Butler WJ, Norton ID, Wilson JS, Apte MV, Pirola RC, Ryan P, Roberts-Thomson IC. Polymorphism in alcohol-metabolizing enzymes, glutathione S-transferases and apolipoprotein E and susceptibility to alcohol-induced cirrhosis and chronic pancreatitis. J Gastroenterol Hepatol. 2002;17:177–182. doi: 10.1046/j.1440-1746.2002.02670.x. [DOI] [PubMed] [Google Scholar]
- 18.Homann N, Stickel F, König IR, Jacobs A, Junghanns K, Benesova M, Schuppan D, Himsel S, Zuber-Jerger I, Hellerbrand C, Ludwig D, Caselmann WH, Seitz HK. Alcohol dehydrogenase 1C*1 allele is a genetic marker for alcohol-associated cancer in heavy drinkers. Int J Cancer. 2006;118:1998–2002. doi: 10.1002/ijc.21583. [DOI] [PubMed] [Google Scholar]
- 19.Khan AJ, Husain Q, Choudhuri G, Parmar D. Association of polymorphism in alcohol dehydrogenase and interaction with other genetic risk factors with alcoholic liver cirrhosis. Drug Alcohol Depend. 2010;109:190–197. doi: 10.1016/j.drugalcdep.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Kim MS, Lee DH, Kang HS, Park HS, Jung S, Lee JW, Kwon KS, Kim PS, Kim HG, Shin YW, Kim YS, Baek I, Lee MS. Genetic polymorphisms of alcohol-metabolizing enzymes and cytokines in patients with alcohol induced pancreatitis and alcoholic liver cirrhosis. Korean J Gastroenterol. 2004;43:355–363. [PubMed] [Google Scholar]
- 21.Lee HC, Lee HS, Jung SH, Yi SY, Jung HK, Yoon JH, Kim CY. Association between polymorphisms of ethanol-metabolizing enzymes and susceptibility to alcoholic cirrhosis in a Korean male population. J Korean Med Sci. 2001;16:745–750. doi: 10.3346/jkms.2001.16.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monzoni A, Masutti F, Saccoccio G, Bellentani S, Tiribelli C, Giacca M. Genetic determinants of ethanol-induced liver damage. Mol Med. 2001;7:255–262. [PMC free article] [PubMed] [Google Scholar]
- 23.Poupon RE, Nalpas B, Coutelle C, Fleury B, Couzigou P, Higueret D. Polymorphism of alcohol dehydrogenase, alcohol and aldehyde dehydrogenase activities: implication in alcoholic cirrhosis in white patients. The French Group for Research on Alcohol and Liver. Hepatology. 1992;15:1017–1022. doi: 10.1002/hep.1840150608. [DOI] [PubMed] [Google Scholar]
- 24.Sun L, König IR, Jacobs A, Seitz HK, Junghanns K, Wagner T, Ludwig D, Jacrobs A, Homann N. Mean corpuscular volume and ADH1C genotype in white patients with alcohol-associated diseases. Alcohol Clin Exp Res. 2005;29:788–793. doi: 10.1097/01.alc.0000163500.81691.74. [DOI] [PubMed] [Google Scholar]
- 25.Vidal F, Lorenzo A, Auguet T, Olona M, Broch M, Gutiérrez C, Aguilar C, Estupiñà P, Santos M, Richart C. Genetic polymorphisms of ADH2, ADH3, CYP4502E1Dra-I and Pst-I, and ALDH2 in Spanish men: lack of association with alcoholism and alcoholic liver disease. J Hepatol. 2004;41:744–750. doi: 10.1016/j.jhep.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Cichoz-Lach H, Partycka J, Nesina I, Celinski K, Słomka M, Wojcierowski J. Genetic polymorphism of alcohol dehydrogenase 3 in alcohol liver cirrhosis and in alcohol chronic pancreatitis. Alcohol. 2006;41:14–7. doi: 10.1093/alcalc/agh225. [DOI] [PubMed] [Google Scholar]
- 27.Cichoz-Lach H, Partycka J, Nesina I, Wojcierowski J, Słomka M, Celiński K. Genetic polymorphism of alcohol dehydrogenase 3 in digestive tract alcohol damage. Hepatogastroenterology. 2007;54:1222–1227. [PubMed] [Google Scholar]
- 28.Lorenzo A, Auguet T, Vidal F, Broch M, Olona M, Gutiérrez C, López-Dupla M, Sirvent JJ, Quer JC, Santos M, Richart C. Polymorphisms of alcohol-metabolizing enzymes and the risk for alcoholism and alcoholic liver disease in Caucasian Spanish women. Drug Alcohol Depend. 2006;84:195–200. doi: 10.1016/j.drugalcdep.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 30.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 31.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 32.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 33.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokoyama A, Tsutsumi E, Imazeki H, Suwa Y, Nakamura C, Yokoyama T. Polymorphisms of alcohol dehydrogenase-1B and aldehyde dehydrogenase-2 and the blood and salivary ethanol and acetaldehyde concentrations of Japanese alcoholic men. Alcohol Clin Exp Res. 2010;34:1246–1256. doi: 10.1111/j.1530-0277.2010.01202.x. [DOI] [PubMed] [Google Scholar]


