Abstract
Background: Dysregulation of miR-675 has been found in a variety of solid tumors. MiR-675 has been suggested as having both oncogenic and tumor suppression properties in cancer. However, there is no evidence whether miR-675 is involved in breast cancer. The objective of this study was to evaluate the expression status of miR-675 and its clinical relevance in breast cancer patients. Methods: The expression level of miR-675 was detected in 100 breast cancer patients and 38 cancer-free controls using real-time quantitative PCR. The clinicopathological characteristics of miR-675 in breast cancer were also investigated. All statistical analyses were performed using SPSS 20.0. Results: The study showed that miR-675 was significantly up-regulated in breast cancer patients compared with controls (P < 0.01). There was no significant difference in age, lymph nodes stage, ER status and PR status between patients with and without miR-675 over-expression (P > 0.05). The frequency of miR-675 over-expression was higher in the patients of histological grade I-II than in others (50% versus 9%, P = 0.011). The expression level of miR-675 had a high correlation with miR-24/93/98/378 in breast cancer patients. Conclusions: Taken together, our study demonstrated that miR-675 in formalin-fixed paraffin-embedded (FFPE) tissues might serve as a good source for biomarker discovery and breast cancer validation.
Keywords: Biomarkers, miR-675, breast cancer, formalin-fixed paraffin-embedded
Introduction
MiRNAs are a class of eukaryotic endogenous small non-coding RNAs that are single-stranded with a length of about 18~25 nt [1]. More than 1000 miRNAs have been identified in the human genome. MiRNAs can regulate the expression of target gene mainly through binding complementary sequences within the 3’-untranslated region (UTR) of its mRNAs, resulting in translational repression or mRNA degradation [2]. MiRNAs are involved in different biologic processes including tumor growth [3], tumor development [4], cell proliferation [5], cell differentiation [6], cell survival [3], tumor migration and invasion [7,8], and apoptosis [9,10].
Dysregulation of miR-675 has been found in a variety of solid tumors. Some studies reported that miR-675 was up-regulated in colorectal cancer [11], hepatocellular cancer [12] and gastric cancer [13], through the regulation of different targets such as Cadherin11 [14], Twist1 (twist basic helix-loop-helix transcription factor 1) [12], RUNX1 (Runt Domain Transcription Factor 1) [15]. While other studies found that miR-675 expression was down-regulated in non-small cell lung cancer [16], adrenal cortical carcinoma [17] and metastatic prostate cancer [18]. It suggested that the expression of miR-675 and its role in different tumors were different. However, limited knowledge is available concerning the role of miR-675 in breast cancer and its effect on the biological characteristics.
Fresh and frozen tissue samples are found to be a suitable source of DNA, RNA and protein for clinical experiment research [19]. However, because of the limitation and ineffectiveness of prospectively collecting fresh and frozen samples as well as current deficiency of bio-banking of patient samples, scientists began to choose alternative stocks of archived samples with potential utility for disease analysis such as formalin-fixed paraffin-embedded (FFPE) tissues [20]. RNA was found to be degraded in FFPE tissue [21]. Surprisingly, due to their shorter lengths, miRNAs remained to be more stable when preserved. Using FFPE tissues, the expression of miRNAs has been successfully analyzed in various kinds of cancers [22]. There is a high association between the miRNA expression level derived from FFPE and matched fresh-frozen tissues [20]. As they carry abundant pathological data, FFPE tissues are found to be ideal for miRNA study and immunohistochemistry. Thereby FFPE tissues may serve as a valuable source for miRNA expression study, biomarker discovery and validation.
More and more evidence has demonstrated that miR-675 is aberrantly expressed in various cancers and may serve as a potential biomarker for cancer detection. But little is known about the expression level of miR-675 in FFPE tissue of breast cancer patients and its potential prognostic value. In this study, we examined the expression of miR-675 in FFPE tissue of breast cancer patients as well as the correlation between miR-675 and miR-24/93/98/378. Subsequently, we investigated potential predictive value of miR-675 as a kind of new molecular biomarker for breast cancer patients.
Methods
Patients and samples
100 FFPE tissue samples of breast cancer patients were collected from the Affiliated People’s Hospital of Jiangsu University in present study. Moreover, FFPE tissue samples were obtained from 38 cancer-free people as controls. All samples were acquired according to the guidelines of The Affiliated People’s Hospital’s protocol including patient consent and specimen collection. The diagnosis and classification of breast cancer patients was according to the Tumor-Node-Metastasis (TNM) system of American Joint Committee on Cancer (AJCC). All cases were diagnosed with histologically and clinically confirmed stage I-II, II, II-III and III breast cancer. The clinical characteristics of patients were listed in Table 1.
Table 1.
Clinicopathological characteristics and expression of miR-675 in breast cancer
| Characteristics | Cases | miR-675 expression level | ||
|---|---|---|---|---|
|
|
||||
| Low | High | P | ||
| Age (years) | ||||
| ≤ 40 | 19 | 18 | 1 | 0.362 |
| 40-55 | 32 | 26 | 6 | |
| > 55 | 49 | 44 | 5 | |
| Histological grade | ||||
| I-II | 8 | 4 | 4 | 0.011 |
| II | 39 | 36 | 3 | |
| II-III | 28 | 26 | 2 | |
| III | 25 | 22 | 3 | |
| N stage | ||||
| 0 | 58 | 53 | 5 | 0.482 |
| 1-3 | 24 | 20 | 4 | |
| 4-9 | 10 | 8 | 2 | |
| > 10 | 8 | 7 | 1 | |
| ER status | ||||
| Negative | 41 | 36 | 5 | 1 |
| Positive | 59 | 52 | 7 | |
| PR status | ||||
| Negative | 44 | 38 | 6 | 0.760 |
| Positive | 56 | 50 | 6 | |
RNA extraction and reverse transcription
Total RNA was isolated from 100 FFPE tissue of breast cancer patients and 38 controls using the RecoverAll™ Total Nucleic Acid Isolation Kit (Ambion, catalog no: AM 1975) and reverse transcribed to cDNA using miScript Reverse Transcription Kit (Qiagen, catalog no. 218061).
Real-time quantitative PCR
Real-time quantitative PCR (RQ-PCR) was performed based on the manufacturer’s instructions using the miScript SYBR green PCR kit (Qiagen, catalog no. 218073) with the manufacturer-provided miScript universal primer and miR-675-specific forward primer: GCGGAGAGGGCCCACAG; RQ-PCR amplification was described in our previous study (Qian et al., 2013).
Statistical analyses
All statistical analyses were performed using SPSS 20.0 software. The difference of miRNA expression levels between patients and controls was compared using the Mann-Whitney test. Spearman correlation coefficient was used to analyze the correlation between miR-675 and miR-24, miR-93, miR-98 or miR-378 expression, respectively. The Pearson χ2 analysis or Fisher exact test was employed to analyze the correlation of miR-675 expression with clinicopathological characteristics. More than 0.54 (determined as the mean plus 3SD) was chosen to define the miR-675 over-expression in breast cancer samples. All values shown were two sided, and a P-value < 0.05 was considered statistically significant.
Results
Expression and correlation of miR-675 in FFPE tissues of breast cancer patients
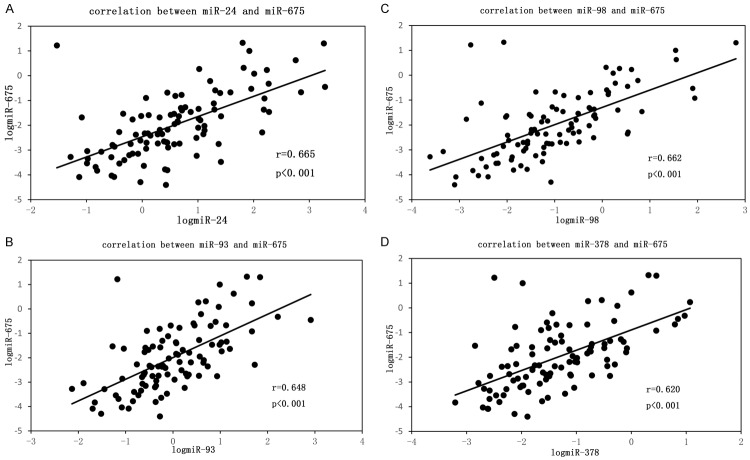
We evaluated the level of miR-675 expression in breast cancer and controls. The expression of miR-675 was found to be higher in FFPE tissues of breast cancer patients than that in controls (P < 0.01) (Figure 1; Table 2). The median of the miR-675 expression level was 0.0073 in breast cancer patients and 0.0021 in controls respectively. Furthermore, correlation analysis showed that the expression level of miR-675 had a high correlation with miR-24/93/98/378 in breast cancer patients; the Spearman correlation scatter plot indicated a high association between miR-675 and miR-24/93/98/378, with r = 0.665 between miR-675 and miR-24 (Figure 2A), r = 0.648 between miR-675 and miR-93 (Figure 2B), r = 0.662 between miR-675 and miR-98 (Figure 2C), and r = 0.620 between miR-675 and miR-378 (Figure 2D). These findings imply that the selected miR-675 might be as a biomarker for breast cancer.
Figure 1.
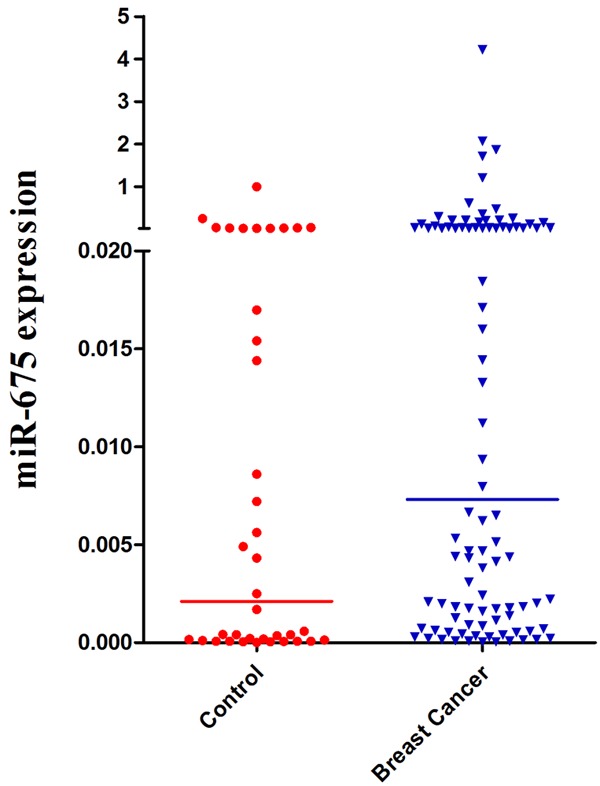
Relative expression levels of miR-675 in breast cancer patients and controls.
Table 2.
Expression level of miR-675 and its diagnostic significance in breast cancer patients
| Median in cancer patients | Median in control | P value | AUC | Cutoffs | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|---|---|---|
| miR-675 | 0.0073 | 0.0021 | < 0.01 | 0.664 | 0.536 | 55 | 58 |
Figure 2.
Correlation between miR-675 and miR-24/93/98/378, the spearman correlation scatter plot indicated association between miR-675 and miR-24/93/98/378, with r = 0.665 between miR-675 and miR-24 (A), r = 0.648 between miR-675 and miR-93 (B), r = 0.662 between miR-675 and miR-98 (C), r = 0.620 between miR-675 and miR-378 (D). miRNA expression levels were presented after log10 transformation.
Correlation of miR-675 expression with clinicopathological characteristics of breast cancer
To investigate whether the up-regulation of miR-675 was related to the clinical progression of breast cancer, we analyzed the correlation between the expression level of miR-675 and the clinicopathological status of breast cancer patients (Table 1). No significant difference could be observed in age, tumor stage, lymph nodes stage, ER (estrogen receptor) status, and PR (progesterone receptor) status between patients with and without miR-675 over-expression (Table 1). However, a statistically significant association between miR-675 expression level and histological grade was observed in this study. The frequency of miR-675 over-expression was higher in the patients of histological grade I-II than in others (50% versus 9%, P = 0.011).
Discussion
Abnormal expression of miRNAs has been revealed in various tumor tissues, dysregulation of miRNAs are found to play an important role in the occurrence and development of different cancers [23-26]. Breast cancer is one of the most malignant tumors in women, which seriously affect human health and even can be fetal. It is suggested to be a heterogeneous neoplasm, involving a variety of profile alterations in both expression of miRNA and mRNA [27]. Large number of studies about the aberrant expression status of various miRNAs and their functions in breast cancer have been reported [27-29]. Over-expression of miR-21 and under-expression of miR-125b have been found in breast cancer patients [30]. Sun Y et al. revealed that serum miR-155 was up-regulated in breast cancer patients [31]. Recent studies demonstrated that miRNAs may serve as ideal potential candidates for the development of therapeutic targets and novel biomarkers.
This study measured and statistically analyzed the expression level of miR-675 in FFPE tissues of 100 breast cancer patients and 38 cases in control. We found that miR-675 showed higher expression level in breast cancer patients when compared with controls (P < 0.01), which might imply that miR-675 might be an onco-miRNA in breast cancer. MiR-24, miR-93, miR-98 and miR-378 are all thought to be onco-miRNAs for their capabilities of enhancing tumor growth [3,26,32,33]. Our previous study found that miR-24/93/98/378 had a high expression in FFPE tissue of breast cancers and the four miRNAs were highly correlated with each other in breast cancer patients (data not shown). In this study, we have revealed that miR-675 is associated with miR-24/93/98/378, respectively. The miR-675 might be thought as a good potential candidate for the development of novel biomarker in breast cancer.
In addition, the relativity between the expression of miR-675 and clinicopathological characteristics of breast cancer patients were also studied. However, there were no significant differences between miR-675 over-expression with clinicopathological characteristics except for histological grade. No significant difference was observed in age, tumor stage, lymph nodes stage, ER status and PR status between patients with and without miR-675 over-expression (P > 0.05, Table 1). The frequency of miR-675 over-expression was dramatically higher in the patients of histological grade I-II than in others (50% versus 9%, P = 0.011), which meant that miR-675 over-expression was much more common in lower histological grade than higher grade. It might implicate that high miR-675 expression can be associated with high degree of malignancy in breast cancer, which are in need of further investigation. According to our observations and analyses of miR-675 expression status and clinical characteristics, the lack of correlation between miR-675 and clinicopathological characteristics might imply that the potential mechanisms of miR-675 over-expression is different from that of other clinic factors in breast cancer, which makes them potential candidates for the diagnosis and prognosis of breast cancer that are independent from other known biomarkers.
FFPE tissues could be long-term stored and they can permanently preserve the tissue structure. MiRNAs were proved to be more stable in FFPE tissues when preserved due to their shorter length. The biomarkers derived from FFPE tissues can provide efficient biological insight because they are linked to pathological and clinical databases. Recent studies revealed that miRNAs obtained from FFPE tissues showed reliable expression levels when compared with frozen tissues [34-36]. Taken together, our study demonstrated that miR-675 in formalin-fixed paraffin-embedded (FFPE) tissues might serve as a good source for biomarker discovery and validation in breast cancer patients.
Conclusions
In conclusion, we suppose that over-expression of miR-675 in FFPE tissues could be a reference for the diagnosis of breast cancer patients, which might conduct as a valuable source for biomarker discovery and breast cancer validation.
Acknowledgements
This study was supported by National Natural Science foundation of China (81172592, 81270630), Science and Technology Special Project in Clinical Medicine of Jiangsu Province (BL2012056), 333 Project of Jiangsu Province (BRA2011085).
Disclosure of conflict of interest
None.
References
- 1.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 2.Seitz H, Youngson N, Lin SP, Dalbert S, Paulsen M, Bachellerie JP, Ferguson-Smith AC, Cavaillé J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet. 2003;34:261–2. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 3.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A. 2007;104:20350–5. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye G, Fu G, Cui S, Zhao S, Bernaudo S, Bai Y, Ding Y, Zhang Y, Yang BB, Peng C. MicroRNA 376c enhances ovarian cancer cell survival by targeting activin receptor-like kinase 7: implications for chemoresistance. J Cell Sci. 2011;124:359–68. doi: 10.1242/jcs.072223. [DOI] [PubMed] [Google Scholar]
- 5.Shatseva T, Lee DY, Deng Z, Yang BB. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J Cell Sci. 2011;124:2826–36. doi: 10.1242/jcs.077529. [DOI] [PubMed] [Google Scholar]
- 6.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–5. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 7.Guo H, Chen Y, Hu X, Qian G, Ge S, Zhang J. The regulation of Toll-like receptor 2 by miR-143 suppresses the invasion and migration of a subset of human colorectal carcinoma cells. Mol Cancer. 2013;12:77. doi: 10.1186/1476-4598-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lv J, Ma L, Chen XL, Huang XH, Wang Q. Downregulation of LncRNAH19 and MiR-675 promotes migration and invasion of human hepatocellular carcinoma cells through AKT/GSK-3beta/Cdc25A signaling pathway. J Huazhong Univ Sci Technolg Med Sci. 2014;34:363–9. doi: 10.1007/s11596-014-1284-2. [DOI] [PubMed] [Google Scholar]
- 9.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 10.Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120:3045–52. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 11.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–8. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez JM, Elahi A, Clark CW, Wang J, Humphries LA, Centeno B, Bloom G, Fuchs BC, Yeatman T, Shibata D. miR-675 mediates downregulation of Twist1 and Rb in AFP-secreting hepatocellular carcinoma. Ann Surg Oncol. 2013;20(Suppl 3):S625–35. doi: 10.1245/s10434-013-3106-3. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–29. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim NH, Choi SH, Lee TR, Lee CH, Lee AY. Cadherin 11, a miR-675 target, induces N-cadherin expression and epithelial-mesenchymal transition in melasma. J Invest Dermatol. 2014;134:2967–76. doi: 10.1038/jid.2014.257. [DOI] [PubMed] [Google Scholar]
- 15.Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315–22. doi: 10.1016/j.bbrc.2013.12.126. [DOI] [PubMed] [Google Scholar]
- 16.He D, Wang J, Zhang C, Shan B, Deng X, Li B, Zhou Y, Chen W, Hong J, Gao Y, Chen Z, Duan C. Down-regulation of miR-675-5p contributes to tumor progression and development by targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol Cancer. 2015;14:73. doi: 10.1186/s12943-015-0342-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitz KJ, Helwig J, Bertram S, Sheu SY, Suttorp AC, Seggewiss J, Willscher E, Walz MK, Worm K, Schmid KW. Differential expression of microRNA-675, microRNA-139-3p and microRNA-335 in benign and malignant adrenocortical tumours. J Clin Pathol. 2011;64:529–35. doi: 10.1136/jcp.2010.085621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, Yu J. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–75. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 19.Kokkat TJ, Patel MS, McGarvey D, LiVolsi VA, Baloch ZW. Archived formalin-fixed paraffin-embedded (FFPE) blocks: A valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv Biobank. 2013;11:101–6. doi: 10.1089/bio.2012.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goswami RS, Waldron L, Machado J, Cervigne NK, Xu W, Reis PP, Bailey DJ, Jurisica I, Crump MR, Kamel-Reid S. Optimization and analysis of a quantitative real-time PCR-based technique to determine microRNA expression in formalin-fixed paraffin-embedded samples. BMC Biotechnol. 2010;10:47. doi: 10.1186/1472-6750-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macabeo-Ong M, Ginzinger DG, Dekker N, McMillan A, Regezi JA, Wong DT, Jordan RC. Effect of duration of fixation on quantitative reverse transcription polymerase chain reaction analyses. Mod Pathol. 2002;15:979–87. doi: 10.1097/01.MP.0000026054.62220.FC. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Lu Y, Chen Y, Lu W, Xie X. MicroRNA profile of paclitaxel-resistant serous ovarian carcinoma based on formalin-fixed paraffin-embedded samples. BMC Cancer. 2013;13:216. doi: 10.1186/1471-2407-13-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takakura S, Mitsutake N, Nakashima M, Namba H, Saenko VA, Rogounovitch TI, Nakazawa Y, Hayashi T, Ohtsuru A, Yamashita S. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008;99:1147–54. doi: 10.1111/j.1349-7006.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79:313–20. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 25.Baumjohann D, Kageyama R, Clingan JM, Morar MM, Patel S, de Kouchkovsky D, Bannard O, Bluestone JA, Matloubian M, Ansel KM, Jeker LT. The microRNA cluster miR-17 approximately 92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat Immunol. 2013;14:840–8. doi: 10.1038/ni.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang L, Deng Z, Shatseva T, Yang J, Peng C, Du WW, Yee AJ, Ang LC, He C, Shan SW, Yang BB. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-beta8. Oncogene. 2011;30:806–21. doi: 10.1038/onc.2010.465. [DOI] [PubMed] [Google Scholar]
- 27.Fu SW, Chen L, Man YG. miRNA Biomarkers in Breast Cancer Detection and Management. J Cancer. 2011;2:116–22. doi: 10.7150/jca.2.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farazi TA, Horlings HM, Ten Hoeve JJ, Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F, van Kouwenhove M, Kreike B, Sie D, Hovestadt V, Wessels LF, van de Vijver MJ, Tuschl T. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443–53. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu C, Li J, Wang X, Song L. Unregulated miR-96 induces cell proliferation in human breast cancer by downregulating transcriptional factor FOXO3a. PLoS One. 2010;5:e15797. doi: 10.1371/journal.pone.0015797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mar-Aguilar F, Luna-Aguirre CM, Moreno-Rocha JC, Araiza-Chavez J, Trevino V, Rodriguez-Padilla C, Reséndez-Pérez D. Differential expression of miR-21, miR-125b and miR-191 in breast cancer tissue. Asia Pac J Clin Oncol. 2013;9:53–9. doi: 10.1111/j.1743-7563.2012.01548.x. [DOI] [PubMed] [Google Scholar]
- 31.Sun Y, Wang M, Lin G, Sun S, Li X, Qi J, Li J. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS One. 2012;7:e47003. doi: 10.1371/journal.pone.0047003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du WW, Fang L, Li M, Yang X, Liang Y, Peng C, Qian W, O’Malley YQ, Askeland RW, Sugg SL, Qian J, Lin J, Jiang Z, Yee AJ, Sefton M, Deng Z, Shan SW, Wang CH, Yang BB. MicroRNA miR-24 enhances tumor invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling. J Cell Sci. 2013;126:1440–53. doi: 10.1242/jcs.118299. [DOI] [PubMed] [Google Scholar]
- 33.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. Rna. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin JY, Deng ZQ, Liu FQ, Qian J, Lin J, Tang Q, Wen XM, Zhou JD, Zhang YY, Zhu XW. Association between mir-24 and mir-378 in formalin-fixed paraffin-embedded tissues of breast cancer. Int J Clin Exp Pathol. 2014;7:4261–7. [PMC free article] [PubMed] [Google Scholar]
- 35.Deng ZQ, Qian J, Liu FQ, Lin J, Shao R, Yin JY, Tang Q, Zhang M, He L. Expression level of miR-93 in formalin-fixed paraffin-embedded tissues of breast cancer patients. Genet Test Mol Biomarkers. 2014;18:366–70. doi: 10.1089/gtmb.2013.0440. [DOI] [PubMed] [Google Scholar]
- 36.Deng ZQ, Yin JY, Tang Q, Liu FQ, Qian J, Lin J, Shao R, Zhang M, He L. Over-expression of miR-98 in FFPE tissues might serve as a valuable source for biomarker discovery in breast cancer patients. Int J Clin Exp Pathol. 2014;7:1166–71. [PMC free article] [PubMed] [Google Scholar]