Abstract
Increasing studies have demonstrated that altered expression of histone deacetylases (HDACs) plays a critical role in the tumorigenesis through up-regulation or down-regulation of key genes involved in cell proliferation, cell-cycle regulation and apoptosis. In the present study, the expression and function of HDAC9 were investigated in osteosarcoma. Quantitative real-time PCR and Western blot analysis found that HDAC9 was up-regulated in osteosarcoma tissues, when compared with that in adjacent normal tissues. In vitro studies further demonstrated that overexpression of HDAC9 in U2OS and MG63 cells promoted cell proliferation and invasion. Using chromatin immunoprecipitation (ChIP) assay, we found that HDAC9 epigenetically repressed p53 transcription through binding to its proximal promoter region. Therefore, our data suggest an important role for HDAC9/p53 regulatory pathway in the osteosarcoma progression.
Keywords: Osteosarcoma, cell proliferation, HDAC9, p53, epigenetics
Introduction
Osteosarcoma (OS) is the most common type of primary malignant bone tumor in adolescents and young adults [1,2]. Although extensive advancements attempt to understand its initiation and progression [3-5], search for its novel markers might provide potential therapeutic targets for cancer treatment.
It has been well-documented that the balance of the activities of histone acetyltransferase (HAT) and histone deacetylase (HDAC) plays a crucial role in the regulation of gene transcription [6-8]. Remarkably, the functions of HDACs in cancer attract recent attention. Firstly, there are a number of studies showing altered expression of HDACs in tumor tissues [9,10]. Besides, HDACs could form a corepressor complex with other transcription factors and bind to the promoter region of tumor suppressor gene to repress its expression [11,12]. Moreover, HDACs were shown to interact with and deacetylate non-histone proteins, such as p53, to regulate the stability and transcriptional activity of these proteins [13,14].
The expression and roles of HDAC9 has been revealed in several types of human cancers by previous reports. For instance, differential expression of HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia [15]. Besides, HDAC9 issignificantly upregulated in high-risk medulloblastoma in comparison with low-risk medulloblastoma, and its expression is associated with poor survival [16]. Moreover, SNP rs10248565 in HDAC9 has been identified as a novel genomic aberration biomarker of lung adenocarcinoma in non-smoking women [17]. However, biological function of HDAC9 in osteosarcoma remains poorly understood.
In the present study, we aim to investigate the possible oncogenic role of HDAC9 in osteosarcoma.
Materials and methods
Human tissue samples
30 parried of osteosarcoma tissues and adjacent non-tumor normal specimens were collected from routine therapeutic surgery at our department. Informed consent was obtained from each patient before surgery. The study protocol was reviewed and approved by the Institutional Review Board of Zhengzhou University Affiliated Cancer Hospital.
Cell culture, transfection and luciferase assays
Osteosarcoma cell lines (U2OS and MG63 cells) were obtained from The Cell Bank of Type Culture Collection of Chinese Academy of Sciences (CAS, Shanghai). Cells were grown in Dulbecco’s modified Eagle’s medium (Invitrogen, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Invitrogen) and maintained at 37°C in a humidified atmosphere with 5% CO2. All transfections were performed using Lipofectamine 2000 reagents (Invitrogen) according to the manufacturer’s instructions. For luciferase assays, cells were seeded in 24-well plates and transfection efficiency was normalized by co-transfecting Simian virus 40 (SV40) plasmids (Promega). Luciferase values were measured using the Dual-Luciferase Reporter Assay System (Promega).
BrdU incorporation and cell invasion assays
A cell proliferation enzyme-linked immunosorbent assay (BrdU kit; Beyotime) was used to analyze the incorporation of BrdU during DNA synthesis following the manufacturer’s protocols. Absorbance was measured at 450 nm in the Spectra Max 190 ELISA reader (Molecular Devices, Sunnyvale, CA). Invasion assays were conducted using a specialized Chemicon invasion chamber which included a 24-well tissue culture plate with 12 cell culture inserts (Millipore, Bedford, MA, USA).
Real-time PCR analysis
Total RNA from tissues and cells was extracted using the TRIzol Kit (Invitrogen) according to the manufacturer’s instructions. Quantitative real-time PCR was performed using SYBR Premix Ex Taq reagents (Takara, Shiga, Japan). Relative expression levels of HDAC9 were calculated using the 2-ΔΔCt method with U6 as the endogenous reference gene.
Western blot
Cells and tissues were harvested and lysed with ice-cold lysis buffer (50 mM Tris-HCl, pH 7.4, 100 mM 2-Mercaptoethanol, 2% w/v SDS, 10% glycerol). After centrifugation at 10,000× g for 10 min at 4°C, proteins in the supernatants were quantified and separated by 10% SDS PAGE. Immunoblots were performed using primary antibodies targeting HDAC9 and P53 (Abcam, Cambridge, Massachusetts, USA). Protein levels were normalized to total GAPDH, using a rabbit anti-GAPDH antibody (Abcam). The proteins were visualized by an ECL chemiluminescence detection kit (Amersham Biosciences, Buckinghamshire, UK).
Statistical analysis
Data were expressed as mean ± standard error of the mean (SE). Analysis was conducted with GraphPad Prism version 6.01 (GraphPad Software). Significance between two groups was analyzed using the unpaired two-tailed t test (*P<0.05, **P<0.01, ***P<0.001).
Results
Up-regulation of HDAC9 in osteosarcoma tissues
To investigate the expression levels of HDAC9 in osteosarcoma tissues, quantitative real-time PCR and western blot analysis were performed. As shown in the Figure 1A and 1B, message RNA (mRNA) and protein levels of HDAC9 was significantly up-regulated in in osteosarcoma tissues, compared with adjacent normal tissues (Figure 1A, 1B).
Figure 1.
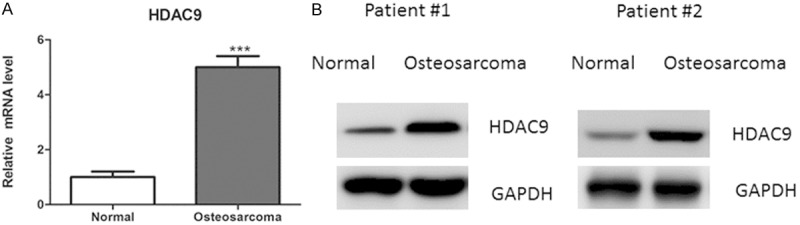
HDAC9 expression in osteosarcoma and normal tissues. A. Relative expression levels of HDAC9 were determined by quantitative real-time PCR analysis in 30 primary osteosarcoma and adjacent normal tissues. B. Representative protein levels of HDAC9 were determined by western blot in osteosarcoma and normal tissues.
HDAC9 promotes osteosarcoma cell proliferation and invasion
Next, to determine the biological role of HDAC9 on osteosarcoma growth, adenovirus containing HDAC9 or empty vector was transfected into U2OS and MG63 cells (Figure 2A, 2B). As expected, forced overexpression of HDAC9 enhanced cell proliferation and invasion abilities (Figure 2C-F). On the other hand, knockdown of HDAC9 using small interfering RNA (siRNA) led to reduced abilities of cell proliferation and invasion in U2OS and MG63 cells (Figure 3A-F).
Figure 2.
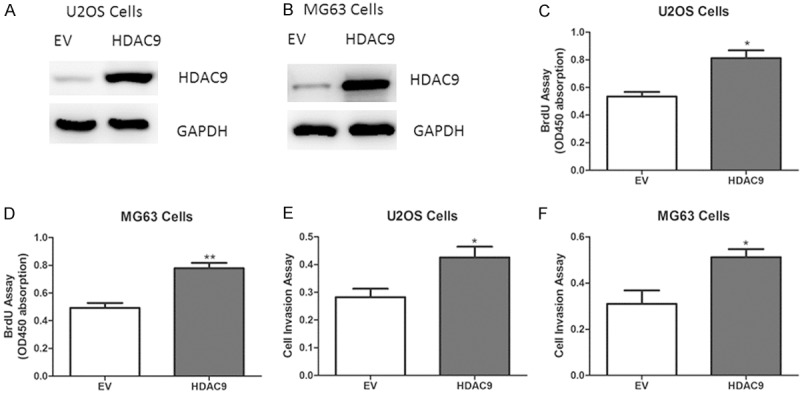
Effect of HDAC9 overexpression on osteosarcoma growth in vitro. (A, B) Representative protein levels of HDAC9 in U2OS and MG63 cells transfected with adenovirus containing HDAC9 or empty vector (EV). (C-F) Cell proliferation (C, D) and invasion (E, F) assays in U2OS and MG63 cells expressing HDAC9 or EV.
Figure 3.
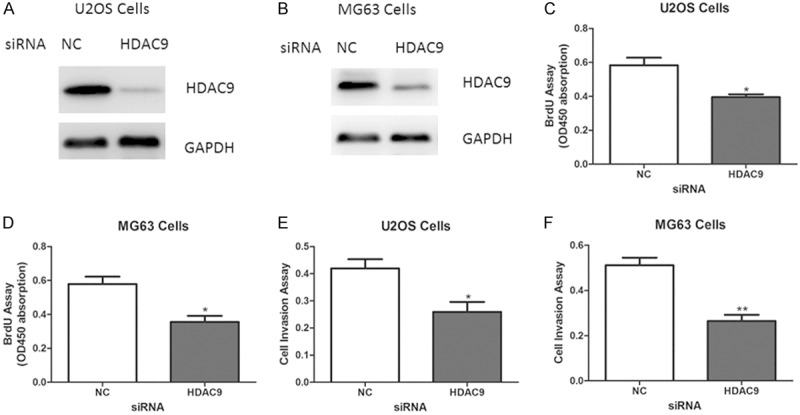
Suppression of HDAC9 inhibits osteosarcoma cell proliferation and invasion. (A, B) Representative protein levels of HDAC9 in U2OS and MG63 cells transfected with siRNA oligos targeting HDAC9 or negative control (NC). (C-F) Cell proliferation (C, D) and invasion (E, F) assays in U2OS and MG63 cells with HDAC9 deficiency.
HDAC9 inhibits p53 transcription
To understand the molecular mechanisms underlying HDAC9-mediated cell proliferation, quantitative real-time PCR analysis of proliferation-related genes was performed. As shown in the Figure 4A and 4B, HDAC9 overexpression resulted in a reduced expression of p53 (Figure 4A, 4B). The down-regulation of p53 was also confirmed by western blot (Figure 4C, 4D). In contrast, knockdown of HDAC9 did the opposite (Figure 4E-H).
Figure 4.
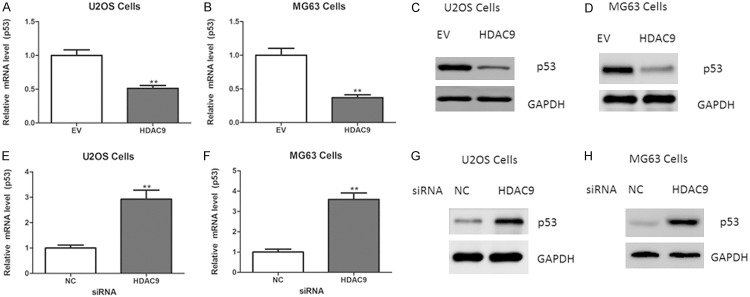
HDAC9 represses p53 expression in osteosarcoma cells. A, B. Relative mRNA levels of p53 in U2OS and MG63 cells transfected with adenovirus containing HDAC9 or empty vector (EV). C, D. Protein levels of p53 in U2OS and MG63 cells transfected with adenovirus containing HDAC9 or empty vector (EV). E, F. Relative mRNA levels of p53 in U2OS and MG63 cells transfected with siRNA oligos targeting HDAC9 or negative control (NC). G, H. Protein levels of p53 in U2OS and MG63 cells transfected with siRNA oligos targeting HDAC9 or negative control (NC).
HDAC9 regulates p53 transcription
We further performed chromatin immunoprecipitation (ChIP) assays using three distinct pairs of primers for the p53 promoter loci (Figure 5A). As expected, the ChIP assays clearly showed that HDAC9 bound to the p53 promoter, and HDAC9 knockdown notably reduced binding of HDAC9 at p53 loci (Figure 5A). Moreover, ablation of HDAC9 increased the acetylated histone H3 at p53 loci (Figure 5B), suggesting that HDAC9 inhibits p53 transcription through regulation of histone acetylation state.
Figure 5.
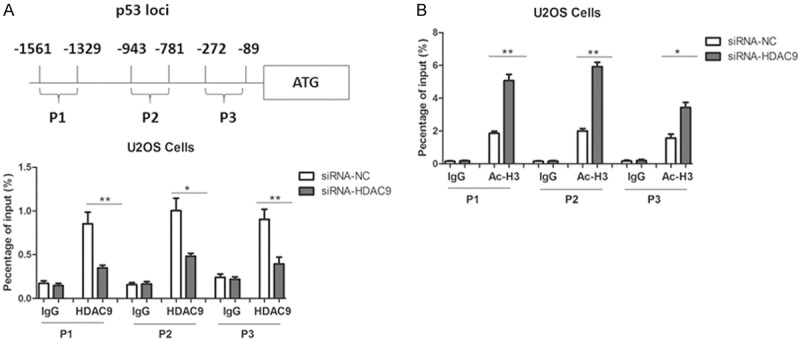
Role of HDAC9 in p53 transcription. A. A schematic representation of the human p53 and primers used for ChIP assays. ChIP assays were performed using the antibody against HDAC9 in U2OS cells. The IgG served as a negative control for ChIP assays. B. ChIP assays using antibodies against acetylated histone H3 (Ac-H3) were performed in U2OS cells.
Discussion
In this study, we identified for the first time the oncogenic role of HDAC9 in osteosarcoma progression. This is supported by multiple lines of evidence. Firstly, HDAC9 expression was increased in osteosarcoma tissues, compared with adjacent normal tissues. Secondly, ectopic overexpression of HDAC9 promoted, whereas its deficiency inhibited cell proliferation and invasion. Furthermore, at the molecular level, we identify p53 as a direct target of HDAC9. However, in vivo studies, such as HDAC9 knockout mice, are still needed to further establish the precise roles of HDAC9 in osteosarcoma development. Moreover, the molecular determinants for the up-regulation of HDAC9 in osteosarcoma remain to be determined.
It has been shown that HDAC1 binds MDM2 in a p53-independent manner and deacetylates p53 at all known acetylated lysines in vivo [18]. Besides, HDAC2 modulates p53 transcriptional activities through regulation of p53-DNA binding activity [14,19]. Moreover, HDAC6 is up-regulated in hepatocellular carcinoma tissues and promotes cell proliferation through promotion of p53 degradation [20]. Therefore, HDACs could regulate the biological function of p53 at the transcriptional and post-transcriptional level.
Taken together, our data highlight an important role of HDAC9 in the regulation of p53 transcription. Given that several preclinical studies have demonstrated that small-molecule HDAC inhibitors could serve as a new class of mechanism-based anti-cancer agent [21-24], our study might provide novel insight for the development of new strategies for cancer treatment.
Disclosure of conflict of interest
None.
References
- 1.Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14:722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 2.Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92. doi: 10.1007/978-3-319-07323-1_4. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Yu XH, Yan YG, Wang C, Wang WJ. PI3K/Akt signaling in osteosarcoma. Clin Chimi Acta. 2015;444:182–192. doi: 10.1016/j.cca.2014.12.041. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Yan YG, Wang C, Zhang SJ, Yu XH, Wang WJ. MicroRNAs in osteosarcoma. Clin Chim Acta. 2015;444:9–17. doi: 10.1016/j.cca.2015.01.025. [DOI] [PubMed] [Google Scholar]
- 5.He JP, Hao Y, Wang XL, Yang XJ, Shao JF, Guo FJ, Feng JX. Review of the molecular pathogenesis of osteosarcoma. Asian Pac J Cancer Prev. 2014;15:5967–5976. doi: 10.7314/apjcp.2014.15.15.5967. [DOI] [PubMed] [Google Scholar]
- 6.Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nature reviews Drug discovery. 2013;12:917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 9.Gong F, Miller KM. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutat Res. 2013;750:23–30. doi: 10.1016/j.mrfmmm.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Tang J, Yan H, Zhuang S. Histone deacetylases as targets for treatment of multiple diseases. Clin Sci. 2013;124:651–662. doi: 10.1042/CS20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoang JJ, Baron S, Volle DH, Lobaccaro JM, Trousson A. Lipids, LXRs and prostate cancer: are HDACs a new link? Biochem Pharmacol. 2013;86:168–174. doi: 10.1016/j.bcp.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Matsuda KI, Mori H, Kawata M. Epigenetic mechanisms are involved in sexual differentiation of the brain. Rev Endocr Metab Disord. 2012;13:163–171. doi: 10.1007/s11154-012-9202-z. [DOI] [PubMed] [Google Scholar]
- 13.Choi S, Reddy P. HDAC inhibition and graft versus host disease. Mol Med. 2011;17:404–416. doi: 10.2119/molmed.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner T, Brand P, Heinzel T, Kramer OH. Histone deacetylase 2 controls p53 and is a critical factor in tumorigenesis. Biochim Biophys Acta. 2014;1846:524–538. doi: 10.1016/j.bbcan.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Moreno DA, Scrideli CA, Cortez MA, de Paula Queiroz R, Valera ET, da Silva Silveira V, Yunes JA, Brandalise SR, Tone LG. Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. Br J Haematol. 2010;150:665–673. doi: 10.1111/j.1365-2141.2010.08301.x. [DOI] [PubMed] [Google Scholar]
- 16.Milde T, Oehme I, Korshunov A, Kopp-Schneider A, Remke M, Northcott P, Deubzer HE, Lodrini M, Taylor MD, von Deimling A, Pfister S, Witt O. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res. 2010;16:3240–3252. doi: 10.1158/1078-0432.CCR-10-0395. [DOI] [PubMed] [Google Scholar]
- 17.Lai LC, Tsai MH, Chen PC, Chen LH, Hsiao JH, Chen SK, Lu TP, Lee JM, Hsu CP, Hsiao CK, Chuang EY. SNP rs10248565 in HDAC9 as a novel genomic aberration biomarker of lung adenocarcinoma in non-smoking women. J Biomed Sci. 2014;21:24. doi: 10.1186/1423-0127-21-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arora A, Gera S, Maheshwari T, Raghav D, Alam MJ, Singh RK, Agarwal SM. The dynamics of stress p53-Mdm2 network regulated by p300 and HDAC1. PLoS One. 2013;8:e52736. doi: 10.1371/journal.pone.0052736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shahbazi J, Scarlett CJ, Norris MD, Liu B, Haber M, Tee AE, Carrier A, Biankin AV, London WB, Marshall GM, Lock RB, Liu T. Histone deacetylase 2 and N-Myc reduce p53 protein phosphorylation at serine 46 by repressing gene transcription of tumor protein 53-induced nuclear protein 1. Oncotarget. 2014;5:4257–4268. doi: 10.18632/oncotarget.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding G, Liu HD, Huang Q, Liang HX, Ding ZH, Liao ZJ, Huang G. HDAC6 promotes hepatocellular carcinoma progression by inhibiting P53 transcriptional activity. FEBS letters. 2013;587:880–886. doi: 10.1016/j.febslet.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Mottamal M, Zheng S, Huang TL, Wang G. Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules. 2015;20:3898–3941. doi: 10.3390/molecules20033898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song SH, Han SW, Bang YJ. Epigenetic-based therapies in cancer: progress to date. Drugs. 2011;71:2391–2403. doi: 10.2165/11596690-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 23.Huffman K, Martinez ED. Pre-clinical studies of epigenetic therapies targeting histone modifiers in lung cancer. Front Oncol. 2013;3:235. doi: 10.3389/fonc.2013.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu T, Zhou L, Zhu W, Wang T, Wang J, Shu Y, Liu P. Effects of treatment with histone deacetylase inhibitors in solid tumors: a review based on 30 clinical trials. Future Oncol. 2013;9:255–269. doi: 10.2217/fon.12.173. [DOI] [PubMed] [Google Scholar]


