Summary
Chemical carcinogenesis is caused mainly by the modulation of important cellular mechanisms and pathways that regulate cell death. Mechanisms of several environmental chemicals that disrupt these important targets conferring resistance to cell death have been discussed.
Abstract
Cell death is a process of dying within biological cells that are ceasing to function. This process is essential in regulating organism development, tissue homeostasis, and to eliminate cells in the body that are irreparably damaged. In general, dysfunction in normal cellular death is tightly linked to cancer progression. Specifically, the up-regulation of pro-survival factors, including oncogenic factors and antiapoptotic signaling pathways, and the down-regulation of pro-apoptotic factors, including tumor suppressive factors, confers resistance to cell death in tumor cells, which supports the emergence of a fully immortalized cellular phenotype. This review considers the potential relevance of ubiquitous environmental chemical exposures that have been shown to disrupt key pathways and mechanisms associated with this sort of dysfunction. Specifically, bisphenol A, chlorothalonil, dibutyl phthalate, dichlorvos, lindane, linuron, methoxychlor and oxyfluorfen are discussed as prototypical chemical disruptors; as their effects relate to resistance to cell death, as constituents within environmental mixtures and as potential contributors to environmental carcinogenesis.
Introduction
Cancer death is one of the major causes of mortality worldwide. According to the World Health Organization, there were ~32.6 million cancer patients in the world in 2012 (http://www.iarc.fr/en/media-centre/pr/2013/pdfs/pr223_E.pdf). The projected figures show that this year alone >14 million new cancer cases will be diagnosed and ~8.2 million cancer estimated deaths within 5 years of diagnosis worldwide. Among these, 57% (8 million) of new cancer cases, 65% (5.3 million) of the cancer deaths and 48% (15.6 million) of the 5 year prevalent cancer cases occurred in the less/under-developed regions of the world (http://www.iarc.fr/en/media-centre/pr/2013/pdfs/pr223_E.pdf). In all cancers, an abnormal and ongoing division of damaged/dysfunctional cells initially leads to the formation of a tumor (initiation), where the immortalized cells that have avoided cell death continue to proliferate in an unregulated manner (progression) and then ultimately invade other tissues at later stages in the disease (metastasis).
The immortalized cellular phenotypes that emerge in most cancers have largely avoided cell death, which can be defined as a terminal failure of a cell to maintain essential life functions, and can be classified according to its morphological appearance, as apoptosis, necrosis, autophagy or mitotic catastrophe. During cell death, numerous enzymes and signaling pathways are modulated [nucleases, distinct classes of proteases (e.g. caspases, calpains, cathepsins and transglutaminases, protein binding signaling intermediates and so on)], which can exhibit immunogenic or non-immunogenic responses (1). Tumor cells are genetically programmed to undergo apoptotic and non-apoptotic death pathways (e.g. necrosis, autophagy, senescence and mitotic catastrophe). Normally, apoptotic resistance is rendered by the up-regulation of antiapoptotic molecules and the down-regulation, inactivation or alteration of pro-apoptotic molecules. However, dysfunction in these cell-death pathways is associated with initiation and progression of tumorigenesis. An increased resistance to apoptotic cell death (involving the inhibition of both intrinsic and extrinsic apoptotic pathways) is therefore an important hallmark for cancer cells.
Several tumor suppressor proteins, such as TP53, recognize DNA damage and activate DNA repair processes. Irreparable DNA damage can induce apoptosis and prevent neoplastic transformation (2) and can also trigger cellular senescence of transformed cells. Regulation of apoptosis is influenced by BCL-2 family members of pro-apoptotic and antiapoptotic factors, death receptors and the caspase network. Alterations of proto-oncogenes, tumor suppressor genes and de-regulation in epigenetic factors such as microRNAs are potent causes of cancer growth. Proto-oncogenes encode proteins that stimulate cell proliferation, inhibit apoptosis or both. They are classified into six broad groups: transcription factors, chromatin remodelers, growth factors, growth factor receptors, signal transducers and apoptosis regulators. Normally, they are activated by genetic alterations (e.g. mutations or gene fusions, amplification during tumor progression or by juxtaposition to enhancer elements into an oncogene) (3–5). These genetic changes can alter oncogene structure or increase/decrease its expression. Similarly, tumor suppressor genes, which are involved in DNA repair, regulation of cell division (cell cycle arrest) and apoptosis, when mutated or inactivated by epigenetic mechanisms can cause cancer (4,5).
In this review, we discuss these mechanisms, their relationship to resistance to apoptosis and the importance of this hallmark characteristic of cancer as a potential enabler of environmental carcinogenesis. In 2011, a non-profit organization called Getting to Know Cancer launched an initiative called ‘The Halifax Project’ with the aim of producing a series of overarching reviews to assess the relevance of biologically disruptive chemicals (i.e. chemicals that are known to have the ability to act in an adverse manner on important cancer-related mechanisms) for carcinogenesis. To that end, our team was specifically tasked to review the hallmark of cancer ‘resistance to cell death’ and its relationships to other hallmarks of cancer. We were also tasked to identify a list of important, prototypical target sites for chemical disruption and a corresponding list of environmental chemicals that have been shown to have the potential to act on these targets. Ultimately, this review was not intended as a means to implicate specific chemicals in environmental carcinogenesis. Rather we undertook this review to explore what is known on this topic to provide a basis for further discussion of this idea and to help us identify future research needs.
To begin, we offer a brief review of several key mechanisms and pathways that are related to resistance to cell death. Specifically, we highlight apoptotic pathways, necrosis and necroptosis, the role of autophagy and the relationship that these mechanisms and pathways have with cancer (Table 1). For those who are seeking more in-depth treatment of these topics, several recent reviews can provide additional information (6,7). In doing so, we also focus on a number of important mechanisms and pathways that are relevant for disruption [i.e. binding to estrogen receptor α (ERα), P53, ErbB-2/HER-2 tyrosine kinase, extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK), MAP kinase, P16/P53, BCL-2/P53, peroxisome proliferator-activated receptor-α (PPAR-α), gap junctional intracellular communication (GJIC), hypersecretion of luteinizing hormone (LH) by gonadotroph cells in pituitary gland]. This list of target sites is not intended to be comprehensive. Other targets exist, including well-known mechanisms such as ALK, CD20/22/79b, MDM2, PD-L1, VEGF, HER receptors, BRAF, Rho-associated protein kinase, fibroblast growth factor-9, cathepsins, cyclooxygenases, prostaglandins and so on. We selected these targets because each of them are actively involved in resistance to cell death and all of them have been shown to be of considerable importance.
Table 1.
Characteristics of apoptosis, autophagy and necrosis pathways
| Morphological and biochemical features and modulators of cell death | Methods of detection | |
|---|---|---|
| Apoptosis | Morphological features: cellular shrinking, condensation and margination of chromatin, nuclear fragmentation and DNA laddering, plasma membrane budding and formation of apoptotic bodies in cytoplasm. Not surrounded by tissue injury of inflammation. Biochemical features: caspase-dependent cell death pathway. Activation: activation of intrinsic apoptotic pathway; BCL-2, c-FLIP, survivin IAP–antisense mRNA technology; recombinant TRAIL for DR4 and/or DR5 receptor; E2F-1 gene therapy; TWEAK (tumor necrosis factor-related weak inducer of apoptosis) is a cytokine belonging to TNF-ligand family for Tweak-receptor inducing apoptosis. Inhibition: natural and synthetic inhibitors of caspases; nitrosylation of caspase 9 or 3; c-Jun–mRNA antisense technology; CEP 1347–inhibitor of JNK signaling blocks Aβ-induced cortical neuron apoptosis. | Microscopic techniques: cellular features by light microscopy, nuclear DNA analysis by fluorescent stains (annexin V), confocal laser microscopy and electron microscopy. Assessment of DNA fragmentation: enzyme-linked immunosorbent assay, terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling assay, comet method, DNA diffusion, immunohistochemistry for single-stranded DNA and gel electrophoresis. Flow cytometry: cell cycle. Laser scanning cytometry: DNA content, phosphatidylserine translocation, inner mitochondrial transmembrane potential and caspase activity. Gene expression: northern blot, RNA protection assay, reverse transcription–polymerase chain reaction and immunohistochemistry. Evaluation of apoptosis-associated proteins: enzyme-linked immunosorbent assay, western blot and electrophoretic mobility shift assay. |
| Autophagy | Morphological features: partial chromatin condensation, no DNA laddering, cell membrane blebbing and formation of more autophagosome. Biochemical features: caspase-independent cell death pathway. Activators: conventional cytotoxic drugs and irradiation; BCR-ABL tyrosine kinase inhibitor–imatinib; anti-EGFR–cetuximab; proteasome inhibitors; TRAIL and histone deacetylase inhibitors; mTOR inhibitors and its analogs; ATP-competitive inhibitors of mTORC1 and mTORC2; dual PI3K-mTOR inhibitor; antidiabetic drug–metformin; serotonin reuptake inhibitor–fluoxetine; norepinephrine reuptake inhibitor–maprotiline; antiepileptic drug–valproic. Inhibitors: antibody against EGFR–cetuximab; Class III PI3K inhibitors-3-methyadenine, wortmannin and LY294002; antimalarial drugs–hydroxychloroquine; vacuolar ATPase–bafilomycin A1; lysosomotropic drug–monensin; microtubule-disrupting agents–taxanes, nocodazole, colchicine, vinca alkaloids; antidepressant drug–clomipramine; antischistome agent–lucanthone (autophagosome degradation) . | Electron microscopy, immunohistochemical staining of microtubule-associated protein 1 light chain 3 (LC3) as a general marker for autophagic membranes, monodansylcadaverine staining of autophagic vacuoles and protein degradation assays. |
| Necrosis | Morphological features: cell size increases, clumping and random degradation of nuclear DNA, cell membrane swelling and rupture, swelling of organelles, gain in cell volume (oncosis), organelle degeneration mitochondrial swelling and increased vacuolation. Activation: hyperactivation of poly(ADP-ribose) polymerase 1 (PARP1) enzyme with depletion of β-nicotinamide adenine dinucleotide and of ATP, hypoxic injury and oxidative stress (ROS/reactive nitrogen species); chetomin–inhibitor of tumor growth by inducing necrosis in vivo; dimethoxy-naphthoquinone–generation of ROS and induces apoptosis or necrosis; myristoleic acid methyl ester–induces apoptosis and necrosis in prostate cancer cells; sterigmatocystin–a mycotoxin inhibits DNA synthesis and causes necrosis. Inhibition: necrox-2 [5-(1,1-dioxo-thiomorpholin-4-ylmethyl)-2-phenyl-1H-indol-7-yl]-(1-methanesulfonyl-piperidin-4-yl)-amine] and necrox-5 [5-(1,1-dioxo-thiomorpholin-4-ylmethyl)-2-phenyl-1H-indol-7-yl]-(tetrahydro-pyran-4-ylmethyl)-amine]–is a cell-permeable necrosis inhibitor selectively locks oxidative stress-induced necrosis with antioxidant property; tyrphostin AG 126–reduces LPS-induced tyrosine phosphorylation of p42MAPK; cyclosporin A–inhibitor of the mitochondrial permeability transition pore (MPTP) and prevents necrosis; IM-54 (indolylmaleimide derivative)–inhibits necrotic cell death induced by H2O2 in promyelocytic leukemia HL-60 cells; PARP inhibitor VIII, PJ34 [2-(dimethylamino)-N-(6-oxo-5,6-dihydrophenanthridin-2-yl)acetamide hydrochloride]–inhibitor of PARP-1 and PARP-2 and inhibits necrosis; IM-54 [2-(1H-indol-3-yl)-3-pentylamino-maleimide]—a selective inhibitor of oxidative stress-induced necrosis. | Electron microscopy; nuclear negative staining; ethidium homodimer III DNA assay; detection of inflammation and damage in surrounding tissues. |
Apoptotic pathways
The extrinsic pathway: death receptor-mediated apoptosis
Receptor-mediated pathways are initiated by death ligands that bind to their specific death receptors, which include TNF-receptor 1, Fas/CD95 and TNF-related apoptosis-inducing ligand (TRAIL) receptor (8). All of these receptors contain the death domain, which is essential for the transduction of an apoptotic signal. After death ligands bind to their receptors, adapter molecules including Fas-associated death domain (FADD) or TNF receptor-1-associated death domain (TRADD) recruit the procaspase-8 for forming the death-inducing signaling complex. This leads to the initiation of the caspase cascade through activation of CASP-8 or -10, followed by subsequent activation of executive caspases such as CASP-3 and -7, and an irreversible commitment to apoptosis (9).
The intrinsic pathway: mitochondria-mediated apoptosis
Mitochondria play a pivotal role in cell survival as well as in apoptotic cell death, and defects in mitochondrial function might contribute to cancer initiation and progression. The mitochondria-mediated intrinsic pathway is initiated by various stimuli, such as high cytoplasmic Ca2+ levels, reactive oxygen species (ROS), ultraviolet irradiation, viral infections or xenobiotics (10). Mitochondrial control of apoptosis is evolutionary conserved and tightly regulated by the BCL-2 proteins divided into 20 pro-apoptotic and antiapoptotic members, which share conserved BCL-2 homology (BH) domains. The antiapoptotic members (BCL-2, BCL-XL, BCL-w, MCL-1, BCL-B) exhibit four BH domains (BH1-4). The pro-apoptotic members are categorized as BH3-only proteins (BAX, BAK, BIK, BAD, BIM, HRK, BCL-G HRK/DP5, NOXA and PUMA/BPC3), as reviewed in ref. 11. In normally proliferating cells, the pro-apoptotic BH3-only proteins are sequestered away from the antiapoptotic BCL-2 proteins. Although the antiapoptotic members such as BCL-2, BCL-XL or BCL-w are integral proteins of the outer mitochondrial membrane, the pro-apoptotic BCL-2 members are located predominantly in the cytoplasm. After an apoptotic signal, the free pro-apoptotic BH3-only proteins associate with BCL-2 on mitochondria. Additionally, pro-apoptotic BAX and BAK undergo conformational changes leading to homo- or oligo-merization at the mitochondrial outer membrane (12). Consequently, this leads to a mitochondrial outer membrane permeabilization, the decisive event that delimits the frontier between survival and death. Upon apoptosis induction, the voltage-dependent anion channel protein plays a critical role in the dissipation of mitochondrial transmembrane potential (13,14). After mitochondrial membrane disruption followed by osmotic swelling, soluble pro-apoptotic mitochondrial intermembrane space proteins like cytochrome c, apoptosis-inducing factor, endonuclease G, second mitochondrial activator of caspases (SMAC/DIABLO) and OMI/HTRA2 are released into cytosol resulting in activation of intrinsic apoptotic signaling cascades. The released cytochrome c, along with apoptosis-activating factor-1 (APAF-1) and procaspase-9, form the cytosolic apoptosome complex, which leads to the activation of CASP9, and in turn triggers the caspase cascade, resulting in apoptotic cell death (15,16). However, inhibitor of apoptosis proteins (IAPs) can directly bind to CASP-3, -6 and -7 and antagonize their proteolytic activities. In contrast, IAPs are inactivated, and caspase activity restored by proteins released from the mitochondria, such as SMAC/DIABLO or HTRA2/OMI (17). The intrinsic pathway might also operate independently of the caspase cascade by utilizing the release of the apoptosis-inducing factor and endonuclease G from mitochondria, and their translocation to the nucleus. Apoptosis-inducing factor is linked to chromatin condensation and the high-molecular-mass chromatin fragments, and after nuclear translocation, endonuclease G elicits DNA fragmentation (15). Because mitochondria-mediated apoptosis plays a critical role in cancer development and in the cellular response to anticancer agents, the significance of mitochondrial DNA mutations in cancer is currently an important area of investigation (18) (Figure 1).
Figure 1.
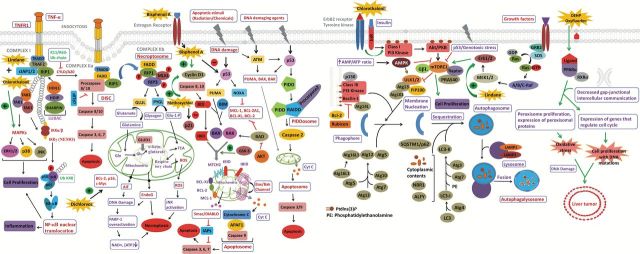
Apoptotic and non-apoptotic signaling pathways and the involvement of anthropogenic chemicals.
The novel PIDDosome-mediated apoptotic pathway
CASP2 was identified as the first apoptotic and the most conserved caspase (19). CASP2 was detected at various compartments in the cell including the nucleus, the Golgi apparatus, endoplasmic reticulum and cytoplasm. Previous studies have shown that CASP2 can be activated by DNA damage induced by anticancer drugs, such as cisplatin and etoposide, or by ultraviolet and γ-irradiation, and it is a critically involved in genotoxic stress-induced apoptosis (19). CASP2 activation leads to the release of cytochrome c, indicating that CASP2 acts upstream of mitochondria-mediated intrinsic pathway (20). Moreover, the treatment of cells with the CASP2 inhibitor and/or small interfering RNA to block CASP2 from inducing the release of cytochrome c is followed by the activation of CASP9 and 3. Similar to other initiator caspases, pro-caspase-2 contains a Caspase Activation and Recruitment Domain at the N-terminus. CASP2 recognizes a pentapeptide VDVAD for cleavage of target proteins, and its known target proteins are BID, PARP, Plakin, Huntingtin and DNA fragmentation factor 45. Because CASP2 is activated by a proximity-induced self-cleavage mechanism, it obtains proximity by forming a PIDDosome, which is composed of three protein components, PIDD (TP53-induced protein with death domain), RAIDD (RIP-associated Ich-1/Ced-3-homolog protein with a death domain) and CASP2, whose interaction supported by their respective death domains. PIDD death domain can also interact with the death domain of receptor-interacting protein-1 kinase implicated in the nuclear factor-κB (NF-κB) activation (21). PIDD appears to act as a molecular switch, controlling the balance between life and death upon DNA damage (Figure 1).
Necrosis and necroptosis
In contrast with apoptosis, necrosis is a genetically controlled process; necrosis involves an uncontrolled and progressive loss of cytoplasmic membrane integrity, a rapid influx of Na+, Ca2+ and water, resulting in cytoplasmic swelling, nuclear pyknosis and the release of lysosomal and granular contents into the surrounding extracellular space (22). Although the molecular mechanisms underlying apoptosis are better understood, little is known about the molecular events leading to necrosis. Necrosis has recently emerged as an important and physiologically relevant signaling process contributing to ovulation, immune defense, death of chondrocytes controlling the longitudinal growth of bones and cellular turnover in the intestine (23). In vivo studies indicated that removal of interdigital cells in the paws of Apaf1−/− mice during embryogenesis occurs by a caspase-independent necrotic-like process (24). However, accumulating evidence by many researchers suggests that necrosis is not just an unregulated and uncontrollable process. Rather, it involves a programmed and actively regulated process (aptly named necroptosis), which is regulated by the kinase activity of RIPK1 and RIPK3 that form the necrosome complex (25). This leads to the plasma membrane permeabilization, release of cell contents and exposure of damage/danger-associated molecular patterns (DAMPs), such as HMGB1, S100 protein, IL33 and mitochondrial DNA. Under normal physiological conditions, autophagy and the caspase-8-FLIPL-FADD platform are apparently gatekeepers preventing necroptosis (26).
The paradoxical role of autophagy in cancer
Autophagy is the basic catabolic mechanism in response to starvation or other stressful conditions whereby unnecessary or dysfunctional misfolded or aggregated proteins and cellular components (e.g. mitochondria, endoplasmic reticulum and peroxisomes) are engulfed within double-membrane vesicles called autophagosomes and are eventually digested by lysosomal enzymes to sustain cellular metabolism (27,28). During macroautophagy, a cytoplasmic cargo is delivered to the lysosome through an autophagosome, which fuses with the lysosome to form an autolysosome. Microautophagy involves the inward folding of the lysosomal membrane, which delivers a small portion of cytoplasm into the lysosomal lumen. Both macro- and micro-autophagy can be either non-selective or selective in the removal of large cellular components and protein aggregates (29). Autophagy involves several key steps for the final degradation of cellular components in lysosomes: (i) initiation and nucleation of phagophore; (ii) expansion and maturation of autophagosomes; (iii) fusion of the autophagosome with the lysosome to form the autolysosome and (iv) execution of autophagy (degradation). These steps are tightly regulated by highly conserved Atg genes and non-Atg genes (30).
Disorders in autophagic signaling pathways are frequently observed in cancer patients. Autophagy has been referred to as a ‘double-edged sword’ because it acts as an activator of tumor cell death (tumor suppression) as well as it plays a part in tumor cell survival during tumor development and in cancer therapy. Impaired autophagy was shown leading to failure of removing damaged protein and organelles, and exerting genomic instability and aneuploidy, which promotes tumorigenesis (31–33). The loss of BECN1 was found in human breast and ovarian cancers (34), whereas Becn1 null mice were shown to be tumor prone (35). In contrast, the BECN1 forced expression can inhibit tumor development. Additionally, sustained p62 (SQSTM1) expression, which results from autophagy defects, was found to be important in the promotion of tumorigenesis through de-regulation of NF-κB expression (33). Tumor cells experience elevated cytotoxic and metabolic stresses (e.g. hypoxia and deprivation of growth factor and oxygen), which can activate autophagy to maintain cellular biosynthesis and survival (28). Recent data indicate that suppression of autophagic proteins inhibited cell growth and conferred or potentiated the induction of cell death, indicating that autophagy contributes to cell survival in human cancer cells, as well as plays a role in adaptive response of tumor cells to anticancer therapies (36). A careful examination of the literature shows that an increased level of autophagic markers in the dying cell might not be the result of increased autophagic flux but due to a blockage of autophagy at its maturation. Therefore, the simple determination of numbers of autophagosomes is insufficient for an overall estimation of autophagic activity. It is necessary to distinguish by performing ‘autophagic flux’ assays whether autophagosome accumulation is due to autophagy induction or, alternatively, a blockade of steps in the downstream of autophagosome formation. Now, it is agreed that the true meaning of ‘autophagic cell death’ should be cell death by autophagy, not cell death with autophagy (Figure 1).
Dysfunctional apoptosis in cancers
The fundamental link between malignancy and apoptosis is exemplified by the ability of oncogenes, such as MYC and RAS, and tumor suppressors, such as TP53 and RB (Retinoblastoma), to actively engage apoptosis as well as the aberrant alterations of apoptosis regulatory proteins such as BCL-2 and c-FLIP in various solid tumors (37–39). Acquired apoptosis resistance is a hallmark of most human cancers. With regard to apoptosis triggers, a variety of signals (irradiation, growth/survival factor depletion, hypoxia, oxidative stress, DNA damage, cell cycle checkpoints defects, telomere malfunction and oncogenic mutations, chemotherapeutic agents and heavy metals) appear to provide the selective pressure needed to alter apoptotic programs during tumor development in support of tumor evolution (40–42). The ability of tumor cells to acquire resistance to apoptosis is a compensatory mechanism, which gives tumor cells a distinct (survival) advantage over normal cells. Defects in apoptosis have been implicated in many events relevant to tumorigenesis: (i) cell accumulation from the imbalance of cell proliferation and cell death or a failure of normal turnover process; (ii) permissive cell survival in the face of antigrowth signals, for example, hypoxia in tumor mass, cell–matrix and cell–cell adherence or contact inhibition; (iii) promoting resistance to the killing mechanisms of immune cell attack and (iv) fostering tumor metastasis by promoting cell survival in the circulation under detachment conditions, also known as anoikis resistance (43). The importance of this sort of dysfunction is underscored by the fact that tumor cells that possess alterations in proteins involved in apoptosis are often resistant to chemotherapy and are more difficult to treat (because anticancer drugs primarily work by inducing apoptosis). Tumor cell survival, unlike the survival of normal cells, is therefore highly dependent on aberrations of apoptosis signaling pathways (37).
Emerging evidence indicates that cancer stem cells (CSCs), the rare subpopulation of undifferentiated tumorigenic cells, are potential driving force for tumor growth and maintenance (44). To date, CSCs have been identified and isolated from various solid tumors including the lung, brain, breast, colon and skin. These CSCs are highly capable of self-renewal and are able to generate a progeny of differentiated cells that constitute a large majority of cells in the tumors (45). Most importantly, CSCs are apoptosis resistant and very likely responsible for tumor resistance to chemotherapy and irradiation (46). This can be attributed to the undifferentiated status of CSCs and to the extrinsic factors such as the tumor microenvironment and adhesion-based interactions, which also support their apoptotic resistance (47). Furthermore, the epithelial/mesenchymal transition (EMT) mechanism has been found to underlie the CSC characteristics that are linked to anoikis resistance (48). Accumulating evidence also suggests that this inherent resistance in CSCs shares similar extrinsic and intrinsic apoptotic pathway as normal stem cells and differentiated cancer cells (49,50).
Regulation of apoptosis in cancer
Evasion of apoptotic pathways allows cells to sustain chronic proliferation, which is a hallmark of cancer. Recently, two working models of apoptosis (both regulated by BCL-2 family and BH3-only proteins) were reviewed (51). The direct model proposes that the activator BH3-only proteins (BIM, BID and PUMA) can directly activate BAX and/or BAK oligomerization in addition to neutralizing BCL-2-like proteins, whereas the sensitizer BH3-only proteins (BAD and NOXA) release activator from activator/pro-survival protein complex. The indirect model suggests that BAX is primed in normal cells by BH3-only protein and bound with BCL-2. In excess of pro-apoptotic signaling, BH3-only proteins compete with BCL-2 allowing oligomerization of BAX and BAK leading to apoptosis (52). The BAX/BAK oligomerization loosens the integrity of mitochondria and culminates with mitochondrial outer membrane permeabilization facilitating the release of cytochrome c into the cytoplasm, which interacts with APAF-1, and leads to the ATP-dependent formation of apoptosome, and the recruitment and activation of the CASP-9, -3 and -7. In the absence of APAF-1 and CASP-9, cytochrome c release itself is not sufficient to induce apoptosis (53–55). Cytochrome c diffusion and death receptor signaling mediates modulation of XIAP by SMAC/DIABLO and OMI/HTRA2, and activation of caspases (56) (Figure 1). Up-regulation of XIAP, survivin and down-regulation of APAF-1 has been observed in several tumors.
Cellular stress and DNA damage are regulated through two tumor suppressor genes TP53, which induces expression of NOXA, PUMA and RB upon various environmental and chemical stresses. Recently, a bona fide tumor suppressor gene neurofibromin 2 (NF2/Merlin) was shown to regulate apoptosis through the Hippo pathway (57). RB integrates outside inhibitory signals, whereas TP53 senses irreparable damage in genomic integrity, intracellular organelles and nucleotides, as well as suboptimal level of glucose, and growth inhibitory signals (58). TP53 activities are tightly regulated by a network of protein–protein interactions, microRNAs and a range of post-translational modifications, including phosphorylation, acetylation, methylation and ubiquitination (59,60). TP53 activity is suppressed by a direct binding of TP53 to murine double minute 2 (MDM2), which targets TP53 for proteasomal degradation. NOXA also induces apoptosis in TP53/TP73-dependent manner in response to DNA damage, whereas PUMA, the most potent pro-apoptotic regulator, induces apoptosis both in a TP53-dependent and -independent fashion (61–63).
Cellular metabolism is a key for the survival of cells, whereas altered metabolism in cells induces either apoptosis or resistance to apoptotic stimuli. Metabolic enzymes and its intermediates from glycolysis, pentose phosphate pathway and tricarboxylic acid cycle have shown deregulated in many cancer types to provide nicotinamide adenine dinucleotide phosphate (NADPH), citrate, acetyl CoA and various other metabolites for high demand of biosynthesis and proliferation (64). Chronic proliferating cells short circuit their metabolic pathways and mostly depend on aerobic glycolysis to sustain the massive biosynthesis of intracellular structures. Various post-translational modification regulates cellular growth especially phosphorylation and acetylation and increase apoptotic sensitivity. Metabolic intermediates also regulate pro- and anti-apoptotic regulators (BCL-2 family protein). Perturbations in acetyl-CoA production may extend to other oncogenic contexts beyond that of BCL-xL (65–67). Redox status of tissues/cells affects their sensitivity to cytochrome c. Reduced glutathione mostly produced by NADPH inactivates cytochrome c, whereas apoptotic agents produce ROS to activate cytochrome C and apoptosis (68). Key regulatory metabolic enzymes, which affect apoptosis (e.g. hexokinase, fructose 2,6-bisphosphate kinase, lactate dehydrogenase M and pyruvate dehydrogenase kinase), are also implicated in cancer (55). Growth factors/cytokines regulate pro-survival signaling by RAS- and PI3K-AKT pathways through cognate receptor tyrosine kinase (RTK). Most human cancers harbor mutations in AKT and PTEN, which leads to AKT activation and resistance to apoptosis (69,70). Death receptor signaling triggers the recruitment of FADD and TRADD adapter proteins to induce dimerization of CASP-8 and subsequent activation of CASP-3 and -7. In some cell types, CASP-8 directly cleaves BH3-only protein BID to localize it to the mitochondria and activate BAX (71).
Additionally, ‘anoikis’, the detachment of cells, is another major regulator of apoptosis. The detachment of adherent cells (loss of critical interaction between the cell and the extracellular cell matrix) leads to apoptosis due to the loss of integrin α-5 or β-5 signaling and the loss of focal adhesion kinase, a reduction of talin–integrin interaction, and of c-Jun N-terminal kinase signaling (72).
Oncogenes, tumor suppressor genes and apoptosis
Human health is continuously challenged by exposure to a wide range of environmental chemicals that affect DNA integrity (73). When DNA repair capacity is exhausted, DNA damage accumulates in cells at a higher level, and this excess damage causes an increased frequency of mutation and/or epigenetic alterations of specific genes (oncogenes and tumor suppressors) resulting in the disruption of the cellular networking that controls cellular homeostasis and leads to cellular transformation and cancer development (74). The inactivation of expression of tumor suppressor genes via genetic and epigenetic changes (DNA hypermethylation, histone deacetylation/methylation and microRNA targeting) often leads to tumor initiation and progression, whereas amplification and overexpression of oncogenes result in the similar tumorigenic phenotype (75). Tumor suppressor ‘driver’ genes include: genes for retinoblastoma protein (RB), tumor protein TP53 (TP53), BRCA1 and 2, PTEN, VHL, APC, CD95, ST5, 7 and 14, YPEL3, whereas ‘driver’ oncogenes include: growth factors (e.g. C-SIS, WNT), RTKs (EGFR, PDGFR, VEGFR, TRK, ERBB2), cytoplasmic tyrosine (SRC, ABL and BTK) and serine/threonine (ATM, MTOR, ERK, PI3KCA, AKT1, 2 and 3, LKB1 and RAF) kinases, transcriptional factors (MYC and E2F), GTP-ases (RAS) and others (CCND1), as reviewed by Lee et al. (74). Discovery of microRNA genes added new members to both tumor suppressor (e.g. miR-34a) and oncogene (e.g. miR-17–92) families (76).
As part of the DNA damage response to genotoxic stress, apoptosis is triggered by chemical-induced DNA lesions and represents a first line of defense allowing the organism to eliminate damaged cells. Notably, cells respond to stress-induced DNA damage by increasing their levels of TP53 (77). The wild-type TP53 prevents cancer formation through the activation of cell cycle arrest or apoptosis via transcriptional regulation of hundreds of specific gene targets or via multiple protein–protein interactions. TP53 and its evolutionary older relatives, TP63 and TP73, exhibit a similar modular structure and share significant structural and functional homologies; however, their tumor suppressive role is not as straightforward as TP53. Genes for all TP53 family members produce proteins with the transactivation domain displaying a tumor suppressive function and proteins without transactivation domain acting as oncoproteins (78). TP53 is mutated in >50% of human cancers, whereas in other cancers, its function is compromised by de-regulation of the TP53 pathway. Both TP63 and TP73 are rarely mutated or epigenetically altered in human cancers. Tp53−/− mice develop tumors with short latency and 100% penetrance (77). Tumor suppressive function for TP73 was confirmed using Tp73−/− mice (79). Tp53+/− and tp63+/+ mice are less cancer prone than Tp53−/− and tp63+/− mice, respectively.
The synergistic effects of the TP53 family members in tumor suppression were highlighted using mice heterozygous for mutations in both TP53 and TP63, or TP53 and TP73 displaying higher tumor burden and metastasis, compared with tp53+/− mice (80). Accumulating data show that TP53 family proteins can regulate cell survival via cell cycle arrest, senescence and apoptosis and are abnormally expressed in different cancer types (breast tumors, acute myeloid leukemia, head and neck tumors, melanoma, renal cell carcinoma, colon, ovarian and lung tumors) suggesting that their differential expression may disrupt the TP53 response and contribute to tumor initiation/progression and linked to cancer prognosis and treatment (78).
Although mutations of TP63 mutations are almost non-existent in human cancers, >80% of primary head and neck squamous cell carcinomas, other squamous cell epithelial malignancies and non-small cell lung cancer retain TP63 expression, where it is often over-expressed and occasionally amplified. The TP63 expression strongly influences the tumor cell response to genotoxic stress (81). TP63 activates death domain receptor- and mitochondria-mediated apoptosis pathways, which are clearly reinforced by concomitant treatment with genotoxic stress. However, ΔNp63α confers resistance to apoptosis via a transcriptional regulation of AKT1, as well as via down-regulation of several microRNAs (miR-181a, -519a and -374a) and up-regulation of miR-630, which targets proteins involved in cell cycle arrest and apoptosis for down-regulation, hence conferring tumor cell chemoresistance (82,83). It is likely that apoptosis sensitivity to genotoxic agents may be determined not only by TP53 but also by TP73 and TP63 function, and its isoforms (84).
The disruption of normal cell death
From a disruption standpoint, the inactivation or attenuation of the TP53 apoptotic response, achieved by mutations or epigenetic alterations, is known to promote cell transformation (77). For example, non-polycyclic aromatic hydrocarbon components present in tobacco smoke condensate are able to attenuate the TP53 apoptotic response, as suggested by studies in mouse epidermal cells (78). The transcription factor C/EBPβ, which is induced by cigarette smoke has also been involved in TP53 repression (85,86). Following a prolonged exposure to environmental chemicals, bulky DNA adducts may not be removed by DNA repair mechanisms but converted into mutations. Subsequent DNA replication cycles may lead to hot spot mutations in key growth regulatory genes, thereby resulting in malfunction of tumor suppressor genes and amplification/overexpression of oncogenes (74).
Similarly, mutated RAS oncogenes were found in the experimental tumors of rodents that had been exposed to chemical or physical compounds, as well as in many human cancers (87). For example, exposure to hydrocarbon solvents has been associated with an increased risk of exocrine pancreatic cancer, the human tumor with the highest prevalence of K-RAS mutations (88). And heterocyclic amines have been implicated in both initiation and maintenance of breast tumorigenesis mediated by upregulated H-RAS expression, ERK pathway activation, NOX1 expression and elevation of ROS (89). Although the sustained activation of the NF-κB transcription factor is another important element involved in chemical tumorigenesis, tobacco, alcohol, high-fat diet, environment pollutants, cancer-causing viruses (human papillomavirus, hepatitis B and C viruses, human immunodeficiency virus and bacteria (Helicobacter pylori), ultraviolet light, ionizing radiation, obesity and oxidative stress are all potent NF-κB stimuli (90,91). The following proteins: pro-inflammatory proteins (cyclooxygenase-2, inducible nitric oxide synthase, TNF, interleukin-8); proliferative/pro-survival factors (bone morphogenetic proteins, stem cell factor, vascular endothelial growth factor, granulocyte–monocyte colony stimulating factor) and antiapoptotic proteins [TRAF-1 and -2, the CASP-8 inhibitor (FLIP), IAPs, XIAP, BCL2 and its homologues and matrix metalloproteinases] are overall involved in tumor promotion, initiation and progression (92).
The critical research gaps for a clear understanding of chemical carcinogenesis include the following:
Understanding how genetic modifications by low-dose environmental mixtures can disrupt/overcome normal cell death.
Understanding the molecular processes and pathways activated/blocked by individual chemicals and mixture of chemicals with disruptive potential.
Understanding the low-dose effects of environmental chemicals (single and mixtures) on cell death within different tissues and organs of human.
Clearly distinguishing the differences between the contributions of both carcinogenic and non-carcinogenic chemicals (individually and in mixtures) in environmental carcinogenesis by experimental methods.
Key target sites for disruption
In this review, we wanted to look at several key target sites that disrupt normal cell death and potentially have relevance for environmental carcinogenesis. It is generally agreed that many cancers arise from a single cell that has accumulated genetic and epigenetic mutations of a few crucial genes of proto-oncogenes and tumor suppressors, and that this is caused by random errors in DNA replication or a reaction of the DNA with free radicals or other chemical species of endogenous or exogenous origin (93). However, we also know that chemicals with disruptive potential are capable of a wide range of additional cellular level effects that are relevant to cancer (94), and the general population now faces ongoing exposures to thousands of environmental chemicals that are present in consumer products, our food, our water and in the air (95). At the same time, regulators worldwide have remained largely focused on the effects of single chemicals while placing very little emphasis on the effects of exposures to mixtures of chemicals in the environment (96). Accordingly, in this review, we emphasize the pivotal and enabling role that resistance to cell death plays in carcinogenesis and we highlight some of the key mechanisms and pathways that can be chemically disrupted (i.e. in a manner that results in dysfunction of normal cell death routines) and that have the potential to be supportive of the emergence of an immortalized cellular phenotype.
To that end, we first identify and review a number of key targets of this nature that have been shown to be active sites for chemical disruption in the past as follows:
Binding to ERα
Given that many anthropogenic agents are xenoestrogens, a considerable amount of environmental health research has focused on ER level disturbances (97). Many xenoestrogens binds to ER and either activates it or inhibits it. ERα activation stimulates cell proliferation and initiates cancer through tumor promotion, whereas the activation of ERβ stimulates terminal cell differentiation and disrupts cancer progression, which is an anticancer effect. For example, many of the organochlorine (OC) pesticides such as lindane or their metabolites fall into the category of xenoestrogens that disrupt endocrine processes by acting as agonists of ERα and/or antagonists of ERβ and by exerting antiandrogenic effects (by binding to androgen receptors). ERα and tumor suppressor protein p53 exert opposing effects on cellular proliferation. ERα’s repression of p53-mediated cell death has been widely investigated, especially in breast cancer (98), but emerging evidence suggests a much more complex role for ERα-controlled pathways in other tumor-related phenomena. ERα interacts with p53 bound to promoters of Survivin and multidrug resistance gene 1(MDR1), and inhibits p53-mediated transcriptional repression of these genes in human cancer cells in vivo. It was found that p53 is necessary for ERα to access the promoters and there is cross-talk between the pathways mediated by ERα and p53 (99). It has been also been shown that an increase of ERα messenger RNA (mRNA) level in ERα-positive breast cancer is associated with de-regulation of metabolism, which produce a complementary effect on cell differentiation and proliferation (100). On the other hand, evidence of ERα’s role in the EMT has also been reported. In endometrial carcinomas and breast cancer, ERα’s activity is negatively associated with the activation of EMT via the Wnt, Sonic Hedgehog and transforming growth factor-β (TGF-β) signaling (101,102). EMT involves the loss of cell–cell adhesion and a consequent increase in mobility and invasiveness. It has been proposed that ERα acts to promote invasive growth in breast cancer cells by a direct, ERα-dependent expression of metastasis-associated genes, such as the MTA-3 protein. It is important to note that a physiological feedback mechanism regulates the efficiency of ERα activation through the state of cell–cell interactions that are mediated by E-cadherin (103). Although EMT promotes a decrease in cellular contacts, it also inhibits ERα transcription thus limiting its own ERα-dependent activation. Although no effects on genetic instability and immune system evasion of systematic ERα activation have been reported, the synergy of action involved in these different (deregulated) pathways may be very important for cancer onset and progression.
Gap junctional intracellular communication
In addition to tumor promotion ability, some environmental chemicals directly or indirectly cause DNA mutations through free radical production (ROS/reactive nitrogen species) and may cause both tumor initiation and tumor promotion by inhibiting GJICs and connexins (Cxs) (104,105). Blockage of GJIC between the normal and the pre-neoplastic cells creates an intra-tissue microenvironment in which tumor-initiated pre-neoplastic cells are isolated from growth controlling factors of normal surrounding cells resulting in clonal expansion (106). Gap junction channels and Cxs control cell apoptosis by facilitating the influx and flux of apoptotic signals between adjacent cells and hemi-channels between the intracellular and extracellular environments. Recently, it has also been demonstrated that Cx proteins in conjunction with their intracytoplasmic localization, may act as signaling effectors that are able to activate the canonical mitochondrial apoptotic pathway (107). Tumor-promoting chemicals such as phenobarbital, dichlorodiphenyltrichloroethane (DDT) and 12-O-tetradecanoylphorbol 13-acetate block apoptosis and also block GJIC, whereas several antitumor chemicals, such as retinoids and dexamethasone, increase GJIC and increase apoptosis. So, it has been hypothesized that GJIC is necessary for apoptosis and blockage of apoptosis with chemicals/carcinogens could therefore promote the initiation of premalignant cells in tumorigenesis (108). For example, knockdown of connexin 43 (Cx43) had an inhibitory effect on GJIC and resulted in a reduction of cell death after treatment with cisplatin and Salmonella (109), and Kang et al. (105) reported on the inhibition of GJICs in normal human breast epithelial cells by pesticides, polychlorinated biphenyls, polybrominated biphenyls and halogenated hydrocarbons (when given as single compounds or as mixtures), and suggested that they may contribute to carcinogenesis through this mechanism.
Peroxisome proliferator-activated receptor-α
PPAR-α receptors are mainly found in the liver and belong to the steroid hormone receptor superfamily that functions as a transcription factor for genes involved in glucose, lipid and amino acid metabolism, and that also induces enzymes involved in the metabolism of xenobiotics. Upon ligand binding, PPARs heterodimerize with the retinoid X receptor and bind to the specific promoter sequence and trigger the expression of target genes (110). A variety of chemicals including certain herbicides and plasticizers induce peroxisome proliferation with increased replicative DNA synthesis, suppression of apoptosis and increased expression of peroxisomal acyl-CoA oxidase in rodent liver and other tissues. In rodents, these peroxisome proliferating chemicals act as non-genotoxic carcinogens that promote the development of tumors (111). Chemicals of industrial importance such as diethylhydroxylamine and chlorinated solvents are peroxisome proliferators (PP) that induce elevated S-phase in rat and mouse and play an important role in hepatocarcinogenicity. The molecular mechanisms of altered expression of cell cycle regulatory proteins resulting in the elevation of S-phase, and suppression of apoptotic cell death and induction of proliferation are evidenced by the activation of PPAR-α and survival signaling by p38 MAPK in hepatocellular carcinomas (112).
Hypersecretion of LH by gonadotroph cells in pituitary gland
Neuroendocrine disruptors are environmental pollutants that are agonists/antagonists or modulators of the synthesis and/or metabolism of neurohormones, neuropeptides and neurotransmitters. Sustained hypersecretion of LH occurs following the disruption of the hypothalamic–pituitary–testicular axis. The tumorigenic response to a chemical in an endocrine tumor is generally dose responsive. As the dose of environmental chemical increases, the extent of perturbation of normal endocrine homeostasis increases resulting in a stronger trophic stimulus to the target cell (113). LH up-regulates the expression of apoptotic inhibitor, survivin in a dose-dependent manner via ERK1/2 signaling pathway and inhibits apoptosis in ovarian epithelial tumors in vitro (114).
p53
As noted previously, p53 is a tumor suppressor gene and has been described as the ‘guardian of the genome’. p53 is a transcriptional activator regulating the expression of Mdm2 (negative regulator of p53) and genes involved in growth arrest (p21, Gadd45 and stratifin), DNA repair (p53R2) and apoptosis (Bax, Apaf-1, PUMA and NoxA). Its activity disrupts the formation of tumors by arresting growth and inducing apoptosis. This 53kDa phosphoprotein induces apoptosis by stimulating BAX and FAS antigen expression, or by repression of BCL-2 expression. p53 mutations are found in most of the tumors and contribute to the complex molecular network events leading to tumor formation. Notably, the progression of cancers which overcome cell death [via the inactivation of tumor suppressor genes (p53) and activation of oncogenes (c-Ha-ras)] after exposures to organophosphorus pesticides is also associated with an increase in genome instability (115). Accordingly, one the most important candidates, as a key regulator of malignant transformation, is P53. Somatic mutations of this gene or perturbations in its pathways are among the most frequent alterations in human cancers (116). Arguably, the most important decision maker in cellular process that unfold in response to every kind of stress and harm, P53 is involved in cell cycle arrest, apoptosis, regulation of metabolism, DNA repair and every pathway connected to them. Its action is therefore opposed to evasion from growth control, genetic instability, sustained proliferative signaling and cellular motility, whereas it can be an important promoter of metabolic changes and even replicative immortality. Cross-talk between P53 pathways and most molecular mechanisms that transduce external signals (to promote or inhibit cell proliferation) is branched and efficient so chemical disruptors that systematically impair P53 can readily produce harmful effects on almost all of the hallmarks involved in malignant transformation.
p16/p53
p16INK4A (p16) and p53 are tumor suppressor genes (antioncogenes). p16 is known as cyclin-dependent kinase (CDK) inhibitor and specifically blocks the activity of CDK4 and CDK6. The binding of 16kDa protein p16INK4A to CDK4 inhibits the phosphorylation of retinoblastoma protein (pRB) and subsequently inhibits the transcription factor (E2F), the release and arrest of the G1 phase of cell cycle and the suppression of cellular proliferation. p16 also inhibits the growth of breast cancer cells by inhibiting the VEGF signaling pathway and angiogenesis. And recently, it has been demonstrated that the anticancerous ability of p16 is additionally attributed to its ability to induce tumor cells to enter senescence. It also induces apoptosis both in vitro and in vivo (117). The functional or structural loss of p16INK4A therefore leads to the cell cycle propagation of genetically damaged/mutated cells and increases the subsequent risk of tumor development. p16 is encoded by INK4a gene and an alternative reading frame of INK4a transcribes to p14ARF, which mediates the link between p16 and p53 pathways. So, loss of the INK4a gene disrupts p16INK4A/CDK4/6/pRB and p14ARF/MDM2/p53 pathways, which controls cell proliferation (118). Notably, the p16 locus was found to be inactivated in many cancers such as lung, breast, melanoma, pancreatic, brain and >80% of squamous cell carcinoma of the head and neck tumors (119). Thus, p16 INK4A, p14 ARF and p53 genes involved in cell cycle pathways are major targets of inactivation in carcinogenesis. Occupational exposure to chemicals and metal dusts form ROS and reactive nitrogen species in humans through oxidant-mediated responses, which causes hypermethylation of p16INK4A and p53 along with the activation of MAP kinase to induce carcinogenesis.
BCL-2/p53
BCL-2 is a proto-oncogene, which regulates cell cycle progression and apoptosis (antiapoptotic), whereas p53 is a tumor suppressor gene. BCL-2 constitutively suppresses p53-dependent apoptosis. The BCL-2/p53 axis requires pro-apoptotic protein (Bax) and the effector molecule (CASP-2) as essential apoptotic mediators following the silencing of Bcl-2 or Bcl-xL. p53 possesses pro-apoptotic properties that appear to be constitutively active despite its suppression by Bcl-2 (120). Both p53 and Bcl-2 are strong predictors of recurrence and survival in rectal cancer (121). And the chemical 7,12-dimethyl benz-(a)-anthracene induces tumor growth in breast cancer that is apparently due to the inactivation of p53 aided by the absence of Bcl-2 (122).
ErbB-2/HER-2 tyrosine kinase
The human epidermal growth factor receptor (EGFR/HER) family consists of ErbB/HER lineage of receptor proteins (ErbB1–4) as it shows similarity to the v-ErbB oncogene of avian erythroblastosis virus (123). The ErbB-2/HER protein tyrosine kinase receptor contains an extracellular domain followed by a single transmembrane segment and intracellular tyrosine kinase activity, which regulates cell growth and differentiation particularly during embryogenesis (124). Overexpression of ErbB2/HER2 is related with cancer. Binding of epidermal growth factor ligands to their cognate ErbB receptors induces homo- or hetero-dimerization of ErbB2 and autophosphorylation of phosphotyrosine residues in the cytoplasmic domain, which serve as docking sites for adaptor proteins to downstream signals for growth and survival. Up-regulation of PI3K/AKT signaling pathway is found in ErbB2+ breast cancers, where it exerts pro-survival effects overcoming cell death (125).
The PI3K/AKT/mammalian target of rapamycin (mTOR) pathway is important for cell growth and survival, and it is also frequently activated in cancer. PI3Ks are a family of intracellular signal transducer enzymes involved in many cellular functions such as cellular growth, proliferation, differentiation and survival playing an important role in tumorigenesis (126). Upon activation of the RTKs by growth factors, PI3Ks convert phosphatidylinositol-4,5-biphosphate into phosphatidylinositol-3,4,5-triphosphate, which provides docking sites for pleckstrin homology domain containing proteins, including phosphoinositide-dependent kinase-1 and protein kinase B. This conversion is mainly controlled by the phosphatase and tensin homolog (PTEN), which dephosphorylates PIP3 into phosphatidylinositol-4,5-biphosphate, thereby regulating the uncontrolled activation of AKT pathway. Loss of PTEN tumor suppressor is common in malignancies and correlates with increased AKT activity. AKT is activated by phosphorylation of Thr308 by phosphoinositide-dependent kinase-1 (PDK1) and Ser473 by the mammalian target of rapamycin complex 2 (mTORC2). Activated AKT phosphorylates glycogen synthase kinase 3, forkhead box transcription factors, BCL-2 family members and the tuberous sclerosis complex (TSC1/2) thereby regulating a range of pathways involved in protein synthesis, proliferation, metabolism and apoptosis (127).
The mTOR pathway is the main target of the rapamycin, a natural compound produced by the Streptomyces hygroscopicus, which displays potent immunosuppressant and antiproliferative properties. The mTOR pathway integrates stimuli from diverse upstream pathways including the PI3K/AKT pathway and is responsible for the synthesis of a wide range of proteins involved in cell growth, proliferation, survival and tumorigenesis. mTOR can act in complex with Raptor (mTORC1) or Rictor (mTORC2) and these complexes regulate entirely different programs in the cell. When activated, mTORC2 activates and stabilizes AKT by its phosphorylation at Ser473, and controls actin cytoskeleton organization and cell migration, whereas mTORC1 increases mRNA translation by phosphorylation of the downstream molecules p70S6K (S6K) and 4E binding protein 1 (4EBP1). Phosphorylation of p70S6K leads to mRNA biogenesis, translation of ribosomal proteins and cellular proliferation. 4EBP1, in the hypophosphorylated state, binds the eukaryotic initiation factor 4E preventing its binding to eIF4G, and thereby to form the translational initiation complex eIF4F. Once phosphorylated, 4EBP1 is unable to bind to eukaryotic initiation factor 4E, which results in increase of translation of proteins related to cell proliferation and viability (128,129).
AKT activation affects components of the apoptosis regulatory machinery, including the BCL-2 family, the caspase family or the function of death domain receptors. AKT directly phosphorylates the BAD protein, which is a pro-apoptotic member of the BCL-2 family, whereas the dephosphorylated BAD promotes apoptosis (130,131). AKT might also prevent apoptosis by phosphorylation and inactivation of glycogen synthase kinase-3, CASP-9 and indirect activation of NF-κB leading to the altered transcription of pro-survival genes (e.g. IAP1, IAP2), as reviewed in refs 132–134. The mTOR pathway has also been linked by several studies to play a role in cell death by apoptosis and autophagy (135). One of the proposed pathways by which mTOR regulates autophagy was discovered in studies from yeast essential autophagy genes (Atgs), as reviewed in ref. 136. Atg1/Atg13/Atg17 complex is required for maximal catalytic activity of mTOR leading to Atg13 phosphorylation, subsequently destabilizing the complex and inactivating Atg1. In the mammalian cells, unc-51-like kinase 1 (ULK1) and focal adhesion kinase interacting protein of 200 kD (FIP200) form the complex with mammalian ATG13. mTORC1 activation correlates with the phosphorylation of ULK1-ATG13-FIP200 complex and inhibition of autophagy. Activation of P70S6k by mTOR may block apoptosis by increasing antiapoptotic BCL-2/BCL-xL protein expression and inactivating the pro-apoptotic protein BAD (137). In human prostate cancer cell lines, ErbB-2 kinase activity was increased by OC insecticides such as lindane, DDT and fungicide chlorothalonil. DDT induces cellular proliferation of the androgen-dependent human prostate cancer cell line LNCaP by phosphorylation of MAP kinase. However, no proliferative effect was induced in androgen-independent PC-3 cell line (138).
Mitogen-activated protein kinase
MAPK are serine/threonine kinases that transduce extracellular signals from a diverse range of stimuli and elicit cellular responses such as proliferation, differentiation, survival, migration, development, inflammatory responses and apoptosis. In mammalian cells, three MAPK families have been characterized namely classical MAPK (ERK), C-Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) and p38 kinase. MAPK pathways involve a series of protein kinase cascades, and each cascade consists of more than three enzymes that are activated in a series: a MAPK kinase kinase (MAPKKK/MAP3K), a MAPK kinase (MAPKK/MAP2K/MEK) and a MAP kinase (MAPK) (139). MAPK has a pleiotropic role in cancer, especially p38 and JNK MAPK pathways that are involved in the cross-talk between autophagy and apoptosis induced by genotoxic stress. p38 MAPK plays a dual role in genotoxic stress-induced apoptosis. Rottlerin-induced apoptosis of HT29 colon carcinoma cells was contributed by the up-regulation of non-steroidal anti-inflammatory drug activated gene-1 (NAG-1) via a p38 MAPK-dependent mechanism (140). However, under certain circumstances, it also involved in mediating resistance to apoptosis. The phosphorylation of p38 significantly increased the resistance to docetaxel-induced apoptosis in prostate cancer cells (141). And the suppression of p38 MAPK reversed the overexpression of micro RNA-214, which is linked to the radiotherapy resistance of non-small-cell lung carcinoma cells (142). It also regulates autophagy both as a positive and negative regulator. Platinum compounds such as E-platinum induced autophagic cell death in gastric carcinoma BGC-823 cells via suppression of mTOR by decreasing phosphorylation of p38 MAPK (143). On the other hand, suppression of the p38 signaling pathway induced autophagic and necroptotic cell death in TNFα-treated L929 cells. JNK MAPK promotes the phosphorylation of c-Jun and activating transcription factor-2 (ATF-2) resulting in the activation of AP-1 and the expression of Fas/FasL signaling pathway proteins, which subsequently activate effector caspase 3 and trigger apoptosis (144). JNK activation is associated with transformation in many oncogene and growth-factor-mediated pathways, whereas p38 MAPK activation involves in cell differentiation processes such as adipocytes, erythroblasts, myoblasts, cardiomyocytes and neurons. Regulation of the cell cycle is critical in cellular proliferation and development of multicellular organisms, and abnormalities in MAPK signaling play a critical role in the development and progression of cancer. MAP kinases are reported to be involved in several pathological conditions such as cancer and other diseases. MEK4/MKK4 is involved in stress-activated pathways such as JNK, and p38 is consistently inactivated by mutation in many cancers including cancers of the ovary, breast, pancreas, bile duct, lung, colon and testes (145).
ERK/MAPK
ERK pathway is a well-characterized MAPK signaling cascade with the Raf-MEK-ERK pathway. The stimulation of RTKs initiates the multistep cascade process resulting in the phosphorylation of p44MAPK (ERK1) and p42MAPK (ERK2) and increasing its enzymatic activity. The activated ERKs translocate into the nucleus and transactivate many transcription factors and regulate expression of genes to promote cell growth, differentiation or mitosis (139). It also regulates post-translational regulation of the assembly of cyclin D-cdk4/6 complexes, which subsequently phosphorylates the RB protein causing the activation of transcription factor E2F and regulates the genes involved in G1/S progression of cell cycle. In human hepatocytes, TGF-β induces apoptosis by the up-regulation of Rac-independent NADPH oxidase NOX4 mediating the production of ROS, which precedes the loss of mitochondrial transmembrane potential, cytochrome c release and caspase activation, for an efficient mitochondrial-dependent apoptosis (146). However, NOX4 up-regulation was inhibited by intracellular antiapoptotic signals such as PI3K and ERK/MAPK pathways. The overactivation of the MEK/ERK pathway in hepatocellular carcinoma HCC cell line confers resistance to TGF-β-induced apoptosis by impairing the up-regulation of the NADPH oxidase NOX4 expression (147). De-regulation of ERK activity is common in cancer leading to proliferation, migration, resistance to apoptosis and loss of differentiated phenotypes. In particular, cancerous mutations are mostly affecting Ras and B-Raf along with the overexpression of EGFR and ERBB2 in the ERK-signaling pathway. ERK signaling also plays a crucial role in disrupting the antiproliferative effects of ligands such as TGF-β (145). OC pesticides or their metabolites (endosulfan, dieldrin and DDE) and p-nonylphenol, a detergent by-product from plastic manufacturing, all produced dose-dependent ERK-1/2 phosphorylation in a pituitary tumor cell line GH3/B6/F10, which expresses high levels of membrane receptors for ER-α (148).
Environmental chemicals that confer resistance to cell death
In this review, we wanted to further consider the possibility that low-dose exposures to combination of environmental chemicals might have a role to play in environmental carcinogenesis. To that end, we developed a list of environmental chemicals that had been shown to act disruptively on the key target sites mentioned previously. Specifically, we sought to identify chemicals that were ubiquitous in the environment and not known to be carcinogenic, or classified as carcinogenic to humans. We focused on bisphenol A (BPA), chlorothalonil, dibutyl phthalate (DBP), diethylhexyl phthalate (DEHP), dichlorvos (DDVP), lindane, methoxychlor (MXC), linuron, and oxyfluorfen. These reported actions of these chemicals on these important target sites are described below.
Bisphenol A
Ubiquitous environmental anthropogenic chemicals such as BPA (4,4ʹ-(propane-2,2-diyl)diphenol) and phthalates are commonly found in consumer products, and act as obesogens by disrupting the metabolic homeostasis pathway. This involves the activation of PPARγ, which is a critical regulator of fat formation and also regulates lipid, glucose and energy in humans. BPA in particular is an estrogenic mimic which does not cause mutations per se, but increases breast cancer incidence (149–151). BPA-exposed to HRBEC cell lines and T47D breast cancer cells showed markedly reduced pro-apoptotic negative regulators of the cell cycle (p53, p21WAF1 and BAX) with concomitant increases in proliferation initiating gene products (proliferating cell nuclear antigen, cyclins, CDKs and phosphorylated pRB). It also induced an increase in the ERα: ERβ ratio (152). In addition, TP53 loss of function promoted activation of mTOR pathway, together with PI3K, AKT and 4EBP1 and, concurrently, PTEN was suppressed which resulted in enhanced cell growth and proliferation, and ultimately breast tumorigenesis (153). Besides increasing the risk of breast cancer, BPA neutralizes the effects of tamoxifen, undermining a widely used preventive measure to control disease. It has been shown that BPA affects the P53 pathway, inducing a prominent evasion of apoptosis coupled by an increased proliferation (152), and the GPER/EGFR/ERK pathway influencing proliferation and migration (154). This action seems to be delivered mainly through a substantial activation of mTOR pathways and a negative regulation of pro-apoptotic proteins like P53, P21 and BAX. In a number of cases, this BPA-induced cellular misbehavior persists even after BPA has been removed thus providing additional evidences of the chronic potential of this chemical disruptor. BPA has also been shown to disrupt double-strand break repair machinery in vivo suggesting that consistent environmental exposure to BPA may severely and dangerously affect the stability of DNA in mammalian cells (155). And BPA exerts a pro-metastatic influence in at least one mouse model of mammary carcinogenesis (156).
Chlorothalonil
Chlorothalonil (2,4,5,6-tetrachloroisophthalonitrile) is a broad-spectrum, non-systemic, OC pesticide (fungicide). It is used to control pathogenic fungi that attack vegetables, fruits, trees and agricultural crops. It is predominantly used on peanuts, potatoes and tomatoes and as an antifungal additive in paints, emulsion and resin. The carcinogenicity of chlorothalonil was evaluated in rodents, and the studies have shown evidence of renal tubular carcinomas and adenomas, and stomach tumors (157). Chlorothalonil up-regulates the expression of ErbB-2 tyrosine kinase and MAP kinase leading to cell proliferation in a prostate cancer cell line (138). Chlorothalonil readily reacts and conjugates with glutathione in the liver, and chlorothalonil metabolites consist of a mixture of di- and tri-glutathione conjugates, cysteine S-conjugates and mercapturic acids. In the proximal tubules of kidney, glutathione conjugates are completely cleaved by enzymes γ-glutamyl transpeptidase and dipeptidases to the cysteine S-conjugates, which are further cleaved by enzyme β-lyases to the corresponding thiol derivatives. The production of thiol derivatives is thought to be responsible for the toxicity seen in the kidneys (158). In a eukaryotic system, chlorothalonil reacted with proteins and decreased cell viability by formation of substituted chlorothalonil-reduced glutathione (GSH) derivatives and inhibition of specific NAD thiol-dependent glycolytic and respiratory enzymes (159). Caspases (cysteine-dependent proteases) and transglutaminase are some of the thiol-dependent enzymes involved in apoptosis. So, inhibition of these thiol-dependent enzymes in tumor-initiated cells (by chlorothalonil) may disrupt apoptotic cell death and aid in tumor survival. Chlorothalonil is considered to be non-genotoxic but classified as ‘likely’ to be a human carcinogen by all routes of exposure (95). It may also act as cytochrome P-450 inducer with the formation of ROS and peroxisome proliferation, which increases the subsequent risk of tumor development (160).
DBP and DEHP
Diesters of phthalic acid are commonly referred to as phthalates. These man-made chemicals are widely used in consumer products, food processing and medical applications. They are measured in residential indoor environments (indoor air and house dust) and also in foods, milk and drinking water. High-molecular-weight phthalates are used as plasticizers in the manufacturing of polyvinyl chloride and low-molecular-weight phthalates are used in making varnishes, lacquers and personal-care products. All of the phthalates have been shown to disrupt reproductive tract development in male rodents in an antiandrogenic manner (161). Phthalate compounds such as DBP, butyl benzyl phthalate and DEHP mimic the function or activity of the endogenous estrogen 17β-estradiol (E2) and bind to ERs. Interestingly, phthalates can mimic estrogen in the inhibition of tamoxifen-induced apoptosis in human breast cancer cell lines by increasing intracellular BCL-2/BAX ratio, which promotes drug resistance to the ER antagonist tamoxifen in breast cancer (162). DEHP also up-regulates the expression of antiapoptotic activating transcription factor-3 (ATF-3) and down-regulates the pro-apoptotic P53 transcription and thereby suppresses apoptotic cell death in fetal mouse genital tubercle (163). It also inhibited apoptosis of Syrian hamster embryo cells (164). DBP induces proliferation and invasiveness of ER-negative breast cancer through AhR/HDAC6/c-Myc signaling pathway (165) and induces cell proliferation of ovarian cancer cells by inducing the expression of cyclin D and cdk-4 (166), whereas butyl benzyl phthalate promotes breast cancer progression by inducing the expression of lymphoid enhancer-binding factor 1 (165). DEHP induces hepatocarcinogenesis in rodents by activating PPARα and peroxisomal genes or cell proliferation and also decreases GJIC with enhanced replicative DNA synthesis (167,168), whereas DEHP and its main metabolite mono(2-ethylhexyl) phthalate induce ROS species and activate nuclear p53 and p21 in a human prostate adenocarcinoma cell line (169).
DDVP
DDVP (2,2-dichlorovinyl dimethyl phosphate) is an organophosphate insecticide used on crops and animals, and to control household pests. It is effective as an external insecticide against flies, aphids, spider mites and caterpillar, and also as anthelmintic in the treatment of parasitic worm infections in dogs, livestock and humans (170). It acts as a cholinesterase inhibitor on the nervous systems of insects. The United States Environmental Protection Agency (USEPA) has classified DDVP as a probable carcinogen, and DDVP administration induced adenomas of the pancreatic acinar in male rats, mononuclear cell leukemia in male rats, mammary gland fibroadenomas in female rats and squamous cell papilloma of the forestomach in both male and female mice (171). DDVP is also both mutagenic and clastogenic, actions that probably involve the alkylation of DNA or protein (172,173), and it generates ROS species, which induce oxidative stress in human erythrocytes in vitro (174). It also significantly induced the levels of DNA repair enzyme, ataxia telangiectasia mutated in primary rat microglial cells (175), and it has been shown to cause cancer in mouse gastric tissues by upregulating the expression of p16, Bcl-2 and c-myc genes. DDVP induces DNA methylation in multiple tissues in an animal toxicity study. Pro-apoptotic gene silencing mediated by DNA hypermethylation causes apoptosis resistance (176) and it is the link between DDVP and cancer risk observed in some epidemiology studies (177). However, its impact on resistance to apoptosis is not entirely clear. For example, it was also reported to cause an increase in the expression of caspase-1 and TNF-α in brain tissues and intracellular caspase-3 in natural killer cells (in a dose- and time-dependent manner) inducing apoptosis (178).
Lindane
Lindane (γ-hexachlorocyclohexane) is a pesticide that has been used heavily in the past. Its long-term use and the dumping of its production waste have resulted in a widespread and persistent environmental presence. Recently, the effects of lindane, as an activator of ERα and a promoter of angiogenesis, have been investigated both in vitro and in vivo (179). It has been demonstrated that this pesticide positively influences endothelial cell proliferation and migration. Lindane strongly potentiates metalloprotease activity and nitric oxide production through the enhancement of eNOS. Lindane also exerts a cytotoxic effect on human peripheral blood lymphocytes (180) and disrupts the autophagic pathway by activating MAPK/ERK pathway. This high constitutive induction of MAPK/ERK pathways impedes the tumor suppressive function and provides a malignant advantage to tumors. Lindane disrupts the autophagic process evidenced by enlarged acidic vesicles labeled with specific autophagic vacuole maturation markers LC3, Rab7 and LAMP1, the conversion of cytosolic form of LC3-I into membrane-bound LC3-II and enhanced formation of the Bcl-xL/Beclin 1 complex.
Lindane also inhibits mitochondrial apoptotic cell death by the up-regulation of Bcl-xL, Bax down-expression, prevention of cytochrome c release and inhibition of caspase-3 and -9 activities in rat hepatocytes. So, the disruption of two pro-survival mechanisms (autophagic and apoptotic pathways) occurs in parallel with necrosis induction (181). Lindane also up-regulates antiapoptotic isoforms of protein kinase C in rat hepatocytes by increasing oxidative stress in a cytochrome P-450 (CYP)-dependent manner. Overall, these events clearly demonstrate that the acute and chronic effects of lindane in vivo with the induction of necrotic cell death and tumor promotion, respectively (182). In vivo studies demonstrated a decline in the activity of tricarboxylic acid cycle dehydrogenase enzymes with the modulation of acid phosphatase and lactic dehydrogenase in hepatocarcinogenesis induced by lindane in mice (179,180). Lindane also activates ERK1/2 and c-Jun cascades in human HaCaT keratinocytes cells, but had no effect on p38 MAPK activation. The activation of ERK1/2 results in the activation of Raf and MEK1/2 as well as activation of protein kinase C. It also stimulated ROS generation, which activated ERK and JNK cascades through ROS-dependent mechanism with no effect on MEK1/2 phosphorylation.
Linuron
Linuron (3-(3,4-dichlorophenyl)-1-methoxy-1-methylurea) is a wide-spectrum herbicide and applied to soils to control pre-emergent and post-emergent broad-leaved and annual grasses amongst cultivated crops and vegetables. It enters humans either through contaminated food or drinking water, or by dermal contact. It is an endocrine disruptor structurally related to the non-steroidal antiandrogen (androgen receptor antagonist), flutamide, which inhibits 5α-reductase enzyme. It produces Leydig cell tumors via an antiandrogenic mechanism, where sustained hypersecretion of LH and increased serum estradiol follow the disruption of hypothalamic–pituitary–testicular axis, and appears to be responsible for the development of dose-dependent increase in Leydig cell hyperplasia and adenomas (113). Linuron showed in vitro influence on the cell growth rate and GJIC on the endothelial cell line F-BAE GM 7373 and demonstrated tumor-promoting activity. The inhibition of GJIC by linuron (between the normal and pre-neoplastic cells) creates an environment in which tumor-initiated pre-neoplastic cells are isolated from several growth regulators and results in clonal expansion. Several tumor-promoting chemicals have been reported to block GJIC and thereby disrupt apoptosis (108). The loss of lymphocytes after exposure to the pesticide may also lead to a severely impaired immunological function (183).
Methoxychlor
MXC (1,1,1-trichloro-2,2-bis(4-methoxyphenyl)ethane) is a DDT-derivative OC pesticide that was developed after the ban of DDT and exhibits antiandrogenic and estrogenic activity. It disrupts prolactin secretion by inhibition of dopamine in the hypothalamus and decreases circulatory LH. MXC blocks the surge in LH and follicle-stimulating hormone secretion during the female reproductive cycle (184). MXC stimulates proliferation and human breast cancer cell growth by the up-regulation of genes that involve cell cycle (cyclin D1), and the down-regulation of genes p21 and Bax affecting G1/S transition and apoptosis, respectively, through ERα signaling (185). MXC reduces fertility in female rodents by causing ovarian atrophy and antral follicle atresia (apoptotic cell death) by inducing oxidative stress through mitochondrial production of ROS (186). MXC induced premature nuclear expression of ER gene in neonatal uterine epithelium of mice (187). MXC itself exhibits ER binding potential and the metabolism of MXC forms monohydroxy and dihydroxy metabolites exhibiting estrogenic activity. Another MXC metabolite 2,2-bis(p-hydroxyphenyl)-1,1,1-trichloroethane exhibits reproductive toxicity by binding to ERα receptor and acts as an AR antagonist (188). Chronic exposure to estrogenic chemicals such as MXC leads to persistent cell proliferation causing the formation of neoplastic lesions. MXC interact with nuclear receptors and activates either pregnane X receptor (PXR) or both PXR and constitutive androstane receptor (CAR) (189). In recent years, researches have revealed most unsuspected roles for PXR and CAR in modulating hormone, lipid and energy homeostasis as well as cancer (190). Activation of both PXR and CAR induces CYP3A, and there is a positive association between CYP3A activity, breast cancer disease genesis and lymph node metastasis (191).
Oxyfluorfen
Oxyfluorfen (2-chloro-α,α,α-trifluoro-p-tolyl 3-ethoxy-4-nitrophenyl ether) is a diphenyl-ether herbicide used to control pre-emergent and post-emergent broadleaf and grass weeds in agriculture. Mostly, handlers (mixers, loaders and applicators) are exposed during the use of liquid or granular formulations of oxyfluorfen. It is rapidly absorbed and excreted unchanged in feces and urine with little remaining in the tissues of humans. The primary toxic effects of oxyfluorfen are related to blood and liver disorders. In rodents, it inhibits protoporphyrinogen IX oxidase enzyme resulting in the inhibition of heme biosynthesis and also induces the symptoms consistent with the expression of human variegate porphyria. The USEPA has classified it as a possible human carcinogen (as an increased incidence of combined hepatocellular adenomas and carcinomas was observed in mice treated with oxyfluorfen). It has also been demonstrated to have the potential to induce mouse liver tumors by non-genotoxic means, but it is not predicted to be carcinogenic in humans (192). Toxicological studies in male mice showed expression of Cyp2b10 and Cyp4a10 transcripts, markers of CAR and PPARα nuclear receptor activation (192). PPs cause hepatomegaly, peroxisome proliferation and increased fatty acid catabolism. Chronic administration of PPs causes liver tumors in rodents (193). Chemicals that interact with PPARα are known to induce or facilitate liver tumors (194). Though the molecular mechanisms involved in PPARα-induced hepatocarcinogenesis has not been fully uncovered, recently, it has been demonstrated that PPs may induce severe liver toxicity causing mortality in mice with hepatocyte-restricted PPARα activation in the absence of ligand (VP16PPARα). Long-term exposure to PPs results in hepatocellular carcinomas in VP16PPARα mice by modulating DNA damage response signaling network especially Chek1 and its checkpoint signaling cascade causing increase in DNA synthesis, cell proliferation and apoptosis suppression (195) (Figure 1).
Cross-hallmark relationships
Given that the carcinogenicity of low-dose exposures to chemical mixtures in any given tissue will likely depend upon simultaneous instigation of several important tumor promotion mechanisms and the disruption of several important defense mechanisms, it was felt that one way of visualizing the potential synergies of combinations of chemicals could involve a thorough review of disruptive actions of each chemical across the full range of mechanisms that are known to be relevant in cancer biology. Accordingly, we undertook a cross-validation activity to illustrate the cross-hallmark relationships that are known for the target sites that we identified and to illustrate the extent that these chemicals are also known to disrupt other mechanisms that are also relevant to carcinogenesis.
These relationships are depicted in Tables 2 and 3. Target sites and chemicals that were not only relevant for resistance to cell death but also known to be relevant for other areas of cancer biology were noted as pro-carcinogenic in the areas where evidence existed. Targets and chemicals that were found to have anticarcinogenic potential in other areas of cancer biology were also highlighted where supporting evidence could be found. In instances where reports on relevant actions in other aspects of cancer biology were mixed (i.e. reports showing both pro-carcinogenic potential and anticarcinogenic potential), this was also noted. Finally, in instances where no literature support was found to document the relevance of a target site or chemical in a particular aspect of cancer’s biology, this was documented as well.
Table 2.
Cross-validation of target pathways
| Target pathways | Deregulated metabolism | Evasion of antigrowth signaling | Angiogenesis | Genetic instability | Immune system evasion | Replicative immortality | Sustained proliferative signaling | Tissue invasion and metastasis | Tumor promoting inflammation | Tumor microenvironment |
|---|---|---|---|---|---|---|---|---|---|---|
| Binding to ERα | +(100) | −(196) | 0 | 0 | 0 | +(197,198) | +(199–201) | +/−(101– 103,202,203) | +(204,205) | +(206) |
| P53 | +(207) | −(208) | −(209) | −(210,211) | +(212,213) | +(214–217) | −(218) | −(219–222) | +(223–225) | +(226) |
| ErbB-2/HER-2 tyrosine kinase | 0 | −(227) | 0 | +(138,228) | 0 | +(229,230) | +(216) | +(231–234) | +(235,236) | +(237) |
| ERK/MAPK | +(196,238,239) | +(240,241) | +(242) | −(243,244) | −(245,246) | +(247) | +(154) | +(248–252) | +(253) | +(254) |
| MAP kinase | +(255) | +(240,241,256) | +(257) | +(138,258) | +/−(246,259) | +(260–262) | +(138) | +(248–250) | +(263,264) | +(254) |
| P16/p53 | 0 | 0 | −(265) | −(266,267) | +(268,269) | +(214,270) | − (271) | − (267,272–275) | +(276) | +(277) |
| Bcl-2/p53 | 0 | +/−(278–287) | +(288,289) | −(290) | +(291–293) | +(229,294) | −(271) | +(289,295,296) | +(297) | +(298) |
| PPAR-α | +(299,300) | −(301–303) | −(304) | 0 | 0 | −(301) | +/−(167,192) | 0 | +(305,306) | +(307) |
| GJIC | 0 | 0 | 0 | 0 | 0 | 0 | −(308) | +/−(309–311) | +(312) | +(313) |
| Hypersecretion of LH by gonadotroph cells in pituitary gland | 0 | 0 | 0 | 0 | 0 | 0 | −(314) | +(315,316) | +(317,318) | 0 |
Target pathways resistance to cell death were cross-validated for effects in other cancer hallmark pathways. Targets that were found to have opposing actions in a particular hallmark (i.e. anticarcinogenic) were denoted using ‘−’, whereas targets that were found to have promoting actions in a particular hallmark (i.e. carcinogenic) were denoted using ‘+’. In instances where reports on relevant actions in other hallmarks were mixed (i.e. reports showing both pro-carcinogenic potential and anticarcinogenic potential), the symbols ‘ +/−’ were used. Finally, in instances where no literature support was found to document the relevance of a target in a particular aspect of cancer’s biology, we denoted using ‘0’.
Table 3.
Cross-validation of disruptors
| Prototypical disruptors | Deregulated metabolism | Evasion of antigrowth signaling | Angiogenesis | Genetic instability | Immune system evasion | Replicative immortality | Sustained proliferative signaling | Tissue invasion and metastasis | Tumor promoting inflammation | Tumor microenvironment |
|---|---|---|---|---|---|---|---|---|---|---|
| BPA | +(319–322) | +(152) | +(323) | +(155,324,325) | 0 | + | +(154) | +(326–328) | +(329) | 0 |
| DBP | All via hormone disruption | 0 | +(165) | +(330) | 0 | 0 | +(165,166) | 0 | +(331) | 0 |
| Chlorothalonil | 0 | +(332) | 0 | +(138,333) | 0 | 0 | +(138) | 0 | +(158) | 0 |
| Lindane | +(179,180,334) | 0 | +(335) | +(243,336) | 0 | 0 | +(199,200) | 0 | +(155) | 0 |
| DDVP | +(337) | +(338) | 0 | +(339,340) | 0 | 0 | 0 | 0 | +(338,341) | 0 |
| MXC | 0 | 0 | 0 | +(342,343) | 0 | 0 | +(201) | 0 | +(344,345) | 0 |
| Oxyfluorfen | +(192) | 0 | + | 0 | 0 | 0 | +(192) | 0 | +(192) | 0 |
| DEHP | +(168,346) | +(169) | 0 | 0 | 0 | 0 | +(167) | 0 | +(347) | 0 |
| Linuronx | +(348) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | +(349,350) | 0 |
Disruptors of resistance to cell death were cross-validated for effects in other cancer hallmark pathways. Disruptors that were found to have opposing actions in a particular hallmark (i.e. anticarcinogenic) were denoted using ‘−’, whereas disruptors that were found to have promoting actions in a particular hallmark (i.e. carcinogenic) were denoted using ‘+’. In instances where reports on relevant actions in other hallmarks were mixed (i.e. reports showing both pro-carcinogenic potential and anticarcinogenic potential), the symbol ‘+/−’ were used. Finally, in instances where no literature support was found to document the relevance of a chemical in a particular aspect of cancer’s biology, were denoted using ‘0’.
Perspective
Cell death is intrinsically connected to different kinds of biological damage caused by environmental pollutants. For example, chemicals that promote genetic instability usually trigger apoptosis (a defensive mechanism intended to prevent functionally compromised cells from harming the system). So, hypothetically speaking, exogenous exposures to combinations of disruptive chemicals that act on the mechanisms described previously (conferring resistance to cell death) could exacerbate the effects of unrepaired cellular damage. The potential for dysfunction in this key safeguard is therefore an important consideration because this sort of genetic instability increases the overall probability of damage and mutations that could support the emergence of a fully immortalized cellular phenotype.
Past studies have indicated that several cancer hallmark processes are impacted by a variety of chemical carcinogens and oncogenes (95,96), and there are various agencies worldwide such as USEPA and International Agency for Research on Cancer working on classifying the environmental chemicals based on the carcinogenic potential with the purpose of protecting human health. However, the effects of environmental chemical mixtures have had much less attention, and in this project, we have looked specifically at a number of prototypical chemicals with disruptive potential that is relevant to apoptosis. The concern that we have relates to the possibility that individual chemicals that are disruptive of these key mechanisms and pathways may have the potential to contribute to environmental carcinogenesis without being carcinogenic per se.
For example, as we noted previously, BPA strikingly impairs TP53 activity and its downstream targets, cell cycle regulators, p21WAF1 and RB, or pro-apoptotic BAX, thereby enhancing the threshold for apoptosis (152). BPA activates mTOR pathway and enhances cell growth and proliferation (351). It also activates PPAR-α (which suppresses apoptosis and enhances cell proliferation), and it inhibits GJIC through a modulation of the gating of gap junction channels, which contributes to tumor formation by increasing intracellular signaling and enhancing proliferation (352). And BPA influences cell proliferation and migration by GPER/EGFR/ERK pathway. But despite a significant and growing body of literature that has documented all of these mechanistic contributions, researchers and regulators have had considerable difficulty proving whether or not BPA has carcinogenic effects in humans (319).
From this perspective, it seems that the longstanding focus on the carcinogenic potential of individual chemicals is really too narrow (given the wide range of environmental chemical exposures that we now face). Instead, it would seem more prudent to focus on mechanistic contributions and anticipated synergies of mixtures of individual constituents that have been shown to individually exert disruptive effects on hallmark cancer processes (at dose levels that are environmentally relevant). In this case, BPA is a good example because it is ubiquitous in the environment and it has also been shown to exert its effects on TP53 at low-dose levels (353). So, even if it cannot be definitively categorized as a human carcinogen, it appears to have potential to play a contributing role in environmental carcinogenesis. Future research will therefore need to illustrate how chemicals that have the potential to produce this sort of a disruptive effect can be experimentally combined with other environmental chemicals that disrupt other hallmark processes to enable carcinogenesis.
We fully recognize that much of the evidence in the toxicological literature that documents the disruptive actions of these chemicals has been produced under a wide range of differing experimental circumstances, and it is not our intent to jump to conclusions about the role that aggregated exposures to mixtures of these chemicals might play in environmental carcinogenesis. But it is our contention that the ubiquitous presence of these (and other) chemicals with disruptive potential needs to be carefully considered, even if these chemicals are not individually carcinogenic. Moreover, researchers who investigate this possibility will also need to consider other mechanisms that are affected by these individual chemicals as well (see Tables 2 and 3). In some cases, dose–response research may reveal thresholds that make these actions unlikely at levels of exposure that are seen in the environment, but to the extent that low-dose effects can been found, these additional disruptive effects may also be important factors to consider.
Conclusions
The disruption of the mechanisms that regulate cell death is fundamentally important to our understanding of environmental carcinogenesis. The enablement of an immortalized cellular phenotype can only occur when many important safeguards have been bypassed. It therefore appears reasonable to consider the effects of ubiquitous environmental chemicals that have been shown to disrupt cell death as it is a very important safeguard. Although a considerable amount of research has been done to characterize the effects that many chemicals have on the mechanisms that are relevant for normal cell death, very little attention has been given to the combined effects of this chemicals on this hallmark of cancer, or the role that these sorts of disruptions at the mechanistic level might serve to contribute to environmental carcinogenesis. In this review, we have identified a number of important targets that are highly relevant for cell death and we have identified a number of ubiquitous environmental chemicals that have been shown to act disruptively on these targets. Future research is needed that looks carefully the role of these prototypical disruptors and other disruptive chemicals that can act on these same mechanisms at levels of exposure that are commonly seen in the environment. Regulators who now focus solely on determining the carcinogenic potential of individual chemicals would be well served to additionally consider the synergies that might occur when chemicals that are disruptive at the mechanistic level are combined with other disruptive chemicals (i.e. those that can enable other complementary processes that are similarly instrumental and enabling in carcinogenesis). To anticipate the sorts of synergies that might be produced, the pleiotropic nature of these chemicals will need to be considered as well. Individual chemicals may produce a range of disruptive effects that are relevant for a multitude of mechanisms, yet individual constituents in any given combination of exposures may not need to be carcinogenic per se. Combinations of these chemicals may produce foundational effects that enable carcinogenesis, so progress in our understanding of this potential will help us to refine our approach to cancer risk assessment.
Funding
This research was supported by Yeungnam University research grants in 2014 to Hyun Ho Park. Po Sing Leung was supported by the Health and Medical Research Fund of Food and Health Bureau, Hong Kong Special Administrative Region, Ref. No: 10110021. Yon Rojanasakul was supported by NIH (R01-ES022968) and NSF (CBET-1434503). Rita Dornetshuber-Fleiss was supported by the Austrian Science Fund (FWF, project number T 451-B18) and the Johanna Mahlke, geb.-Obermann-Stiftung. Clement Yedjou was supported by National Institutes of Health (Grant No. NIMHD-G12MD007581, Grant No. NIGMS- P20GM103476).
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AIF
apoptosis-inducing factor
- APAF
apoptosis-activating factor-1
- BH
BCL-2 homology
- BPA
bisphenol A
- CAR
constitutive androstane receptor
- CDK
cyclin-dependent kinase
- CSCs
cancer stem cells
- DBP
dibutyl phthalate
- DD
death domain
- DDT
dichlorodiphenyltrichloroethane
- DEHP
diethylhexyl phthalate
- DISC
death-inducing signaling complex
- 4EBP1
4E binding protein 1
- EGFR
epidermal growth factor receptor
- ERK
extracellular signal-regulated kinase
- FADD
Fas-associated death domain protein
- FLIP
FADD-like apoptosis regulator
- GJIC
gap junctional intracellular communication
- IAP
inhibitor of apoptosis protein
- JNK
C-Jun N-terminal kinase
- LH
luteinizing hormone
- MDM2
murine double minute 2
- mRNA
messenger RNA
- mtDNA
mitochondrial DNA
- mTOR
mammalian target of rapamycin
- MXC
methoxychlor
- NADPH
nicotinamide adenine dinucleotide phosphate
- NF-κB
nuclear factor-κB
- PI3K
phosphoinositide 3-kinase
- PIDD
TP53-induced protein with death domain
- PP
peroxisome proliferators
- PPAR-α
peroxisome proliferator-activated receptor-α
- PTEN
phosphatase and tensin homolog
- PXR
pregnane X receptor
- RAIDD
RIP-associated Ich-1/Ced-3-homologue protein with a death domain
- RB
retinoblastoma
- RIP
receptor-interacting protein
- ROS
reactive oxygen species
- RTK
receptor tyrosine kinase
- SMAC
second mitochondrial activator of caspases
- TGF-β
transforming growth factor-β
- TNF
tumor necrosis factor
- TP
tumor protein
- TRADD
TNF receptor-1-associated death domain
- TRAIL
TNF-related apoptosis apoptosis-inducing ligand receptor
- XIAP
X-linked inhibitor of apoptosis protein
References
- 1. Nicholson D.W., et al. (2003) Apoptosis. Life and death decisions. Science, 299, 214–215. [DOI] [PubMed] [Google Scholar]
- 2. Fridman J.S., et al. (2003) Control of apoptosis by p53. Oncogene, 22, 9030–9040. [DOI] [PubMed] [Google Scholar]
- 3. Shortt J., et al. (2012) Oncogenes in cell survival and cell death. Cold Spring Harb. Perspect. Biol., 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Orr B., et al. (2013) A double-edged sword: how oncogenes and tumor suppressor genes can contribute to chromosomal instability. Front. Oncol., 3, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Croce C.M. (2008) Oncogenes and cancer. N. Engl. J. Med., 358, 502–511. [DOI] [PubMed] [Google Scholar]
- 6. Hetz C.A., et al. (2005) Beyond apoptosis: nonapoptotic cell death in physiology and disease. Biochem. Cell Biol., 83, 579–588. [DOI] [PubMed] [Google Scholar]
- 7. de Bruin E.C., et al. (2008) Apoptosis and non-apoptotic deaths in cancer development and treatment response. Cancer Treat. Rev., 34, 737–749. [DOI] [PubMed] [Google Scholar]
- 8. Elmore S. (2007) Apoptosis: a review of programmed cell death. Toxicol. Pathol., 35, 495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Falschlehner C., et al. (2007) TRAIL signalling: decisions between life and death. Int. J. Biochem. Cell Biol., 39, 1462–1475. [DOI] [PubMed] [Google Scholar]
- 10. Suh D.H., et al. (2013) Mitochondrial permeability transition pore as a selective target for anti-cancer therapy. Front. Oncol., 3, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mayer B., et al. (2003) Mitochondrial regulation of apoptosis. News Physiol. Sci., 18, 89–94. [DOI] [PubMed] [Google Scholar]
- 12. Adams J.M. (2003) Ways of dying: multiple pathways to apoptosis. Genes Dev., 17, 2481–2495. [DOI] [PubMed] [Google Scholar]
- 13. Keinan N., et al. (2010) Oligomerization of the mitochondrial protein voltage-dependent anion channel is coupled to the induction of apoptosis. Mol. Cell. Biol., 30, 5698–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vander Heiden M.G., et al. (2001) Bcl-xL promotes the open configuration of the voltage-dependent anion channel and metabolite passage through the outer mitochondrial membrane. J. Biol. Chem., 276, 19414–19419. [DOI] [PubMed] [Google Scholar]
- 15. Bröker L.E., et al. (2005) Cell death independent of caspases: a review. Clin. Cancer Res., 11, 3155–3162. [DOI] [PubMed] [Google Scholar]
- 16. Vermeulen K., et al. (2005) Apoptosis: mechanisms and relevance in cancer. Ann. Hematol., 84, 627–639. [DOI] [PubMed] [Google Scholar]
- 17. Yang Y.L., et al. (2000) The IAP family: endogenous caspase inhibitors with multiple biological activities. Cell Res., 10, 169–177. [DOI] [PubMed] [Google Scholar]
- 18. Carew J.S., et al. (2002) Mitochondrial defects in cancer. Mol. Cancer, 1, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park H.H., et al. (2007) Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell, 128, 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim I.R., et al. (2009) DNA damage- and stress-induced apoptosis occurs independently of PIDD. Apoptosis, 14, 1039–1049. [DOI] [PubMed] [Google Scholar]
- 21. Park H.H. (2011) Structural analyses of death domains and their interactions. Apoptosis, 16, 209–220. [DOI] [PubMed] [Google Scholar]
- 22. Van Cruchten S., et al. (2002) Morphological and biochemical aspects of apoptosis, oncosis and necrosis. Anat. Histol. Embryol., 31, 214–223. [DOI] [PubMed] [Google Scholar]
- 23. Jäättelä M., et al. (2003) Caspase-independent cell death in T lymphocytes. Nat. Immunol., 4, 416–423. [DOI] [PubMed] [Google Scholar]
- 24. Chautan M., et al. (1999) Interdigital cell death can occur through a necrotic and caspase-independent pathway. Curr. Biol., 9, 967–970. [DOI] [PubMed] [Google Scholar]
- 25. Galluzzi L., et al. (2008) Necroptosis: a specialized pathway of programmed necrosis. Cell, 135, 1161–1163. [DOI] [PubMed] [Google Scholar]
- 26. Kaczmarek A., et al. (2013) Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity, 38, 209–223. [DOI] [PubMed] [Google Scholar]
- 27. Mehrpour M., et al. (2010) Autophagy in health and disease. 1. Regulation and significance of autophagy: an overview. Am. J. Physiol. Cell Physiol., 298, C776–C785. [DOI] [PubMed] [Google Scholar]
- 28. Degenhardt K., et al. (2006) Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell, 10, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Glick D., et al. (2010) Autophagy: cellular and molecular mechanisms. J. Pathol., 221, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. He C., et al. (2009) Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet., 43, 67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karantza-Wadsworth V., et al. (2007) Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev., 21, 1621–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mathew R., et al. (2007) Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev., 21, 1367–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mathew R., et al. (2009) Autophagy suppresses tumorigenesis through elimination of p62. Cell, 137, 1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aita V.M., et al. (1999) Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics, 59, 59–65. [DOI] [PubMed] [Google Scholar]
- 35. Qu X., et al. (2003) Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest., 112, 1809–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu Y.L., et al. (2012) Tumor cell autophagy as an adaptive response mediating resistance to treatments such as antiangiogenic therapy. Cancer Res., 72, 4294–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fesik S.W. (2005) Promoting apoptosis as a strategy for cancer drug discovery. Nat. Rev. Cancer, 5, 876–885. [DOI] [PubMed] [Google Scholar]
- 38. Lowe S.W., et al. (2000) Apoptosis in cancer. Carcinogenesis, 21, 485–495. [DOI] [PubMed] [Google Scholar]
- 39. Plati J., et al. (2011) Apoptotic cell signaling in cancer progression and therapy. Integr. Biol. (Camb)., 3, 279–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dewey W.C., et al. (1995) Radiation-induced apoptosis: relevance to radiotherapy. Int. J. Radiat. Oncol. Biol. Phys., 33, 781–796. [DOI] [PubMed] [Google Scholar]
- 41. Bal W., et al. (2002) Induction of oxidative DNA damage by carcinogenic metals. Toxicol. Lett., 127, 55–62. [DOI] [PubMed] [Google Scholar]
- 42. Johnstone R.W., et al. (2002) Apoptosis: a link between cancer genetics and chemotherapy. Cell, 108, 153–164. [DOI] [PubMed] [Google Scholar]
- 43. Stenner-Liewen F., et al. (2003) Apoptosis and cancer: basic mechanisms and therapeutic opportunities in the postgenomic era. Cancer Res., 63. [Google Scholar]
- 44. Dalerba P., et al. (2007) Cancer stem cells: models and concepts. Annu. Rev. Med., 58, 267–284. [DOI] [PubMed] [Google Scholar]
- 45. Levina V., et al. (2008) Drug-selected human lung cancer stem cells: cytokine network, tumorigenic and metastatic properties. PLoS One, 3, e3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. D’Andrea F.P. (2012) Intrinsic radiation resistance of mesenchymal cancer stem cells and implications for treatment response in a murine sarcoma model. Dan. Med. J., 59, B4388. [PubMed] [Google Scholar]
- 47. Kruyt F.A., et al. (2010) Apoptosis and cancer stem cells: implications for apoptosis targeted therapy. Biochem. Pharmacol., 80, 423–430. [DOI] [PubMed] [Google Scholar]
- 48. Frisch S.M., et al. (2013) Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. J. Cell Sci., 126(Pt 1), 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu G., et al. (2006) Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer, 5, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zobalova R., et al. (2008) CD133-positive cells are resistant to TRAIL due to up-regulation of FLIP. Biochem. Biophys. Res. Commun., 373, 567–571. [DOI] [PubMed] [Google Scholar]
- 51. Ouyang L., et al. (2012) Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif., 45, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Strasser A., et al. (2011) Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J., 30, 3667–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fiandalo M.V., et al. (2012) Caspase control: protagonists of cancer cell apoptosis. Exp. Oncol., 34, 165–175. [PMC free article] [PubMed] [Google Scholar]
- 54. Reubold T.F., et al. (2012) A molecular view on signal transduction by the apoptosome. Cell Signal., 24, 1420–1425. [DOI] [PubMed] [Google Scholar]
- 55. Galluzzi L., et al. (2012) Mitochondria: master regulators of danger signalling. Nat. Rev. Mol. Cell Biol., 13, 780–788. [DOI] [PubMed] [Google Scholar]
- 56. de Almagro M.C., et al. (2012) The inhibitor of apoptosis (IAP) proteins are critical regulators of signaling pathways and targets for anti-cancer therapy. Exp. Oncol., 34, 200–211. [PubMed] [Google Scholar]
- 57. Harvey K.F., et al. (2013) The Hippo pathway and human cancer. Nat. Rev. Cancer, 13, 246–257. [DOI] [PubMed] [Google Scholar]
- 58. Sherr C.J., et al. (2002) The RB and p53 pathways in cancer. Cancer Cell, 2, 103–112. [DOI] [PubMed] [Google Scholar]
- 59. Galluzzi L., et al. (2011) Mitochondrial liaisons of p53. Antioxid. Redox Signal., 15, 1691–1714. [DOI] [PubMed] [Google Scholar]
- 60. Matoba S., et al. (2006) p53 regulates mitochondrial respiration. Science, 312, 1650–1653. [DOI] [PubMed] [Google Scholar]
- 61. Kim J.Y., et al. (2004) BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia-inducible factor 1alpha. J. Exp. Med., 199, 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yu J., et al. (2008) PUMA, a potent killer with or without p53. Oncogene, 27(suppl 1), S71–S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hikisz P., et al. (2012) PUMA, a critical mediator of cell death–one decade on from its discovery. Cell. Mol. Biol. Lett., 17, 646–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Icard P., et al. (2012) A global view of the biochemical pathways involved in the regulation of the metabolism of cancer cells. Biochim. Biophys. Acta, 1826, 423–433. [DOI] [PubMed] [Google Scholar]
- 65. Zhao S., et al. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science, 327, 1000–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yi C.H., et al. (2011) Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell, 146, 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gnoni G.V., et al. (2009) The mitochondrial citrate carrier: metabolic role and regulation of its activity and expression. IUBMB Life, 61, 987–994. [DOI] [PubMed] [Google Scholar]
- 68. Vaughn A.E., et al. (2008) Glucose metabolism inhibits apoptosis in neurons and cancer cells by redox inactivation of cytochrome c. Nat. Cell Biol., 10, 1477–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Delbridge A.R., et al. (2012) The role of the apoptotic machinery in tumor suppression. Cold Spring Harb. Perspect. Biol., 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Labbé D.P., et al. (2012) Protein tyrosine phosphatases in cancer: friends and foes! Prog. Mol. Biol. Transl. Sci., 106, 253–306. [DOI] [PubMed] [Google Scholar]
- 71. Sayers T.J. (2011) Targeting the extrinsic apoptosis signaling pathway for cancer therapy. Cancer Immunol. Immunother., 60, 1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Frisch S.M., et al. (1996) A role for Jun-N-terminal kinase in anoikis; suppression by bcl-2 and crmA. J. Cell Biol., 135, 1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schottenfeld D., et al. (2013) Current perspective on the global and United States cancer burden attributable to lifestyle and environmental risk factors. Annu. Rev. Public Health, 34, 97–117. [DOI] [PubMed] [Google Scholar]
- 74. Lee E.Y., et al. (2010) Oncogenes and tumor suppressor genes. Cold Spring Harb. Perspect. Biol., 2, a003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Reddy K.L., et al. (2013) Higher order chromatin organization in cancer. Semin. Cancer Biol., 23, 109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Iorio M.V., et al. (2012) microRNA involvement in human cancer. Carcinogenesis, 33, 1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vousden K.H., et al. (2002) Live or let die: the cell’s response to p53. Nat. Rev. Cancer, 2, 594–604. [DOI] [PubMed] [Google Scholar]
- 78. Murray-Zmijewski F., et al. (2006) p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ., 13, 962–972. [DOI] [PubMed] [Google Scholar]
- 79. Rosenbluth J.M., et al. (2008) The jury is in: p73 is a tumor suppressor after all. Genes Dev., 22, 2591–2595. [DOI] [PubMed] [Google Scholar]
- 80. Flores E.R., et al. (2005) Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell, 7, 363–373. [DOI] [PubMed] [Google Scholar]
- 81. Maddocks O.D., et al. (2011) Metabolic regulation by p53. J. Mol. Med. (Berl), 89, 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Huang Y., et al. (2011) Phospho-ΔNp63α is a key regulator of the cisplatin-induced microRNAome in cancer cells. Cell Death Differ., 18, 1220–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sen T., et al. (2011) DeltaNp63alpha confers tumor cell resistance to cisplatin through the AKT1 transcriptional regulation. Cancer Res., 71, 1167–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ory B., et al. (2011) A microRNA-dependent circuit controlling p63/p73 homeostasis: p53 family cross-talk meets therapeutic opportunity. Oncotarget, 2, 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Xi S., et al. (2010) Cigarette smoke induces C/EBP-β-mediated activation of miR-31 in normal human respiratory epithelia and lung cancer cells. PLoS One, 5, e13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ewing S.J., et al. (2008) C/EBPbeta represses p53 to promote cell survival downstream of DNA damage independent of oncogenic Ras and p19(Arf). Cell Death Differ., 15, 1734–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Balmain A., et al. (1984) Activation of the mouse cellular Harvey-ras gene in chemically induced benign skin papillomas. Nature, 307, 658–660. [DOI] [PubMed] [Google Scholar]
- 88. Alguacil J., et al. (2002) Occupational exposure to organic solvents and K-ras mutations in exocrine pancreatic cancer. Carcinogenesis, 23, 101–106. [DOI] [PubMed] [Google Scholar]
- 89. Zarbl H., et al. (1985) Direct mutagenesis of Ha-ras-1 oncogenes by N-nitroso-N-methylurea during initiation of mammary carcinogenesis in rats. Nature, 315, 382–385. [DOI] [PubMed] [Google Scholar]
- 90. Shishodia S., et al. (2003) Curcumin (diferuloylmethane) down-regulates cigarette smoke-induced NF-kappaB activation through inhibition of IkappaBalpha kinase in human lung epithelial cells: correlation with suppression of COX-2, MMP-9 and cyclin D1. Carcinogenesis, 24, 1269–1279. [DOI] [PubMed] [Google Scholar]
- 91. Srivastava S.K., et al. (2004) Cell cycle arrest, apoptosis induction and inhibition of nuclear factor kappa B activation in anti-proliferative activity of benzyl isothiocyanate against human pancreatic cancer cells. Carcinogenesis, 25, 1701–1709. [DOI] [PubMed] [Google Scholar]
- 92. Romano S., et al. (2011) FKBP51 and the NF-κB regulatory pathway in cancer. Curr. Opin. Pharmacol., 11, 288–293. [DOI] [PubMed] [Google Scholar]
- 93. Hanahan D., et al. (2000) The hallmarks of cancer. Cell, 100, 57–70. [DOI] [PubMed] [Google Scholar]
- 94. Trosko J.E., et al. (2010) A paradigm shift is required for the risk assessment of potential human health after exposure to low level chemical exposures: a response to the toxicity testing in the 21st century report. Int. J. Toxicol., 29, 344–357. [DOI] [PubMed] [Google Scholar]
- 95. Kleinstreuer N.C., et al. (2013) In vitro perturbations of targets in cancer hallmark processes predict rodent chemical carcinogenesis. Toxicol. Sci., 131, 40–55. [DOI] [PubMed] [Google Scholar]
- 96. Kortenkamp A. (2014) Low dose mixture effects of endocrine disrupters and their implications for regulatory thresholds in chemical risk assessment. Curr. Opin. Pharmacol., 19, 105–111. [DOI] [PubMed] [Google Scholar]
- 97. Fucic A., et al. (2012) Environmental exposure to xenoestrogens and oestrogen related cancers: reproductive system, breast, lung, kidney, pancreas, and brain. Environ. Health, 11(suppl 1), S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bailey S.T., et al. (2012) Estrogen receptor prevents p53-dependent apoptosis in breast cancer. Proc. Natl Acad. Sci. USA, 109, 18060–18065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sayeed A., et al. (2007) Estrogen receptor alpha inhibits p53-mediated transcriptional repression: implications for the regulation of apoptosis. Cancer Res., 67, 7746–7755. [DOI] [PubMed] [Google Scholar]
- 100. Al-Dhaheri M., et al. (2011) CARM1 is an important determinant of ERα-dependent breast cancer cell differentiation and proliferation in breast cancer cells. Cancer Res., 71, 2118–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wik E., et al. (2013) Lack of estrogen receptor-α is associated with epithelial-mesenchymal transition and PI3K alterations in endometrial carcinoma. Clin. Cancer Res., 19, 1094–1105. [DOI] [PubMed] [Google Scholar]
- 102. Dhasarathy A., et al. (2007) The transcription factor snail mediates epithelial to mesenchymal transitions by repression of estrogen receptor-alpha. Mol. Endocrinol., 21, 2907–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Huet G., et al. (2008) Loss of E-cadherin-mediated cell contacts reduces estrogen receptor alpha (ER alpha) transcriptional efficiency by affecting the respective contribution exerted by AF1 and AF2 transactivation functions. Biochem. Biophys. Res. Commun., 365, 304–309. [DOI] [PubMed] [Google Scholar]
- 104. Rivedal E., et al. (2005) Connexin43 synthesis, phosphorylation, and degradation in regulation of transient inhibition of gap junction intercellular communication by the phorbol ester TPA in rat liver epithelial cells. Exp. Cell Res., 302, 143–152. [DOI] [PubMed] [Google Scholar]
- 105. Kang K.S., et al. (1996) Inhibition of gap junctional intercellular communication in normal human breast epithelial cells after treatment with pesticides, PCBs, and PBBs, alone or in mixtures. Environ. Health Perspect., 104, 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Klaunig J.E., et al. (1990) Gap-junctional intercellular communication and murine hepatic carcinogenesis. Prog. Clin. Biol. Res., 331, 277–291. [PubMed] [Google Scholar]
- 107. Carette D., et al. (2014) Connexin a check-point component of cell apoptosis in normal and physiopathological conditions. Biochimie, 101, 1–9. [DOI] [PubMed] [Google Scholar]
- 108. Trosko J.E., et al. (1994) The role of modulated gap junctional intercellular communication in epigenetic toxicology. Risk Anal., 14, 303–312. [DOI] [PubMed] [Google Scholar]
- 109. Chang W.W., et al. (2013) Salmonella enhance chemosensitivity in tumor through connexin 43 upregulation. Int. J. Cancer, 133, 1926–1935. [DOI] [PubMed] [Google Scholar]
- 110. Abu Aboud O., et al. (2013) Inhibition of PPARα induces cell cycle arrest and apoptosis, and synergizes with glycolysis inhibition in kidney cancer cells. PLoS One, 8, e71115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Klaunig J.E., et al. (2003) PPARalpha agonist-induced rodent tumors: modes of action and human relevance. Crit. Rev. Toxicol., 33, 655–780. [DOI] [PubMed] [Google Scholar]
- 112. Roberts R.A. (2002) Evidence for cross talk between PPARalpha and p38 MAP kinase. Toxicol. Sci., 68, 270–274. [DOI] [PubMed] [Google Scholar]
- 113. Cook J.C., et al. (1993) Investigation of a mechanism for Leydig cell tumorigenesis by linuron in rats. Toxicol. Appl. Pharmacol., 119, 195–204. [DOI] [PubMed] [Google Scholar]
- 114. Zhang Z., et al. (2011) Luteinizing hormone upregulates survivin and inhibits apoptosis in ovarian epithelial tumors. Eur. J. Obstet. Gynecol. Reprod. Biol., 155, 69–74. [DOI] [PubMed] [Google Scholar]
- 115. Calaf G.M., et al. (2009) Organophosphorous pesticides and estrogen induce transformation of breast cells affecting p53 and c-Ha-ras genes. Int. J. Oncol., 35, 1061–1068. [DOI] [PubMed] [Google Scholar]
- 116. Petitjean A., et al. (2007) TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene, 26, 2157–2165. [DOI] [PubMed] [Google Scholar]
- 117. Lu Y., et al. (2012) Inhibition of breast tumor cell growth by ectopic expression of p16/INK4A via combined effects of cell cycle arrest, senescence and apoptotic induction, and angiogenesis inhibition. J. Cancer, 3, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ruas M., et al. (1998) The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim. Biophys. Acta, 1378, F115–F177. [DOI] [PubMed] [Google Scholar]
- 119. Geisler S.A., et al. (2002) p16 and p53 Protein expression as prognostic indicators of survival and disease recurrence from head and neck cancer. Clin. Cancer Res., 8, 3445–3453. [PubMed] [Google Scholar]
- 120. Jiang M., et al. (2003) Bcl-2 constitutively suppresses p53-dependent apoptosis in colorectal cancer cells. Genes Dev., 17, 832–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Schwandner O., et al. (2000) p53 and Bcl-2 as significant predictors of recurrence and survival in rectal cancer. Eur. J. Cancer, 36, 348–356. [DOI] [PubMed] [Google Scholar]
- 122. Miliaras S., et al. (2011) The role of P53 and Bcl-2 proteins in 7, 12-dimethylbenz-(a)-anthracene-induced tumor growth. J. Biol. Regul. Homeost. Agents, 25, 359–364. [PubMed] [Google Scholar]
- 123. Centelles J.J. (2012) General aspects of colorectal cancer. ISRN Oncol., 2012, 139268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Roskoski R., Jr (2014) The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res., 79, 34–74. [DOI] [PubMed] [Google Scholar]
- 125. Chen F.L., et al. (2008) Acquired resistance to small molecule ErbB2 tyrosine kinase inhibitors. Clin. Cancer Res., 14, 6730–6734. [DOI] [PubMed] [Google Scholar]
- 126. Willems L., et al. (2012) PI3K and mTOR signaling pathways in cancer: new data on targeted therapies. Curr. Oncol. Rep., 14, 129–138. [DOI] [PubMed] [Google Scholar]
- 127. Laplante M., et al. (2009) mTOR signaling at a glance. J. Cell Sci., 122(Pt 20), 3589–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Graff J.R., et al. (2008) Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res., 68, 631–634. [DOI] [PubMed] [Google Scholar]
- 129. Jia Y., et al. (2012) Cap-dependent translation initiation factor eIF4E: an emerging anticancer drug target. Med. Res. Rev., 32, 786–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Blume-Jensen P., et al. (1998) The kit receptor promotes cell survival via activation of PI 3-kinase and subsequent Akt-mediated phosphorylation of Bad on Ser136. Curr. Biol., 8, 779–782. [DOI] [PubMed] [Google Scholar]
- 131. Downward J. (2004) PI 3-kinase, Akt and cell survival. Semin. Cell Dev. Biol., 15, 177–182. [DOI] [PubMed] [Google Scholar]
- 132. Nicholson K.M., et al. (2002) The protein kinase B/Akt signalling pathway in human malignancy. Cell. Signal., 14, 381–395. [DOI] [PubMed] [Google Scholar]
- 133. Romashkova J.A., et al. (1999) NF-kappaB is a target of AKT in anti-apoptotic PDGF signalling. Nature, 401, 86–90. [DOI] [PubMed] [Google Scholar]
- 134. Cardone M.H., et al. (1998) Regulation of cell death protease caspase-9 by phosphorylation. Science, 282, 1318–1321. [DOI] [PubMed] [Google Scholar]
- 135. Maiese K., et al. (2012) Targeting disease through novel pathways of apoptosis and autophagy. Expert Opin. Ther. Targets, 16, 1203–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Tsukada M., et al. (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett., 333, 169–174. [DOI] [PubMed] [Google Scholar]
- 137. Pastor M.D., et al. (2009) mTOR/S6 kinase pathway contributes to astrocyte survival during ischemia. J. Biol. Chem., 284, 22067–22078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Tessier D.M., et al. (2001) Increased ErbB-2 tyrosine kinase activity, MAPK phosphorylation, and cell proliferation in the prostate cancer cell line LNCaP following treatment by select pesticides. Toxicol. Sci., 60, 38–43. [DOI] [PubMed] [Google Scholar]
- 139. Zhang W., et al. (2002) MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res., 12, 9–18. [DOI] [PubMed] [Google Scholar]
- 140. Lim J.H., et al. (2012) Rottlerin induces apoptosis of HT29 colon carcinoma cells through NAG-1 upregulation via an ERK and p38 MAPK-dependent and PKC δ-independent mechanism. Chem. Biol. Interact., 197, 1–7. [DOI] [PubMed] [Google Scholar]
- 141. Gan L., et al. (2011) Resistance to docetaxel-induced apoptosis in prostate cancer cells by p38/p53/p21 signaling. Prostate, 71, 1158–1166. [DOI] [PubMed] [Google Scholar]
- 142. Salim H., et al. (2012) miRNA-214 modulates radiotherapy response of non-small cell lung cancer cells through regulation of p38MAPK, apoptosis and senescence. Br. J. Cancer, 107, 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hu C., et al. (2012) E Platinum, a newly synthesized platinum compound, induces autophagy via inhibiting phosphorylation of mTOR in gastric carcinoma BGC-823 cells. Toxicol. Lett., 210, 78–86. [DOI] [PubMed] [Google Scholar]
- 144. Sui X., et al. (2014) p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett., 344, 174–179. [DOI] [PubMed] [Google Scholar]
- 145. Dhillon A.S., et al. (2007) MAP kinase signalling pathways in cancer. Oncogene, 26, 3279–3290. [DOI] [PubMed] [Google Scholar]
- 146. Carmona-Cuenca I., et al. (2008) Upregulation of the NADPH oxidase NOX4 by TGF-beta in hepatocytes is required for its pro-apoptotic activity. J. Hepatol., 49, 965–976. [DOI] [PubMed] [Google Scholar]
- 147. Caja L., et al. (2009) Overactivation of the MEK/ERK pathway in liver tumor cells confers resistance to TGF-{beta}-induced cell death through impairing up-regulation of the NADPH oxidase NOX4. Cancer Res., 69, 7595–7602. [DOI] [PubMed] [Google Scholar]
- 148. Bulayeva N.N., et al. (2004) Xenoestrogen-induced ERK-1 and ERK-2 activation via multiple membrane-initiated signaling pathways. Environ. Health Perspect., 112, 1481–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Andersen M., et al. (1978) Mutagenic action of aromatic epoxy resins. Nature, 276, 391–392. [DOI] [PubMed] [Google Scholar]
- 150. Ashby J., et al. (1988) Chemical structure, Salmonella mutagenicity and extent of carcinogenicity as indicators of genotoxic carcinogenesis among 222 chemicals tested in rodents by the U.S. NCI/NTP. Mutat. Res., 204, 17–115. [DOI] [PubMed] [Google Scholar]
- 151. Masuda S., et al. (2005) Changes in the mutagenic and estrogenic activities of bisphenol A upon treatment with nitrite. Mutat. Res., 585, 137–146. [DOI] [PubMed] [Google Scholar]
- 152. Dairkee S.H., et al. (2013) Bisphenol-A-induced inactivation of the p53 axis underlying deregulation of proliferation kinetics, and cell death in non-malignant human breast epithelial cells. Carcinogenesis, 34, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Goodson W.H., III, et al. (2011) Activation of the mTOR pathway by low levels of xenoestrogens in breast epithelial cells from high-risk women. Carcinogenesis, 32, 1724–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Pupo M., et al. (2012) Bisphenol A induces gene expression changes and proliferative effects through GPER in breast cancer cells and cancer-associated fibroblasts. Environ. Health Perspect., 120, 1177–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Allard P., et al. (2010) Bisphenol A impairs the double-strand break repair machinery in the germline and causes chromosome abnormalities. Proc. Natl Acad. Sci. USA, 107, 20405–20410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Jenkins S., et al. (2011) Chronic oral exposure to bisphenol A results in a nonmonotonic dose response in mammary carcinogenesis and metastasis in MMTV-erbB2 mice. Environ. Health Perspect., 119, 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Mozzachio A.M., et al. (2008) Chlorothalonil exposure and cancer incidence among pesticide applicator participants in the agricultural health study. Environ. Res., 108, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Wilkinson C.F., et al. (1996) A mechanistic interpretation of the oncogenicity of chlorothalonil in rodents and an assessment of human relevance. Regul. Toxicol. Pharmacol., 24(1 Pt 1), 69–84. [DOI] [PubMed] [Google Scholar]
- 159. Leung-Toung R., et al. (2006) Thiol proteases: inhibitors and potential therapeutic targets. Curr. Med. Chem., 13, 547–581. [DOI] [PubMed] [Google Scholar]
- 160. Rakitsky V.N., et al. (2000) Nongenotoxic (epigenetic) carcinogens: pesticides as an example. A critical review. Teratog. Carcinog. Mutagen., 20, 229–240. [PubMed] [Google Scholar]
- 161. Swan S.H. (2008) Environmental phthalate exposure in relation to reproductive outcomes and other health endpoints in humans. Environ. Res., 108, 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Kim I.Y., et al. (2004) Phthalates inhibit tamoxifen-induced apoptosis in MCF-7 human breast cancer cells. J. Toxicol. Environ. Health. A, 67, 2025–2035. [DOI] [PubMed] [Google Scholar]
- 163. Liu X., et al. (2009) Di-(2-ethylhexyl) phthalate upregulates ATF3 expression and suppresses apoptosis in mouse genital tubercle. J. Occup. Health, 51, 57–63. [DOI] [PubMed] [Google Scholar]
- 164. Maire M.A., et al. (2005) Di-(2-ethylhexyl)phthalate (DEHP) increases Bcl-2/Bax ratio and modifies c-myc expression in Syrian hamster embryo (SHE) cells. Toxicol. Lett., 158, 237–245. [DOI] [PubMed] [Google Scholar]
- 165. Hsieh T.H., et al. (2012) Phthalates induce proliferation and invasiveness of estrogen receptor-negative breast cancer through the AhR/HDAC6/c-Myc signaling pathway. FASEB J., 26, 778–787. [DOI] [PubMed] [Google Scholar]
- 166. Park M.A., et al. (2012) Cell growth of BG-1 ovarian cancer cells is promoted by di-n-butyl phthalate and hexabromocyclododecane via upregulation of the cyclin D and cyclin-dependent kinase-4 genes. Mol. Med. Rep., 5, 761–766. [DOI] [PubMed] [Google Scholar]
- 167. Rusyn I., et al. (2006) Modes of action and species-specific effects of di-(2-ethylhexyl)phthalate in the liver. Crit. Rev. Toxicol., 36, 459–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Isenberg J.S., et al. (2001) Reversibility and persistence of di-2-ethylhexyl phthalate (DEHP)- and phenobarbital-induced hepatocellular changes in rodents. Toxicol. Sci., 64, 192–199. [DOI] [PubMed] [Google Scholar]
- 169. Erkekoğlu P., et al. (2011) Induction of ROS, p53, p21 in DEHP- and MEHP-exposed LNCaP cells-protection by selenium compounds. Food Chem. Toxicol., 49, 1565–1571. [DOI] [PubMed] [Google Scholar]
- 170. Reuber M.D. (1981) Carcinogenicity of dichlorvos. Clin. Toxicol., 18, 47–84. [DOI] [PubMed] [Google Scholar]
- 171. Koutros S., et al. (2008) Dichlorvos exposure and human cancer risk: results from the Agricultural Health Study. Cancer Causes Control, 19, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Page A.C., et al. (1972) Metabolic fate of dichlorvos in swine. Arch. Toxikol., 30, 19–27. [DOI] [PubMed] [Google Scholar]
- 173. Chan P.C., et al. (1991) Carcinogenesis studies of dichlorvos in Fischer rats and B6C3F1 mice. Jpn. J. Cancer Res., 82, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Eroğlu S., et al. (2013) Protective role of vitamins C and E in dichlorvos-induced oxidative stress in human erythrocytes in vitro . Biol. Res., 46, 33–38. [DOI] [PubMed] [Google Scholar]
- 175. Sunkaria A., et al. (2013) Dichlorvos-induced cell cycle arrest and DNA damage repair activation in primary rat microglial cells. J. Neurosci. Res., 91, 444–452. [DOI] [PubMed] [Google Scholar]
- 176. Hervouet E., et al. (2013) DNA methylation and apoptosis resistance in cancer cells. Cells, 2, 545–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Alavanja M.C., et al. (2005) Pesticides and human cancers. Cancer Invest., 23, 700–711. [DOI] [PubMed] [Google Scholar]
- 178. Li Q., et al. (2007) Organophosphorus pesticides induce apoptosis in human NK cells. Toxicology, 239, 89–95. [DOI] [PubMed] [Google Scholar]
- 179. Bhatt D.K., et al. (2009) Modulation of tricarboxylic acid cycle dehydrogenases during hepatocarcinogenesis induced by hexachlorocyclohexane in mice. Exp. Toxicol. Pathol., 61, 325–332. [DOI] [PubMed] [Google Scholar]
- 180. Bhatt D.K., et al. (2012) Modulation of acid phosphatase and lactic dehydrogenase in hexachlorocyclohexane-induced hepatocarcinogenesis in mice. J. Biochem. Mol. Toxicol., 26, 439–444. [DOI] [PubMed] [Google Scholar]
- 181. Zucchini-Pascal N., et al. (2009) Lindane and cell death: at the crossroads between apoptosis, necrosis and autophagy. Toxicology, 256, 32–41. [DOI] [PubMed] [Google Scholar]
- 182. Zucchini-Pascal N., et al. (2011) Molecular investigation of the effects of lindane in rat hepatocytes: microarray and mechanistic studies. Food Chem. Toxicol., 49, 3128–3135. [DOI] [PubMed] [Google Scholar]
- 183. Roloff B.D., et al. (1992) Cytogenetic studies of herbicide interactions in vitro and in vivo using atrazine and linuron. Arch. Environ. Contam. Toxicol., 22, 267–271. [DOI] [PubMed] [Google Scholar]
- 184. Gao X., et al. (2000) Effects of polychlorinated dibenzofurans, biphenyls, and their mixture with dibenzo-p-dioxins on ovulation in the gonadotropin-primed immature rat: support for the toxic equivalency concept. Toxicol. Appl. Pharmacol., 163, 115–124. [DOI] [PubMed] [Google Scholar]
- 185. Kim J.Y., et al. (2014) Methoxychlor and triclosan stimulates ovarian cancer growth by regulating cell cycle- and apoptosis-related genes via an estrogen receptor-dependent pathway. Environ. Toxicol. Pharmacol., 37, 1264–1274. [DOI] [PubMed] [Google Scholar]
- 186. Gupta R.K., et al. (2006) Methoxychlor causes mitochondrial dysfunction and oxidative damage in the mouse ovary. Toxicol. Appl. Pharmacol., 216, 436–445. [DOI] [PubMed] [Google Scholar]
- 187. Eroschenko V.P., et al. (1996) Estradiol or methoxychlor stimulates estrogen receptor (ER) expression in uteri. Reprod. Toxicol., 10, 265–271. [DOI] [PubMed] [Google Scholar]
- 188. Gray L.E., et al. (2001) Effects of environmental antiandrogens on reproductive development in experimental animals. Hum. Reprod. Update, 7, 248–264. [DOI] [PubMed] [Google Scholar]
- 189. Kretschmer X.C., et al. (2005) CAR and PXR: xenosensors of endocrine disrupters? Chem. Biol. Interact., 155, 111–128. [DOI] [PubMed] [Google Scholar]
- 190. di Masi A., et al. (2009) Nuclear receptors CAR and PXR: molecular, functional, and biomedical aspects. Mol. Aspects Med., 30, 297–343. [DOI] [PubMed] [Google Scholar]
- 191. Huang P., et al. (2003) Relationship between CYP3A activity and breast cancer susceptibility in Chinese Han women. Eur. J. Clin. Pharmacol., 59, 471–476. [DOI] [PubMed] [Google Scholar]
- 192. Stagg N.J., et al. (2012) Assessment of possible carcinogenicity of oxyfluorfen to humans using mode of action analysis of rodent liver effects. Toxicol. Sci., 128, 334–345. [DOI] [PubMed] [Google Scholar]
- 193. Reddy J.K., et al. (1980) Hypolipidaemic hepatic peroxisome proliferators form a novel class of chemical carcinogens. Nature, 283, 397–398. [DOI] [PubMed] [Google Scholar]
- 194. Corton J. (2010) Cancer risk assessment: chemical carcinogenesis, hazard evaluation, and risk quantification. In Hsu C.H., Stedeford T. (eds). John Wiley & Sons, Hoboken, NJ, pp. 439–467. [Google Scholar]
- 195. Qu A., et al. (2010) PPARalpha-dependent activation of cell cycle control and DNA repair genes in hepatic nonparenchymal cells. Toxicol. Sci., 118, 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Guenther M.K., et al. (2013) Synthetic lethal interaction between PI3K/Akt/mTOR and Ras/MEK/ERK pathway inhibition in rhabdomyosarcoma. Cancer Lett., 337, 200–209. [DOI] [PubMed] [Google Scholar]
- 197. Daniel M., et al. (2012) Regulation of the human catalytic subunit of telomerase (hTERT). Gene, 498, 135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Kyo S., et al. (2008) Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci., 99, 1528–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Clere N., et al. (2012) Estrogen receptor alpha as a key target of organochlorines to promote angiogenesis. Angiogenesis, 15, 745–760. [DOI] [PubMed] [Google Scholar]
- 200. Silva E., et al. (2010) Cross-talk between non-genomic and genomic signalling pathways–distinct effect profiles of environmental estrogens. Toxicol. Appl. Pharmacol., 245, 160–170. [DOI] [PubMed] [Google Scholar]
- 201. Lee H.R., et al. (2012) Treatment with bisphenol A and methoxychlor results in the growth of human breast cancer cells and alteration of the expression of cell cycle-related genes, cyclin D1 and p21, via an estrogen receptor-dependent signaling pathway. Int. J. Mol. Med., 29, 883–890. [DOI] [PubMed] [Google Scholar]
- 202. Park S.H., et al. (2008) Estrogen regulates Snail and Slug in the down-regulation of E-cadherin and induces metastatic potential of ovarian cancer cells through estrogen receptor alpha. Mol. Endocrinol., 22, 2085–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Giretti M.S., et al. (2008) Extra-nuclear signalling of estrogen receptor to breast cancer cytoskeletal remodelling, migration and invasion. PLoS One, 3, e2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Eritja N., et al. (2012) ERα-mediated repression of pro-inflammatory cytokine expression by glucocorticoids reveals a crucial role for TNFα and IL1α in lumen formation and maintenance. J. Cell Sci., 125(Pt 8), 1929–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Soreq H. (1985) The biosynthesis of biologically active proteins in mRNA-microinjected Xenopus oocytes. CRC Crit. Rev. Biochem., 18, 199–238. [DOI] [PubMed] [Google Scholar]
- 206. Grubisha M.J., et al. (2013) Local endocrine, paracrine and redox signaling networks impact estrogen and androgen crosstalk in the prostate cancer microenvironment. Steroids, 78, 538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Kim H.R., et al. (2013) p53 regulates glucose metabolism by miR-34a. Biochem. Biophys. Res. Commun., 437, 225–231. [DOI] [PubMed] [Google Scholar]
- 208. Vousden K.H., et al. (2009) Blinded by the light: the growing complexity of p53. Cell, 137, 413–431. [DOI] [PubMed] [Google Scholar]
- 209. Teodoro J.G., et al. (2006) p53-mediated inhibition of angiogenesis through up-regulation of a collagen prolyl hydroxylase. Science, 313, 968–971. [DOI] [PubMed] [Google Scholar]
- 210. Hanel W., et al. (2012) Links between mutant p53 and genomic instability. J. Cell. Biochem., 113, 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Harris C.C., et al. (1993) Clinical implications of the p53 tumor-suppressor gene. N. Engl. J. Med., 329, 1318–1327. [DOI] [PubMed] [Google Scholar]
- 212. Niebler M., et al. (2013) Post-translational control of IL-1β via the human papillomavirus type 16 E6 oncoprotein: a novel mechanism of innate immune escape mediated by the E3-ubiquitin ligase E6-AP and p53. PLoS Pathog., 9, e1003536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Gildener-Leapman N., et al. (2013) Promising systemic immunotherapies in head and neck squamous cell carcinoma. Oral Oncol., 49, 1089–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Sperka T., et al. (2012) DNA damage checkpoints in stem cells, ageing and cancer. Nat. Rev. Mol. Cell Biol., 13, 579–590. [DOI] [PubMed] [Google Scholar]
- 215. Qian Y., et al. (2013) Senescence regulation by the p53 protein family. Methods Mol. Biol., 965, 37–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Lu W., et al. (2013) Telomeres-structure, function, and regulation. Exp. Cell Res., 319, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Artandi S.E., et al. (2010) Telomeres and telomerase in cancer. Carcinogenesis, 31, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Lee J.S., et al. (2014) A novel tumor-promoting role for nuclear factor IA in glioblastomas is mediated through negative regulation of p53, p21, and PAI1. Neuro. Oncol., 16, 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Muller P.A., et al. (2011) p53 and its mutants in tumor cell migration and invasion. J. Cell Biol., 192, 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Goh A.M., et al. (2011) The role of mutant p53 in human cancer. J. Pathol., 223, 116–126. [DOI] [PubMed] [Google Scholar]
- 221. Roger L., et al. (2006) Control of cell migration: a tumour suppressor function for p53? Biol. Cell, 98, 141–152. [DOI] [PubMed] [Google Scholar]
- 222. Goh H.S., et al. (1999) p53 point mutation and survival in colorectal cancer patients: effect of disease dissemination and tumour location. Int. J. Oncol., 15, 491–498. [DOI] [PubMed] [Google Scholar]
- 223. Luo M., et al. (2013) Two-stage model of chemically induced hepatocellular carcinoma in mouse. Oncol. Res., 20, 517–528. [DOI] [PubMed] [Google Scholar]
- 224. Cooks T., et al. (2013) Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell, 23, 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Hussain S.P., et al. (2007) Inflammation and cancer: an ancient link with novel potentials. Int. J. Cancer, 121, 2373–2380. [DOI] [PubMed] [Google Scholar]
- 226. Lorin S., et al. (2013) Autophagy regulation and its role in cancer. Semin. Cancer Biol., 23, 361–379. [DOI] [PubMed] [Google Scholar]
- 227. Tsai C.M., et al. (1996) Correlations between intrinsic chemoresistance and HER-2/neu gene expression, p53 gene mutations, and cell proliferation characteristics in non-small cell lung cancer cell lines. Cancer Res., 56, 206–209. [PubMed] [Google Scholar]
- 228. Arriola E., et al. (2008) Genomic analysis of the HER2/TOP2A amplicon in breast cancer and breast cancer cell lines. Lab. Invest., 88, 491–503. [DOI] [PubMed] [Google Scholar]
- 229. Biroccio A., et al. (2004) Telomerase as a new target for the treatment of hormone-refractory prostate cancer. Endocr. Relat. Cancer, 11, 407–421. [DOI] [PubMed] [Google Scholar]
- 230. Vageli D., et al. (2009) Transcriptional activation of hTERT in breast carcinomas by the Her2-ER81-related pathway. Oncol. Res., 17, 413–423. [DOI] [PubMed] [Google Scholar]
- 231. Fuchs I.B., et al. (2002) The prognostic significance of epithelial-mesenchymal transition in breast cancer. Anticancer Res., 22(6A), 3415–3419. [PubMed] [Google Scholar]
- 232. Ortega-Cava C.F., et al. (2011) Continuous requirement of ErbB2 kinase activity for loss of cell polarity and lumen formation in a novel ErbB2/Neu-driven murine cell line model of metastatic breast cancer. J. Carcinog., 10, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Xi C., et al. (2012) Heat shock factor Hsf1 cooperates with ErbB2 (Her2/Neu) protein to promote mammary tumorigenesis and metastasis. J. Biol. Chem., 287, 35646–35657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Moreira Sousa C., et al. (2013) The Huntington disease protein accelerates breast tumour development and metastasis through ErbB2/HER2 signalling. EMBO Mol. Med., 5, 309–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Yuan H., et al. (2013) PPARδ induces estrogen receptor-positive mammary neoplasia through an inflammatory and metabolic phenotype linked to mTOR activation. Cancer Res., 73, 4349–4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Mohammed Z.M., et al. (2012) The relationship between components of tumour inflammatory cell infiltrate and clinicopathological factors and survival in patients with primary operable invasive ductal breast cancer. Br. J. Cancer, 107, 864–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Olson O.C., et al. (2013) Microenvironment-mediated resistance to anticancer therapies. Cell Res., 23, 179–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Min J.W., et al. (2013) INPP4B-mediated tumor resistance is associated with modulation of glucose metabolism via hexokinase 2 regulation in laryngeal cancer cells. Biochem. Biophys. Res. Commun., 440, 137–142. [DOI] [PubMed] [Google Scholar]
- 239. Kim J.G., et al. (2013) Association between phosphorylated AMP-activated protein kinase and MAPK3/1 expression and prognosis for patients with gastric cancer. Oncology, 85, 78–85. [DOI] [PubMed] [Google Scholar]
- 240. Agarwal M.L., et al. (2001) Regulation of p53 expression by the RAS-MAP kinase pathway. Oncogene, 20, 2527–2536. [DOI] [PubMed] [Google Scholar]
- 241. Wu G.S. (2004) The functional interactions between the p53 and MAPK signaling pathways. Cancer Biol. Ther., 3, 156–161. [DOI] [PubMed] [Google Scholar]
- 242. Murphy D.A., et al. (2006) Inhibition of tumor endothelial ERK activation, angiogenesis, and tumor growth by sorafenib (BAY43-9006). Am. J. Pathol., 169, 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Corcelle E., et al. (2006) Disruption of autophagy at the maturation step by the carcinogen lindane is associated with the sustained mitogen-activated protein kinase/extracellular signal-regulated kinase activity. Cancer Res., 66, 6861–6870. [DOI] [PubMed] [Google Scholar]
- 244. Lin Y.W., et al. (2003) Persistent activation of ERK1/2 by lead acetate increases nucleotide excision repair synthesis and confers anti-cytotoxicity and anti-mutagenicity. Carcinogenesis, 24, 53–61. [DOI] [PubMed] [Google Scholar]
- 245. Wang S., et al. (2006) Tumor evasion of the immune system: inhibiting p38 MAPK signaling restores the function of dendritic cells in multiple myeloma. Blood, 107, 2432–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Duan S.G., et al. (2010) The role of MAPK-ERK pathway in 67-kDa laminin receptor-induced FasL expression in human cholangiocarcinoma cells. Dig. Dis. Sci., 55, 2844–2852. [DOI] [PubMed] [Google Scholar]
- 247. Bermudez Y., et al. (2008) Pyk2/ERK ½ mediate Sp1- and c-Myc-dependent induction of telomerase activity by epidermal growth factor. Growth Factors, 26, 1–11. [DOI] [PubMed] [Google Scholar]
- 248. Krueger J.S., et al. (2001) Temporal and quantitative regulation of mitogen-activated protein kinase (MAPK) modulates cell motility and invasion. Oncogene, 20, 4209–4218. [DOI] [PubMed] [Google Scholar]
- 249. Davies M., et al. (2005) Induction of an epithelial to mesenchymal transition in human immortal and malignant keratinocytes by TGF-beta1 involves MAPK, Smad and AP-1 signalling pathways. J. Cell. Biochem., 95, 918–931. [DOI] [PubMed] [Google Scholar]
- 250. Reddy K.B., et al. (2003) Role of MAP kinase in tumor progression and invasion. Cancer Metastasis Rev., 22, 395–403. [DOI] [PubMed] [Google Scholar]
- 251. Weng C.J., et al. (2008) Lucidenic acid inhibits PMA-induced invasion of human hepatoma cells through inactivating MAPK/ERK signal transduction pathway and reducing binding activities of NF-kappaB and AP-1. Carcinogenesis, 29, 147–156. [DOI] [PubMed] [Google Scholar]
- 252. Wegiel B., et al. (2009) Cystatin C is downregulated in prostate cancer and modulates invasion of prostate cancer cells via MAPK/Erk and androgen receptor pathways. PLoS One, 4, e7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Wang L.N., et al. (2011) Cancer-induced bone pain sequentially activates the ERK/MAPK pathway in different cell types in the rat spinal cord. Mol. Pain, 7, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Zhang Y., et al. (2013) Interleukin-6 is required for pancreatic cancer progression by promoting MAPK signaling activation and oxidative stress resistance. Cancer Res., 73, 6359–6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Weinberg F., et al. (2010) Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl Acad. Sci. USA, 107, 8788–8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Lee S.W., et al. (2000) Sustained activation of Ras/Raf/mitogen-activated protein kinase cascade by the tumor suppressor p53. Proc. Natl Acad. Sci. USA, 97, 8302–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Berra E., et al. (2000) Signaling angiogenesis via p42/p44 MAP kinase and hypoxia. Biochem. Pharmacol., 60, 1171–1178. [DOI] [PubMed] [Google Scholar]
- 258. Saavedra H.I., et al. (2000) The RAS oncogene induces genomic instability in thyroid PCCL3 cells via the MAPK pathway. Oncogene, 19, 3948–3954. [DOI] [PubMed] [Google Scholar]
- 259. Gong W., et al. (2011) Paclitaxel induced B7-H1 expression in cancer cells via the MAPK pathway. J. Chemother., 23, 295–299. [DOI] [PubMed] [Google Scholar]
- 260. Inui T., et al. (2001) Telomerase activation and MAPK pathways in regenerating hepatocytes. Hum. Cell, 14, 275–282. [PubMed] [Google Scholar]
- 261. Matsuo T., et al. (2012) Correlation between p38 mitogen-activated protein kinase and human telomerase reverse transcriptase in sarcomas. J. Exp. Clin. Cancer Res., 31, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Ge Z., et al. (2006) Mitogen-activated protein kinase cascade-mediated histone H3 phosphorylation is critical for telomerase reverse transcriptase expression/telomerase activation induced by proliferation. Mol. Cell. Biol., 26, 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Olejarz W., et al. (2014) Mycophenolic acid attenuates the tumour necrosis factor-α-mediated proinflammatory response in endothelial cells by blocking the MAPK/NF-κB and ROS pathways. Eur. J. Clin. Invest., 44, 54–64. [DOI] [PubMed] [Google Scholar]
- 264. Ji R.R., et al. (2009) MAP kinase and pain. Brain Res. Rev., 60, 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Li L., et al. (2010) Inhibition of hypoxia-induced cell motility by p16 in MDA-MB-231 breast cancer cells. J. Cancer, 1, 126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. McDermott K.M., et al. (2006) p16(INK4a) prevents centrosome dysfunction and genomic instability in primary cells. PLoS Biol., 4, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Wang P., et al. (2013) Methylation of p16 CpG islands correlated with metastasis and aggressiveness in papillary thyroid carcinoma. J. Chin. Med. Assoc., 76, 135–139. [DOI] [PubMed] [Google Scholar]
- 268. Montes C.L., et al. (2008) Tumor-induced senescent T cells with suppressor function: a potential form of tumor immune evasion. Cancer Res., 68, 870–879. [DOI] [PubMed] [Google Scholar]
- 269. Stoll E.A., et al. (2013) The impact of age on oncogenic potential: tumor-initiating cells and the brain microenvironment. Aging Cell, 12, 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Rayess H., et al. (2012) Cellular senescence and tumor suppressor gene p16. Int. J. Cancer, 130, 1715–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Iannone F., et al. (2005) Increased Bcl-2/p53 ratio in human osteoarthritic cartilage: a possible role in regulation of chondrocyte metabolism. Ann. Rheum. Dis., 64, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Kawabuchi B., et al. (1999) p16 inactivation in small-sized lung adenocarcinoma: its association with poor prognosis. Int. J. Cancer, 84, 49–53. [DOI] [PubMed] [Google Scholar]
- 273. Hui A.M., et al. (2000) Loss of p16(INK4) protein, alone and together with loss of retinoblastoma protein, correlate with hepatocellular carcinoma progression. Cancer Lett., 154, 93–99. [DOI] [PubMed] [Google Scholar]
- 274. Tada T., et al. (2003) Reduced p16 expression correlates with lymphatic invasion in colorectal cancers. Hepatogastroenterology, 50, 1756–1760. [PubMed] [Google Scholar]
- 275. Wang C.H., et al. (2006) p16 inhibits matrix metalloproteinase-2 expression via suppression of Sp1-mediated gene transcription. J. Cell. Physiol., 208, 246–252. [DOI] [PubMed] [Google Scholar]
- 276. Achyut B.R., et al. (2013) Inflammation-mediated genetic and epigenetic alterations drive cancer development in the neighboring epithelium upon stromal abrogation of TGF-β signaling. PLoS Genet., 9, e1003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Amelio I., et al. (2011) Cell death pathology: cross-talk with autophagy and its clinical implications. Biochem. Biophys. Res. Commun., 414, 277–281. [DOI] [PubMed] [Google Scholar]
- 278. Decary S., et al. (2002) The retinoblastoma protein binds the promoter of the survival gene bcl-2 and regulates its transcription in epithelial cells through transcription factor AP-2. Mol. Cell. Biol., 22, 7877–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Huang H., et al. (2004) Androgens repress Bcl-2 expression via activation of the retinoblastoma (RB) protein in prostate cancer cells. Oncogene, 23, 2161–2176. [DOI] [PubMed] [Google Scholar]
- 280. Crescenzi E., et al. (2001) Bcl-2 exerts a pRb-mediated cell cycle inhibitory function in HEC1B endometrial carcinoma cells. Gynecol. Oncol., 81, 184–192. [DOI] [PubMed] [Google Scholar]
- 281. Ha J.H., et al. (2013) Dual-site interactions of p53 protein transactivation domain with anti-apoptotic Bcl-2 family proteins reveal a highly convergent mechanism of divergent p53 pathways. J. Biol. Chem., 288, 7387–7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Staibano S., et al. (2001) Interaction between bcl-2 and P53 in neoplastic progression of basal cell carcinoma of the head and neck. Anticancer Res., 21(6A), 3757–3764. [PubMed] [Google Scholar]
- 283. Esposito F., et al. (2012) High-mobility group A1 protein inhibits p53-mediated intrinsic apoptosis by interacting with Bcl-2 at mitochondria. Cell Death Dis., 3, e383. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 284. Bharatham N., et al. (2011) Molecular basis of Bcl-X(L)-p53 interaction: insights from molecular dynamics simulations. PLoS One, 6, e26014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Basu A., et al. (1998) The relationship between BcI2, Bax and p53: consequences for cell cycle progression and cell death. Mol. Hum. Reprod., 4, 1099–1109. [DOI] [PubMed] [Google Scholar]
- 286. Dogu Y., et al. (2009) Mathematical model of a network of interaction between p53 and Bcl-2 during genotoxic-induced apoptosis. Biophys. Chem., 143, 44–54. [DOI] [PubMed] [Google Scholar]
- 287. Tomita Y., et al. (2006) WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J. Biol. Chem., 281, 8600–8606. [DOI] [PubMed] [Google Scholar]
- 288. Biroccio A., et al. (2000) Bcl-2 overexpression and hypoxia synergistically act to modulate vascular endothelial growth factor expression and in vivo angiogenesis in a breast carcinoma line. FASEB J., 14, 652–660. [DOI] [PubMed] [Google Scholar]
- 289. Zuo J., et al. (2010) Bcl-2 overexpression induces a partial epithelial to mesenchymal transition and promotes squamous carcinoma cell invasion and metastasis. Mol. Cancer Res., 8, 170–182. [DOI] [PubMed] [Google Scholar]
- 290. Nelson D.A., et al. (2004) Hypoxia and defective apoptosis drive genomic instability and tumorigenesis. Genes Dev., 18, 2095–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Zheng J.M., et al. (2005) [bcl-2 expression in small cell lung cancer: a mechanism for apoptosis antagonism and immune evasion]. Di Yi Jun Yi Da Xue Xue Bao, 25, 1537–1539. [PubMed] [Google Scholar]
- 292. Huerta S., et al. (2007) Modification of gene products involved in resistance to apoptosis in metastatic colon cancer cells: roles of Fas, Apaf-1, NFkappaB, IAPs, Smac/DIABLO, and AIF. J. Surg. Res., 142, 184–194. [DOI] [PubMed] [Google Scholar]
- 293. Zuo J., et al. (2011) Epstein-Barr virus evades CD4+ T cell responses in lytic cycle through BZLF1-mediated downregulation of CD74 and the cooperation of vBcl-2. PLoS Pathog., 7, e1002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Mandal M., et al. (1997) Bcl-2 modulates telomerase activity. J. Biol. Chem., 272, 14183–14187. [DOI] [PubMed] [Google Scholar]
- 295. Wang X., et al. (2007) Oestrogen signalling inhibits invasive phenotype by repressing RelB and its target BCL2. Nat. Cell Biol., 9, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Sun T., et al. (2011) Promotion of tumor cell metastasis and vasculogenic mimicry by way of transcription coactivation by Bcl-2 and Twist1: a study of hepatocellular carcinoma. Hepatology, 54, 1690–1706. [DOI] [PubMed] [Google Scholar]
- 297. Chung G.T., et al. (2013) Constitutive activation of distinct NF-κB signals in EBV-associated nasopharyngeal carcinoma. J. Pathol., 231, 311–322. [DOI] [PubMed] [Google Scholar]
- 298. Singh V., et al. (2009) A tumour stage-dependent evolution of drug resistant T cell lymphoma: role of soluble mediators of tumour and host origin. Leuk. Res., 33, 700–709. [DOI] [PubMed] [Google Scholar]
- 299. Wang B., et al. (2012) Stat3-mediated activation of microRNA-23a suppresses gluconeogenesis in hepatocellular carcinoma by down-regulating glucose-6-phosphatase and peroxisome proliferator-activated receptor gamma, coactivator 1 alpha. Hepatology, 56, 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Kloetzel M., et al. (2013) Trans fatty acids affect cellular viability of human intestinal Caco-2 cells and activate peroxisome proliferator-activated receptors. Nutr. Cancer, 65, 139–146. [DOI] [PubMed] [Google Scholar]
- 301. Gizard F., et al. (2008) The PPARalpha/p16INK4a pathway inhibits vascular smooth muscle cell proliferation by repressing cell cycle-dependent telomerase activation. Circ. Res., 103, 1155–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Gizard F., et al. (2005) PPAR alpha inhibits vascular smooth muscle cell proliferation underlying intimal hyperplasia by inducing the tumor suppressor p16INK4a. J. Clin. Invest., 115, 3228–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Gopinathan L., et al. (2009) Regulation of peroxisome proliferator-activated receptor-alpha by MDM2. Toxicol. Sci., 108, 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Pozzi A., et al. (2008) PPARalpha ligands as antitumorigenic and antiangiogenic Agents. PPAR Res., 2008, 906542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Moraes L.A., et al. (2006) Peroxisome proliferator-activated receptors and inflammation. Pharmacol. Ther., 110, 371–385. [DOI] [PubMed] [Google Scholar]
- 306. Tang C.C., et al. (2013) Hepatoprotective effect of mulberry water extracts on ethanol-induced liver injury via anti-inflammation and inhibition of lipogenesis in C57BL/6J mice. Food Chem. Toxicol., 62, 786–796. [DOI] [PubMed] [Google Scholar]
- 307. Goldring M.B., et al. (2004) The regulation of chondrocyte function by proinflammatory mediators: prostaglandins and nitric oxide. Clin. Orthop. Relat. Res., S37–S46. [DOI] [PubMed] [Google Scholar]
- 308. Spath C., et al. (2013) Inverse relationship between tumor proliferation markers and connexin expression in a malignant cardiac tumor originating from mesenchymal stem cell engineered tissue in a Rat in vivo model. Front. Pharmacol., 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Yano T., et al. (2006) Connexin 32 as an anti-invasive and anti-metastatic gene in renal cell carcinoma. Biol. Pharm. Bull., 29, 1991–1994. [DOI] [PubMed] [Google Scholar]
- 310. Li Q., et al. (2007) Cytoplasmic accumulation of connexin32 protein enhances motility and metastatic ability of human hepatoma cells in vitro and in vivo . Int. J. Cancer, 121, 536–546. [DOI] [PubMed] [Google Scholar]
- 311. Sato H., et al. (2007) Regulation of renal cell carcinoma cell proliferation, invasion and metastasis by connexin 32 gene. J. Membr. Biol., 216, 17–21. [DOI] [PubMed] [Google Scholar]
- 312. Trosko J.E., et al. (2006) Adult stem cell theory of the multi-stage, multi-mechanism theory of carcinogenesis: role of inflammation on the promotion of initiated stem cells. Contrib. Microbiol., 13, 45–65. [DOI] [PubMed] [Google Scholar]
- 313. Corn P.G. (2012) The tumor microenvironment in prostate cancer: elucidating molecular pathways for therapy development. Cancer Manag. Res., 4, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Cui J., et al. (2011) MicroRNA expression and regulation in human ovarian carcinoma cells by luteinizing hormone. PLoS One, 6, e21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Choi J.H., et al. (2006) Gonadotropins activate proteolysis and increase invasion through protein kinase A and phosphatidylinositol 3-kinase pathways in human epithelial ovarian cancer cells. Cancer Res., 66, 3912–3920. [DOI] [PubMed] [Google Scholar]
- 316. Lau M.T., et al. (2010) Gonadotropins induce tumor cell migration and invasion by increasing cyclooxygenases expression and prostaglandin E(2) production in human ovarian cancer cells. Endocrinology, 151, 2985–2993. [DOI] [PubMed] [Google Scholar]
- 317. Herman A.P., et al. (2013) LPS-induced inflammation potentiates the IL-1β-mediated reduction of LH secretion from the anterior pituitary explants. Clin. Dev. Immunol., 2013, 926937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Sand E., et al. (2013) Gonadotropin-releasing hormone analog buserelin causes neuronal loss in rat gastrointestinal tract. Cell Tissue Res., 351, 521–534. [DOI] [PubMed] [Google Scholar]
- 319. Fillon M. (2012) Getting it right: BPA and the difficulty proving environmental cancer risks. J. Natl Cancer Inst., 104, 652–655. [DOI] [PubMed] [Google Scholar]
- 320. Jiang Y., et al. (2014) Prenatal exposure to bisphenol A at the reference dose impairs mitochondria in the heart of neonatal rats. J. Appl. Toxicol., 34, 1012–1022. [DOI] [PubMed] [Google Scholar]
- 321. Lee H.S., et al. (2012) Set, a putative oncogene, as a biomarker for prenatal exposure to bisphenol A. Asian Pac. J. Cancer Prev., 13, 2711–2715. [DOI] [PubMed] [Google Scholar]
- 322. Betancourt A.M., et al. (2012) Altered carcinogenesis and proteome in mammary glands of rats after prepubertal exposures to the hormonally active chemicals bisphenol a and genistein. J. Nutr., 142, 1382S–1388S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Durando M., et al. (2011) Prenatal exposure to bisphenol A promotes angiogenesis and alters steroid-mediated responses in the mammary glands of cycling rats. J. Steroid Biochem. Mol. Biol., 127, 35–43. [DOI] [PubMed] [Google Scholar]
- 324. Goloubkova T., et al. (2000) Effects of xenoestrogen bisphenol A on uterine and pituitary weight, serum prolactin levels and immunoreactive prolactin cells in ovariectomized Wistar rats. Arch. Toxicol., 74, 92–98. [DOI] [PubMed] [Google Scholar]
- 325. Kurosawa T., et al. (2002) The activity of bisphenol A depends on both the estrogen receptor subtype and the cell type. Endocr. J., 49, 465–471. [DOI] [PubMed] [Google Scholar]
- 326. Zhu H., et al. (2010) Environmental endocrine disruptors promote invasion and metastasis of SK-N-SH human neuroblastoma cells. Oncol. Rep., 23, 129–139. [PubMed] [Google Scholar]
- 327. Derouiche S., et al. (2013) Bisphenol A stimulates human prostate cancer cell migration via remodelling of calcium signalling. Springerplus, 2, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Tenev T., et al. (2011) The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol. Cell, 43, 432–448. [DOI] [PubMed] [Google Scholar]
- 329. Hassan Z.K., et al. (2012) Bisphenol A induces hepatotoxicity through oxidative stress in rat model. Oxid. Med. Cell. Longev., 2012, 194829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Kleinsasser N.H., et al. (2000) Phthalates demonstrate genotoxicity on human mucosa of the upper aerodigestive tract. Environ. Mol. Mutagen., 35, 9–12. [DOI] [PubMed] [Google Scholar]
- 331. Li X., et al. (2014) Dibutyl phthalate-induced neurotoxicity in the brain of immature and mature rat offspring. Brain Dev., 36, 653–660. [DOI] [PubMed] [Google Scholar]
- 332. Pariseau J., et al. (2011) Effects of pesticide compounds (chlorothalonil and mancozeb) and benzo[a]pyrene mixture on aryl hydrocarbon receptor, p53 and ubiquitin gene expression levels in haemocytes of soft-shell clams (Mya arenaria). Ecotoxicology, 20, 1765–1772. [DOI] [PubMed] [Google Scholar]
- 333. Lebailly P., et al. (1997) Assessment of DNA damage induced in vitro by etoposide and two fungicides (carbendazim and chlorothalonil) in human lymphocytes with the comet assay. Mutat. Res., 375, 205–217. [DOI] [PubMed] [Google Scholar]
- 334. Nagda G., et al. (2011) Alleviation of lindane induced toxicity in testis of Swiss mice (Mus musculus) by combined treatment with vitamin C, vitamin E and alpha-lipoic acid. Indian J. Exp. Biol., 49, 191–199. [PubMed] [Google Scholar]
- 335. Bharathi S.P., et al. (2013) Role of pesticides in the induction of tumor angiogenesis. Anticancer Res., 33, 231–240. [PubMed] [Google Scholar]
- 336. Bhunya S.P., et al. (1992) Genotoxic potential of the organochlorine insecticide lindane (gamma-BHC): an in vivo study in chicks. Mutat. Res., 272, 175–181. [DOI] [PubMed] [Google Scholar]
- 337. Isoda H., et al. (2005) Effects of organophosphorous pesticides used in china on various mammalian cells. Environ. Sci., 12, 9–19. [PubMed] [Google Scholar]
- 338. Nilufer Yonguc G., et al. (2012) Caspase 1, caspase 3, TNF-alpha, p53, and Hif1-alpha gene expression status of the brain tissues and hippocampal neuron loss in short-term dichlorvos exposed rats. Mol. Biol. Rep., 39, 10355–10360. [DOI] [PubMed] [Google Scholar]
- 339. Wang Q.L., et al. (2013) Risk assessment of mouse gastric tissue cancer induced by dichlorvos and dimethoate. Oncol. Lett., 5, 1385–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Rishi K.K., et al. (1995) Chromosome aberration test for the insecticide, dichlorvos, on fish chromosomes. Mutat. Res., 344, 1–4. [DOI] [PubMed] [Google Scholar]
- 341. Sunkaria A., et al. (2012) Dichlorvos exposure results in activation induced apoptotic cell death in primary rat microglia. Chem. Res. Toxicol., 25, 1762–1770. [DOI] [PubMed] [Google Scholar]
- 342. Laws S.C., et al. (2000) Estrogenic activity of octylphenol, nonylphenol, bisphenol A and methoxychlor in rats. Toxicol. Sci., 54, 154–167. [DOI] [PubMed] [Google Scholar]
- 343. Hodgson A.V., et al. (1998) Estrogen-induced microsatellite DNA alterations are associated with Syrian hamster kidney tumorigenesis. Carcinogenesis, 19, 2169–2172. [DOI] [PubMed] [Google Scholar]
- 344. Nishino R., et al. (2013) Prior oral exposure to environmental immunosuppressive chemicals methoxychlor, parathion, or piperonyl butoxide aggravates allergic airway inflammation in NC/Nga mice. Toxicology, 309, 1–8. [DOI] [PubMed] [Google Scholar]
- 345. Fukuyama T., et al. (2012) Role of regulatory T cells in the induction of atopic dermatitis by immunosuppressive chemicals. Toxicol. Lett., 213, 392–401. [DOI] [PubMed] [Google Scholar]
- 346. Rao K.N., et al. (1997) Hepatic hyperplasia and cancer in rats: metabolic alterations associated with cell growth. Gastroenterology, 113, 238–248. [DOI] [PubMed] [Google Scholar]
- 347. Moushumi Priya A., et al. (2012) Induction of apoptosis and cell cycle arrest by Bis (2-ethylhexyl) phthalate produced by marine Bacillus pumilus MB 40. Chem. Biol. Interact., 195, 133–143. [DOI] [PubMed] [Google Scholar]
- 348. Pasquini R., et al. (1994) Assay of linuron and a pesticide mixture commonly found in the Italian diet, for promoting activity in rat liver carcinogenesis. Pharmacol. Toxicol., 75, 170–176. [DOI] [PubMed] [Google Scholar]
- 349. Mazzoleni G., et al. (1994) Influence of the herbicide Linuron on growth rate and gap-junctional intercellular communication of cultured endothelial cells. J. Environ. Pathol. Toxicol. Oncol., 13, 1–10. [PubMed] [Google Scholar]
- 350. Barshteĭn I.u.A., et al. (1990) [An immunological and morphological study of focal staphylococcal infection against a background of long-term exposure to the herbicide linuron]. Mikrobiol. Zh., 52, 52–59. [PubMed] [Google Scholar]
- 351. Dairkee S.H., et al. (2008) Bisphenol A induces a profile of tumor aggressiveness in high-risk cells from breast cancer patients. Cancer Res., 68, 2076–2080. [DOI] [PubMed] [Google Scholar]
- 352. Lee I.K., et al. (2007) Inhibitory effect of bisphenol A on gap junctional intercellular communication in an epithelial cell line of rat mammary tissue. Arch. Pharm. Res., 30, 337–343. [DOI] [PubMed] [Google Scholar]
- 353. Weng Y.I., et al. (2010) Epigenetic influences of low-dose bisphenol A in primary human breast epithelial cells. Toxicol. Appl. Pharmacol., 248, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]


