Abstract
To evaluate the identification method using the microRNA markers miR10b and miR135b to distinguish semen stains from menstrual blood, peripheral blood, vaginal fluid and so on body fluid stains. The expression levels of miR10b and miR35b in semen stains and menstrual blood and so on were detected utilizing a real-time quantitative PCR technique with a specific fluorescence-labeled TaqMan probe. RNU6b was used as the internal reference gene; the difference in their expression was analyzed, and the specificity, sensitivity, and detection capability of the techniques were evaluated. The expression of miR10b and miR135b in semen stains was significantly higher than that of other body fluid stains, with a mean value of ΔCт from-6 to-7. However, it ranged from-2 to-4 for other body fluid stains. The initial criteria for judging which semen stains can be identified were determined by analyzing the research results. When the threshold value was set to 0.04, the CT value could be detected in the target genes miR10b, miR135b and in the internal reference gene RNU6b, and CT values are<40, ΔCT[10b-U6]<-5.5, and ΔCT[135b-U6]<-6, respectively, and the semen stain could be identified. The expression levels of miR10b and miR135b are higher in semen with strong tissue specificity; thus, they can be used to differentiate semen stains from other body fluid stains in forensic science.
Introduction
MicroRNAs (miRNAs) are a type of endogenous non-coding RNA that has a regulatory function in eukaryotes. The length of miRNAs molecule is approximately 20 to 25 nucleotides. They are generated by Dicer enzyme shearing single-stranded RNA precursors, which contain approximately 70 to 90 nucleotides with a hairpin structure [1, 2]. The miRNAs participate in a variety of regulatory pathways, including growth, virus defense, haematopoiesis, organogenesis, cell proliferation, apoptosis, and fat metabolism, and so on. Recently, they have also been shown to play an important role in tumorigenesis and other related fields. Therefore, the study and identification of miRNAs has become a hot topic in biology [3–5]. It has been found that the nucleotide sequence of a miRNA is short and that its structure is relatively stable, and a large number of studies have demonstrated that miRNAs do not easily undergo degradation. This particular feature has important significance and may aid in detection with problematic forensic evidence samples, particularly for degraded and old samples. As a new type of body fluid identification marker, miRNAs can be used to identify body fluids stains with difficult degradation characteristics [6–9]. It has been reported that the miRNA markers miR10b and miR135b are found in the seminal plasma or in exfoliated cells of the male reproductive tract, with an expression level higher than that of other body fluids [7]. This study was performed to evaluate the use of miR10b and miR135b to identify semen stains from among other bodily fluids stains. These miRNA markers may lead to a novel semen stain identification method.
Materials and Methods
Semen specimens of forty adult males (with normal semen) and five adult male patients with azoospermia were collected. Samples were obtained from the first Affiliated Hospital, Sun Yat-Sen University. The semen samples were used to create semen stains according to forensic operational processes. The 100μl mix liquefied semen was taken, and smeared on the sterile gauze, natural airing, including preparation at 25°C under dry and dark conditions and drying under natural conditions. They were stored at 25°C in dry and dark conditions for one day, one month, two months, three months, six months, or twelve months. Furthermore, RNA was extracted from all semen stains with TRIzol Reagent (Invitrogen, Carlsbad, USA) according to manufacturer’s instructions. In addition, ten samples each of blood, vaginal fluid and menstrual blood were obtained; blood and vaginal fluid stains were made in the same manner as used for the semen stains; menstrual blood stain was made by clipping 1 cm2 sanitary towel. Samples were obtained from the Center of Medicine Expertise of Sun Yat-Sen University. Finally, the RNA of the all samples including 45 semen, 10 blood, vaginal fluid and menstrual blood was extracted and quantified by UV-2450 (Shimadzu, Japan). All volunteers were anonymous and specimens were obtained under informed consent. The primer and probe sequences of the target genes miR10b, miR135b and the reference gene RNU6b were obtained from the miRBase database (http://www.mirbase.org/index.shtml/); a real-time quantitative PCR kit (TaqMan MicroRNA Assay kit) was purchased from the U.S Applied Biosystems (ABI, Foster city, USA).
The reverse transcription and RT-qPCR reactions were performed according to the TaqMan MicroRNA Assay kit manual, to take 1.5μl RNA template carried out the reverse transcription (see S1 Table). The Assay-ID of hsa-miR-10b marker: hsa002218_m1 (primer: RT2218, TaqMan probe: TM2218); The Assay-ID of hsa-miR-135b marker: hsa002261_m1 (primer: RT2261, TaqMan probe: TM2261. The expression levels of miR10b, miR135b, and RNU6b in 10 semen samples which be extracted from 40 samples randomly, 10 menstrual blood, 10 peripheral blood, 10 vaginal fluid stains and negative control were detected duplicate 3 times using RT-qPCR, the protocol for the RT-qPCR components in each tube: Premix Ex Taq (2×) 10μl, TaqMan Micro RNA Assay kit Real-time primer-probe (20×), 1.0μl, ROX Reference Dye II (50×) 0.4μl, ddH2O 6.6μl, Reverse transcription product 2.0μl, the parameters of cycles 50°C 2min, 95°C 10min, then 95°C 15s, 60°C 60s, 50 cycles at ABI 7500 thermal cycler, and the differences in expression levels were compared. The sensitivity was tested with a serial dilution RNA of semen stains sample at the same time, the concentration from 100ng to 0.001ng/ml. The reliability of method was tested with known 30 normal semen stains, 5 no sperm semen samples and 10 old each kind of semen samples, 1 day, 1 month, 3 months, 6 months and 1 years respectively. Then, those samples were analyzed respectively with RT-qPCR and compared Ct values.
The test results were analyzed using the SPSS version 17.0 statistical software package. (Chicago, IL, USA). The means and the standard deviations of the expression of miR10b, miR135b and RNU6b in different body fluids were calculated. The differences were compared, and judgment criteria were established, the level of significance was p< 0.05 for all statistical analyses.
Ethics Statement: The research protocol was approved by the Human Subjects Committee at the Zhongshan School of Medicine, Sun Yat-sen University. Written informed consent was attained by all participants or guardians involved in the study. The research protocol was also approved by the Animal Subjects Committee at the Zhongshan School of Medicine, Sun Yat-sen University.
Results
1. The Expression of miR10b, miR135b, and RNU6b in Body Fluid Stains
The fluorescence detection threshold of miR10b, miR135b, and RNU6b was set at 0.04; at this point, the amplification curves of miR10b, miR135b, and RNU6b are in the exponential growth phase. The expression levels of miR10b, miR135b, and RNU6b were detected to obtain a CT value of body fluid stains. The ΔCT values reflect the CT value of the target gene minus the CT value of the reference gene. The results are shown in Table 1.
Table 1. The CT and ΔCт values of miRNA markers that tested by RT-PCR in different body fluid stains.
| Type | Sample | Cт | ΔCт | |||
|---|---|---|---|---|---|---|
| samples | name | miR10b | miR135b | RNU6b | [10b-U6] | [135b-U6] |
| Normal sperm semen stains* | S01 | 28.827 | 29.705 | 36.982 | -8.155 | -7.277 |
| S03 | 29.14 | 29.045 | 37.419 | -8.279 | -8.374 | |
| S04 | 26.566 | 27.748 | 35.775 | -9.209 | -8.027 | |
| S07 | 26.649 | 25.618 | 35.473 | -8.825 | -9.856 | |
| S09 | 25.954 | 25.285 | 33.68 | -7.726 | -8.395 | |
| S10 | 32.516 | 32.489 | 38.772 | -6.256 | -6.283 | |
| S13 | 26.688 | 26.576 | 35.75 | -9.062 | -9.174 | |
| S21 | 25.848 | 25.261 | 33.475 | -7.627 | -8.214 | |
| S22 | 25.287 | 23.917 | 33.045 | -7.758 | -9.128 | |
| S29 | 26.476 | 25.791 | 34.876 | -8.4 | -9.085 | |
| No sperm semen stains | AS01 | 29.055 | 27.51 | 36.106 | -7.051 | -8.596 |
| AS02 | 26.584 | 26.484 | 33.291 | -6.707 | -6.807 | |
| AS03 | 28.772 | 30.083 | 38.248 | -9.476 | -8.165 | |
| AS04 | 29.816 | 29.867 | 38.112 | -8.296 | -8.245 | |
| AS05 | 27.357 | 27.056 | 35.498 | -8.141 | -8.442 | |
| V01 | 28.068 | 32.027 | 32.872 | -4.804 | -0.845 | |
| V02 | 33.313 | 30.698 | 38.898 | -5.585 | -8.2 | |
| V03 | 29.985 | 30.258 | 29.767 | 0.218 | 0.491 | |
| Vaginal swabs stains | V04 | 29.003 | 27.803 | 31.359 | -2.356 | -3.556 |
| V05 | 29.216 | 27.608 | 30.518 | -1.302 | -2.91 | |
| V06 | 30.491 | 29.436 | 32.303 | -1.812 | -2.867 | |
| V07 | 32.664 | 33.74 | 31.575 | 1.089 | 2.165 | |
| V08 | 34.622 | 30.747 | 36.553 | -1.931 | -5.806 | |
| V09 | 33.395 | 32.983 | 34.925 | -1.53 | -1.942 | |
| V10 | 29.411 | 28.32 | 32.122 | -2.711 | -3.802 | |
| PB01 | 36.916 | 35.959 | 34.708 | 2.207 | 1.251 | |
| PB02 | 39.968 | 37.302 | 38.709 | 1.259 | -1.41 | |
| PB03 | 33.534 | 33.29 | 32.125 | 1.409 | 1.165 | |
| Peripheral marks stains | PB04 | 33.238 | 32.984 | 31.225 | 2.014 | 1.759 |
| PB05 | 35.688 | 33.91 | 33.411 | 2.277 | 0.499 | |
| PB06 | 33.314 | 32.905 | 32.574 | 0.74 | 0.331 | |
| PB07 | 33.939 | 32.237 | 32.349 | 1.59 | -0.112 | |
| PB08 | 34.88 | 34.609 | 33.287 | 1.593 | 1.322 | |
| PB09 | 32.542 | 33.395 | 32.3 | 0.242 | 1.094 | |
| PB10 | 32.143 | 31.823 | 31.175 | 0.968 | 0.648 | |
| MB01 | 27.306 | 29.261 | 33.737 | -6.431 | -8.914 | |
| MB02 | 26.976 | 28.567 | 35.349 | -8.373 | -4.476 | |
| MB03 | 27.637 | 29.395 | 32.292 | -4.655 | -6.781 | |
| Menstrual bloodstains stains | MB04 | 28.628 | 30.555 | 33.425 | -4.797 | -2.896 |
| MB05 | 27.517 | 28.033 | 34.158 | -6.641 | -2.871 | |
| MB06 | 29.707 | 30.755 | 34.712 | -5.005 | -6.125 | |
| MB07 | 25.812 | 26.475 | 29.489 | -3.678 | -3.957 | |
| MB08 | 26.452 | 27.65 | 32.646 | -6.193 | -3.015 | |
| MB09 | 26.267 | 27.375 | 31.625 | -5.358 | -4.995 | |
| MB10 | 26.243 | 25.95 | 35.157 | -6.431 | -4.25 | |
| NTC** | N | undetermined | 40.642 | undetermined | —— | —— |
Note *: The tested 10 semen samples were extracted from 40 normal semen samples randomly.
** NTC: negative control.
2. The expression levels of miR10b, miR135b, and RNU6b, the specificity and sensitivity of the method
2.1 The expression level of miR10b, miR135b, and RNU6b in different body fluids
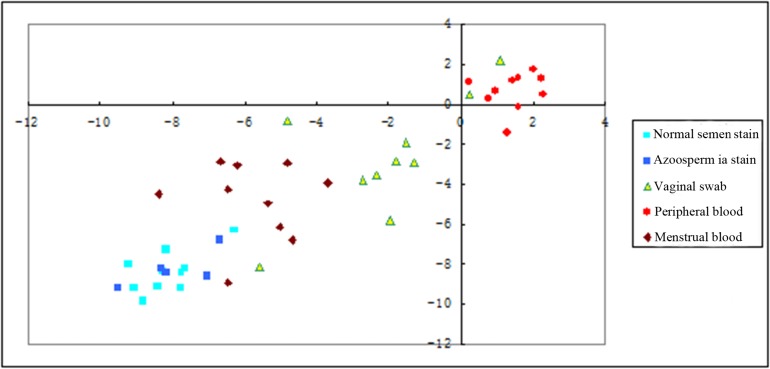
The value of ΔCT [10b-U6] was used as the abscissa axis, and the value of ΔCT [135b-U6] was used as the longitudinal axis; the point distribution graph was obtained according to the detection results for different body fluid stains (Fig 1). The figure have show that the scatter-point distributions of the peripheral blood, semen stains, vaginal swabs, and menstrual bloodstains have obvious different. The scattered points for semen stains, vaginal swabs, and menstrual bloodstains were in the same quadrant, but there were characteristic cluster in different fluid stains. The azoosperma (semen stains without sperm) and normal semen stains were found to be distributed in the same area, with no significant differences in location,(mean ΔCT-8.1297V-7.9342, p>0.05; -8.3813V-8.051, p>0.05) (see S2 Table).
Fig 1. The two-dimensional scatter plots was created with ΔC t values.
The ΔC t values were used to create two-dimensional (2D) scatter plots to determine whether differentiation of each body fluid would be possible. Significantly, distinct clustering of each body fluid was observed in each assay clearly separated from the other body fluid expression data. Individual body fluid data points are represented by colored different shape: brilliant green, normal semen stains; blue, azoospermia stains; green triangle, vaginal swab; red, peripheral blood; brown, menstrual blood.
2.2 The specificity of the method
The histogram was created by comparing the relative expression levels (Relative Quantitation, RQ) of miR10b and miR135b in different body fluids stains (Fig 2), which showed that the expression levels of miR10b and miR135b in semen stains were higher than that in the other body fluid stains. To compared Ct between semen stain and others body fluid (vaginal swabs, Va; peripheral blood, Pe; menstrual bloodstains, Me) with statistical method to two marke, mean △Ct at m10b-8.03195v-2.0724(Va), p<0.001; 1.4299(Pe), p<0.001–5.7562(Me), p<0.002; at 135b-8.2172v-2.7272(Va), p<0.001; 0.6547(Pe), p<0.001; -4.8282(Me), p<0.003 (see S3 Table).
Fig 2. The expression of two miRNA marks in different body fluid.
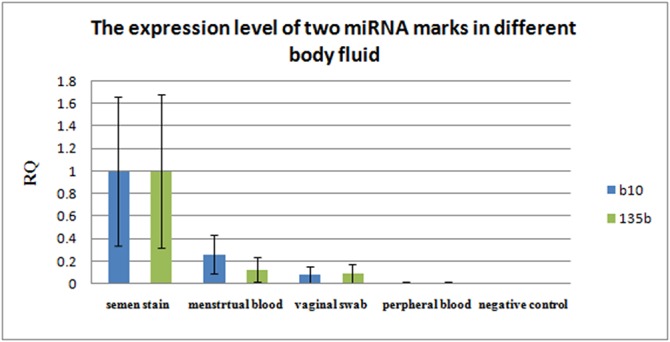
The level of expression are on the X axis, and the relative quantitation (RQ) is on the Y axis. RQ value of 10b and 135b is the highest in semen stain, almost 100%, next, it is in menstrual blood and vaginal swab stains. RQ value is the lowest in peripheral blood except negative control.
2.3 The sensitivity of the method
The results for the sensitivity of the method are shown in Table 2, which were obtained after the semen samples were a series diluted.
Table 2. The results of miRNA RT-qPCR with a series of diluted RNA templates.
| Total RNA | Cт | ΔCт | +/- | |||
|---|---|---|---|---|---|---|
| (ng) | miR10b | miR135b | RNU6b | [10b-U6] | [135b-U6] | |
| 100 | 27.392 | 26.29 | 35.942 | -8.55 | -9.652 | + |
| 10 | 30.327 | 29.68 | 38.968 | -8.641 | -9.288 | + |
| 1 | 31.788 | 33.449 | undetermined | —— | —— | Failed* |
| 0.1 | 32.199 | 36.752 | undetermined | —— | —— | Failed |
| 0.01 | 31.938 | 49.227 | undetermined | —— | —— | Failed |
| 0.001 | 32.381 | undetermined | undetermined | —— | —— | Failed |
| NTC** | undetermined | undetermined | undetermined | —— | —— | Failed |
Note *: Failed: When test results of reference gene is undetermined, it is considered that RNA extraction is unsuccessful or the concentration of RNA is too low to be amplified.
** NTC: negative control.
3. The Reliability and Detection Capability of the Method
Semen stains stored at different times were detected. The results are shown in Table 3. At 25°C, there are no significant differences in the test results.
Table 3. The summarized results of miRNA RT-qPCR with old semen stains.
| Storage time at room temperature | Number of samples | positive |
|---|---|---|
| 1 day | 10 | 10 |
| 1 month | 10 | 7 |
| 3 months | 10 | 9 |
| 6 months | 10 | 8 |
| 1 year | 10 | 8 |
Thirty samples of normal semen stains and five samples of no sperm semen stains were detected. The tested results of those samples were confirmed with the standards defined by the research. The results are 31 confirmed and 3 exclude in 35 semen stains samples, 10 blood, 9 vaginal swabs and 9 menstrual bloodstains exclude. The results were normal, with the exception that the tested gene was not detected in one sample; the results, therefore, suggest that the method is reliable. The old semen samples which were kept in reserve within 1 year could be tested basically (see S4 Table).
Discussion
MiRNA fragments are short and degrade relatively slowly [5]. Their distributions have the tissue specificity; as such, their unique molecular characteristics may be used as a new way of identifying body fluid stains in forensics. This has important implications regarding the detection of problematic samples (i.e. degraded or old samples) and therefore has been a subject of interest by forensic researchers [6–14].
The results demonstrate that the expression levels of the target genes miR10b and miR135b were much higher than that of the reference gene RNU6b in semen stains. The highest levels of expression of miR10b and miR135b were in semen stains, as the levels were higher than those found in menstrual bloodstains and in vaginal swabs. It had been reported that level of expression of miR10b and miR135b are higher in semen stains than in saliva stains [7]. By analyzing the results of this study and comparing the differences in the expression of the target genes using statistical methods, criteria can be derived to distinguish a semen stain from other body fluid stains. The standards were defined as follows: when using the RT-qPCR technique to detect an miRNA marker, the threshold value should be set to 0.04; the derived CT value can be detected in the target genes miR10b and miR135b and in the internal reference gene RNU6b, and CT values are<40, when ΔCT [10b-U6]<-5.5, and ΔCT [135b-U6]<-6, respectively the semen stains can be confirmed. The confirmation standards was derived from the research results by Statistical method and samples test. A test stain can then be compared to the semen stain. In this study, 35 semen stain samples were analyzed. The results were compared to this standard, with the results falling within the positive range 31 except 3 exclude sperm and 1 RNA degradation or extract unsuccessful. Furthermore, the five semen stains without sperm were analyzed using the same test methods and criteria; the results were also within the positive range. In the same way, ten vaginal swab stain, ten menstrual bloodstains and ten peripheral bloodstains were analyzed, the results have not been found to fall within the positive range except a sample of menstrual bloodstains (MB1) and vaginal swabs stains (V2)(Table 2). The result of the two samples was exceptional, and the reasons were relatively complicated. One of a reason was the error caused by operator during the experiment, another perhaps was abnormal expression of the sample itself, such as, a influence was brought about by some unknown disease. It needs to be further researched. Therefore, the technology can be used to distinguish normal semen stains and without sperm stains from other bodily fluids. These results are concordant those of Hanson’s [7], who suggested that the expression levels of miR10b and miR135b were lower than that of RNU6b in a semen stain, but theirs expression levels were higher than other body fluid stains. Accordingly, the expression levels of miR10b and miR135b were suitable for the specificity markers of miRNA to confirm semen stains [7, 11]. However, this experimental result demonstrates that the expression levels of miR10b and miR135b were much higher than that of the reference gene RNU6b in semen; these results are inadequacy conformity with Hansons’ conclusions. Why did the results of this study were partly different from Hanson? The real reason have not been figure out, maybe was condition of experiment, or sequence of probe and primer, or taken sample different. It needs to be research further.
Although most of the semen stains and non-semen stains can be distinguished by the use of the miRNA RT-qPCR technique with a strict positive standard, the false-positive and negative rates when using this technique are still higher than that of the mRNA RT-qPCR technique. This is one particular drawback that is worthy of further attention and research. Theoretically, detecting miRNA could have higher sensitivity than mRNA because of its wider distribution and shorter fragments [10, 12, 14]. Actually, our experimental results illustrated also that the detection sensitivity of the miRNA RT-qPCR techniques to detect a semen stain is within the 10ng level, which is lower than that of the mRNA RT-qPCR technique(data do not be shown). But, by analyzing the experimental data, it was found that miRNA cannot be detected in low concentration semen stains; this was mainly due to the fact that the internal reference gene of RNU6b was not detected. miR10b and miR135b have good amplification curves, with a CT value that is negatively correlated with the template concentration. The sensitivity of the miRNA RT-qPCR technique to detect semen stains will increase if an internal control gene with a relatively higher expression level is used.
In this experiment, the old semen stains were identified using the semen stain confirmation miRNA RT-qPCR system after one day, one month, three months, six months, and one year at 25°C (under dark and dry conditions). The results demonstrated that the positive rates of old stain preservation at one year were 80%. The samples were stored for different times, but the expression levels of miR10b, miR135b, and RNU6b did not significantly vary among different research times. Therefore, it was concluded from the studied results that the stability of miRNA markers was better than that of mRNA (About comparing the stability and sensitivity of miRNA markers and mRNA markers, please read detailed data http://202.116.65.75/c/portal/layout?pl_id=PUB.1001.219 for reference, 《A study of specific mRNA and microRNA markers in seminal stains》 author: Tianyu Xue, a master's thesis.) As such, this technique is more suitable for the identification of old or degraded body fluid stain samples. The further experiment is necessary when the samples were laid up for more lengthy time.
In short, the use of quantitative PCR technology makes it possible to use the expression of miR10b and miR135b as a detection marker in semen stains, especially when used for old or degraded samples.
Supporting Information
(DOC)
(DOC)
(DOC)
(DOC)
Acknowledgments
The authors would like to thank Chen Yong and Chen Wei Hong of the forensic department of Sun Yat-Sen University for helping with the experiment. The authors would also like to thank Zeng Yan Hong of First Affiliated Hospital for providing the specimens.
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
This research is supported by the National Natural Science Foundation (30400519), the Scientific Research Foundation of Ministry of Education (2008-890), and the application innovation program of the Ministry of Public Security (2007YYCXGDST079).
References
- 1. Beuvink I, Kolb FA, Budach W, Garnier A, Lange J, Natt F, et al. A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs. Nucleic Acids Res. 2007; 35: e52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. MiRBase: tools for microRNA genomics. Nucleic Acids Res. 2008; 36: D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005; 33: e179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit. Rev. Biochem. Mol. Biol. 2013; 48: 51–68. 10.3109/10409238.2012.738643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer . Curr Genomics. 2010; 11:537–561. 10.2174/138920210793175895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Virkler K, Lednev IK. Analysis of body fluids for forensic purposes: from laboratory testing to non-destructive rapid confirmatory identification at a crime scene. Forensic Sci. 2009; 188:1–17. [DOI] [PubMed] [Google Scholar]
- 7. Hanson EK, Lubenow H, Ballantyne J. Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal Biochem.2009; 387:303–314. 10.1016/j.ab.2009.01.037 [DOI] [PubMed] [Google Scholar]
- 8. Gao LL, Li YY, Yan JW, Liu YC. Application and progress of RNA in forensic science. Fa Yi Xue Za Zhi.2011; 27:455–459. [PubMed] [Google Scholar]
- 9. Uchimoto ML, Beasley E, Coult N, Omelia EJ, World D, Williams G. Considering the effect of stem-loop reverse transcription and real-time PCR analysis of blood and saliva specific microRNA markers upon mixed body fluid stains. Forensic Sci Int Genet.2013; 7: 418–421. 10.1016/j.fsigen.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 10. Zhu J, Feng X, Lou J, Li W, Li S, Zhu H, et al. Accurate Quantification of microRNA via Single Strand Displacement Reaction on DNA Origami Motif. PLoS One.2013; 8:e69856 10.1371/journal.pone.0069856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hanson EK, Ballantyne J. Circulating microRNA for the identification of forensically relevant body fluids. Methods Mol Biol.2013; 1024:221–234. 10.1007/978-1-62703-453-1_18 [DOI] [PubMed] [Google Scholar]
- 12. An JH, Shin KJ, Yang WI, Lee HY. Body fluid identification in forensics. BMB Rep. 2012; 45: 545–553. [DOI] [PubMed] [Google Scholar]
- 13. Zubakov D, Boersma AW, Choi Y, van Kuijk PF, Wiemer EA, Kayser M. MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int J Legal Med.2010; 124:217–226. 10.1007/s00414-009-0402-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Z, Zhang J, Luo H, Ye Y, Yan J, Hou Y. Screening and confirmation of microRNA markers for forensic body fluid identification. Forensic Sci Int Genet.2013; 7:116–123. 10.1016/j.fsigen.2012.07.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.