Abstract
Since its inception in 1992, Functional Magnetic Resonance Imaging (fMRI) has become an indispensible tool for studying cognition in both the healthy and dysfunctional brain. FMRI monitors changes in the oxygenation of brain tissue resulting from altered metabolism consequent to a task-based evoked neural response or from spontaneous fluctuations in neural activity in the absence of conscious mentation (the “resting state”). Task-based studies have revealed neural correlates of a large number of important cognitive processes, while fMRI studies performed in the resting state have demonstrated brain-wide networks that result from brain regions with synchronized, apparently spontaneous activity. In this article, we review the methods used to acquire and analyze fMRI signals.
Keywords: fMRI, BOLD, methods, resting-state
1 Introduction
Functional Magnetic Resonance Imaging (fMRI) is a neuroimaging tool that employs MRI to image dynamic changes in brain tissue that are caused by changes in neural metabolism. Alterations of neural activity may be caused by asking the subject to perform a task designed to target a specific cognitive process, or can occur spontaneously while the subject is resting in the absence of conscious mentation (i.e., in the “resting state”). Both types of studies- task-based and resting state, have become indispensible tools for studying cognition in healthy as well as diseased brains, and tens of thousands of studies have been published (>150,000 listed in http://www.ncbi.nlm.nih.gov/pubmed under “fMRI; brain”) in the 2+ decades since nearly simultaneous introduction of the technique by three independent groups (Bandettini, Wong et al. 1992, Kwong, Belliveau et al. 1992, Ogawa, Tank et al. 1992).
The MR contrast mechanism used for virtually all fMRI relies on blood oxygenation level dependent (BOLD) changes in brain tissue, exhibited when a brain region experiences altered levels of oxygen consumption consequent to up- or down-regulated metabolic activity caused, e.g., by performing a cognitive task (Ogawa, Tank et al. 1992). When there is a local increase in neural (and glial) activity, concomitant increases in aerobic and anaerobic oxygen consumption trigger increased delivery of fully oxygenated hemoglobin through vasodilatory (Raichle, Grubb et al. 1976, Roland and Larsen 1976, Sokoloff, Reivich et al. 1977, Fox, Raichle et al. 1988, Malonek and Grinvald 1996) processes that increase Cerebral Blood Flow (CBF) to the region. For reasons that are still not fully understood (Fox and Raichle 1986, Frahm, Merboldt et al. 1994, Buxton, Wong et al. 1998), oxygen supply transiently exceeds demand, which results in a net increase in local oxygenation for several seconds (Fig. 1). Thus, the endogenous deoxyhemoglobin (Hb) is dynamically replaced with oxyhemoglobin (HbO2), and is accompanied by a transient increase in intravascular blood volume, resulting in a change in oxygenation state. Because Hb is paramagnetic while HbO2 is diamagnetic, the change in state from paramagnetic to diamagnetic results in a decrease in R2 and R2* relaxivity rates (Thulborn, Waterton et al. 1982, Ogawa, Lee et al. 1990). Thus, an MRI sequence with T2 (1/R2) or T2* (1/R2*) weighting can demonstrate BOLD contrast, and therefore signal neural activity changes through this hemodynamically driven process.
Figure 1.
BOLD contrast results from hemodynamically driven changes in blood oxygen level due to the difference in magnetic state of oxygenated hemoglobin (diamagnetic) molecules versus deoxygenated hemoglobin (paramagnetic) in capillaries and surrounding tissue. During activation (B) increased blood flow and blood volume causes reduction in endogenous Hb, increasing the T2*-weighted MRI signal relative to the baseline state (A).
The microgradients in magnetic field that surround vessels and capillaries filled with Hb result in two forms of BOLD contrast (Bandettini, Wong et al. 1994, Weisskoff, Zuo et al. 1994). The first is due to intravoxel dephasing, which is most prominent near larger vessels, and which causes T2* weighted signal loss. This contrast increases linearly with magnetic field strength and is readily observed with gradient recalled echo (GRE) imaging. The second type of contrast is due to diffusion of spins through the microgradients, causing a reduction in T2-weighted signal detected by spin echo (SE) MRI. The diffusion mechanism is most prominent when the distance the spins diffuse during the signal acquisition is comparable to the spatial extent of the microgradients, which thereby tunes this mechanism to be most sensitive to detecting BOLD contrast in capillaries (Weisskoff, Zuo et al. 1994). Diffusion contrast is proportional to the square of the magnetic field strength. Therefore, as the field is increased, the weighting of T2 contrast increases relative to T2* weighted contrast, with the result that in fields of 4T and higher BOLD contrast is more localized to tissue than to the larger veins when SE acquisitions are employed (Yacoub, Shmuel et al. 2001). By contrast, with GRE acquisitions at 7T the T2* of veins is so short the venous contribution becomes small and diffusion weighting from tissue microstructure dominates (Geissler, Fischmeister et al. 2013). Because of this, SE acquisitions are to be preferred at 7T, although SE methods have higher RF power deposition (Specific Absorption Rate, SAR), which may reduce the number of slices that can be collected.
Gradient recalled acquisitions suffer signal loss from static magnetic field distortions that are caused by magnetic susceptibility differences at air-tissue interfaces, for example in frontal orbital or lateral parietal brain regions. These gradients in magnetic field (~9 ppm difference in susceptibility between air and brain tissue) are large enough to cause signal dropout artifacts from intravoxel dephasing in GRE acquisitions. Spin echo methods refocus the static field heterogeneities, and therefore do not have signal dropout. The relationships between contrast, artifacts and field strength are summarized in Table 1.
Table 1.
Relationships between field strength for GRE & SE acquisition type, BOLD contrast mechanism, dropout severity (IvD = intravoxel dephasing, Diff = diffusion)
| B0 | Acquisition | BOLD Contrast | Dropout Artifact |
|---|---|---|---|
| ≤3T | GRE | (veins : IvD) > (tissue : Dif) | ↑ |
| SE | (tissue : Diff) | - | |
| 7T | GRE | (tissue : Diff) ↑ | ↑↑ |
| SE | (tissue : Diff) ↑ | - | |
At this time, 7T and higher field magnets are not in widespread use, so that the majority of fMRI studies are performed at 3T (in which T2 and T2*-weighted contrasts are comparable or 1.5T, which is mostly sensitive to BOLD contrast in the draining veins (Kruger and Glover 2001, Kruger, Kastrup et al. 2001). Therefore, it would be wise to avoid 1.5T for neuropsychological studies whenever possible, to obtain the most accurate depiction of cognitive processes.
As we have indicated, there are two primary types of fMRI studies- those in which a cognitive task is used to modulate specific neuronal activity, and resting state studies. In either case, a dynamic series of T2*-weighted scans is acquired, resulting in (Kruger, Kastrup et al. 2001) a time series of signals for every brain voxel. These time series are submitted to various levels of correction and denoising (preprocessing steps) before model- or data-driven analyses are applied to obtain maps of activity. Because BOLD signals are tiny- typically a few percent or less- such analyses use statistical methods to discern false from true activation at a given confidence level.
This article reviews the methods employed to acquire and process BOLD fMRI data, with which to draw inferences regarding neural processes. We will not examine other methods often used in conjunction with fMRI such as Diffusion Tensor Imaging (DTI) (Le Bihan, Breton et al. 1986), which can depict or summarize structure of white matter, or Arterial Spin Labeling (ASL) (Williams, Detre et al. 1992), used to map the CBF either in stasis or during task manipulation. Similarly, despite the increasing interest in combining fMRI with other imaging modalities in order to obtain complementary information in the spatiotemporal (e.g., Electorencephalography (EEG) (Teplan 2002), or metabolic (e.g., Near Infrared Spectroscopy (NIRS) (Ferrari, Giannini et al. 1985) and Positron Emission Tomography (PET) (Raichle 1989) domains, these topics are beyond the scope of this review.
2. The fMRI experiment
2.1 Task-based fMRI
In task-based fMRI, time series data are compared against a hypothesized model of neural function based upon the cognitive task being performed. Through the use of statistical inference the hypothesis can be accepted or rejected for every voxel. In this way, a map of those brain regions that respond to the task is constructed, and can be further tested against phenotypical or genotypical models or parametric manipulations of the task, e.g., difficulty.
The typical fMRI experiment employs sensory stimuli to cue the participant to perform a behavioral task while BOLD contrast images are acquired for a fixed duration of minutes (see Fig. 2). Such stimuli can be visual, auditory or of other forms depending on the desired behavioral manipulation. In all cases, the task design employs a modulation of the behavior being studied within each scan (state A – experimental and state B – control in Fig. 2) so that the range of BOLD contrast elicited by the manipulation between experiment and control conditions is captured within one scan. This is important because MRI signal intensities are subject to drifts from instrument instability or changes in participant habitus or physiology that are unrelated to the effect of interest, which makes it difficult to develop accurate estimates of BOLD contrast changes from separate scans.
Figure 2.
Task-based fMRI experiment acquires a time series of images while participant performs cognitive manipulation that causes a change between brain states A and B. The functional map depicts those regions that were more metabolically activated in state A than B, using a statistical test to demonstrate significant signal differences in each voxel.
Task designs are commonly of the “Block Trial” type, “Event-related (ER) Trial” type (Dale and Buckner 1997, Dale 1999, Miezin, Maccotta et al. 2000, Ollinger, Corbetta et al. 2001, Liu 2012) or “Mixed Trials” (Chawla, Rees et al. 1999, Visscher, Miezin et al. 2003, Petersen and Dubis 2012), as shown in Fig. 3. In each case the effect size is inferred from the difference in BOLD contrast between the two states. However, note that because the measured signal is a hemodynamic response to changes in local brain metabolism and therefore only an indirect measure of neural responses, the hemodynamic process itself must be considered in the design of the signal model used to test for activation. A typical Hemodynamic Response Function (HRF), i.e., the BOLD signal obtained from a single brief activation event, is shown in Fig. 4 (Friston, Josephs et al. 1998). The HRF has the characteristic of a low-pass temporal filter, and under the linear assumption for BOLD contrast (Boynton, Engel et al. 1996, Dale and Buckner 1997), it must be convolved with the task design vector to provide the regressor that is used to test for significant activation in any voxel’s time series (see statistical analysis of task data below). As seen by the example in Fig. 4, the HRF’s filtering action can significantly attenuate short duration activity of event-related designs. It has been shown that Block designs are optimum for detecting an activation, while ER designs are most efficient for characterizing the time course of activation, and Mixed designs lie in between (Liu, Frank et al. 2001). Thus, when it is desired to simply decide whether a hypothesized activation occurs in a brain region, the Block design is most effective, but an ER design should be employed when more detailed characteristics of the neural response to the cognitive manipulation are desired (Dale 1999, Birn, Cox et al. 2002, Petersen and Dubis 2012). Of course, there are many variations on these basic designs, and some additional considerations for experimental design are described in (Huettel 2008).
Figure 3.
Types of basic task designs, showing (A) Block design, (B) Event-Related (ER) design and (C) Mixed design. The “on” and “off” levels indicate that the stimulus is either presented for the cognitive manipulation being tested or for a control condition, respectively. Block designs provide maximum detection sensitivity, while ER designs optimize the ability to characterize the time course of BOLD responses. Mixed designs can accomplish signal detection as well as response quantification.
Figure 4.
Hemodynamic Response Function (A) is convolved with an event-related design (B) to derive a regressor (C) with which to model an ER experiment. The regressor can be used either with convolution analysis or in a general linear model.
From the preceding description, statistical inferences regarding task-based brain function are drawn by testing for BOLD signal variations that significantly correlate with a hypothesized model. Therefore, any signal fluctuations unrelated to the effect of interest will degrade the power of the test because of the added unrelated variance. Examples include thermal noise (Edelstein, Glover et al. 1986), and physiological noise resulting from cardiovascular pulsatility or quasi-periodic respiration effects (Hu, Le et al. 1995, Glover, Li et al. 2000), as well as from longer-term vaso-dilatory effects (Birn, Diamond et al. 2006, Shmueli, van Gelderen et al. 2007, Chang, Cunningham et al. 2009). Corrections for these effects are discussed later as preprocessing steps. Of course, there can be cases (e.g., aversive pictures) where the task manipulation causes the participant to alter cardiac function or respiration, which can become a direct confound by causing BOLD signal changes stemming solely from the vascular response of changes in physiology (Birn, Murphy et al. 2009, Chang, Metzger et al. 2013). See later discussion of Preprocessing.
Other considerations important when setting up psychiatric fMRI experiments include possible confounds of medications and hormones. Many medications alter the brain’s vascular function, which in turn causes changes in BOLD response that can result in group differences when comparing medicated experimental populations against healthy controls. For example, even the relatively benign agent caffeine causes elevation in resting CBF and results in reduced BOLD responses due to reduced vascular reserve (Liu, Behzadi et al. 2004). Thus, it may be difficult to perform fMRI studies aiming to investigate cognitive effects of pharmacological treatments because of possible BOLD signal changes unrelated to the neural processes being explored.
2.2 Resting state fMRI
In the resting state (RS) case, the implicit hypothesis is that there are distinct brain regions whose fluctuations are temporally synchronized, and thereby are connected as nodes of networks, such as the Default Mode Network (Greicius, Krasnow et al. 2003, Buckner, Andrews-Hanna et al. 2008). Multiple networks are regularly observed (Damoiseaux, Rombouts et al. 2006, Smith, Fox et al. 2009). The acquisition of RS data is similar to that of task-basked studies, described below. The subject is typically prompted to remain still and avoid targeted mentation, while maintaining eyes open or closed for the scan duration. The latter instruction is important because it has been shown that FC differs between eyes open and closed (e.g., (Patriat, Molloy et al. 2013)). Typically heart rate and respiration data are collected for physiological denoising (see section 4.1.7 physiological noise correction).
3. Acquisition of fMRI data
FMRI scan sequences typically employ single-shot acquisitions using EPI (Mansfield 1977) or Spiral-in/out (Glover and Law 2001) k-space trajectories. Slices are typically acquired in an interleaved order (e.g., 1,3,5, …2,4,6,…), which diminishes “slice-bleed” effects (Bernstein, King et al. 2004), but which must be accounted for during “slice-timing” corrections. The main issues to be controlled during data acquisition are tradeoffs between spatial and temporal resolution, signal dropout in frontal and parietal regions and subject motion. In general, as the spatial resolution is increased, the duration of the readout increases, which makes it more sensitive to signal loss from magnetic susceptibility effects near heterogeneous brain regions. Acceleration using parallel imaging (multiple receiver coil channels) (Sodickson and Manning 1997, Pruessmann, Weiger et al. 1999, Griswold, Jakob et al. 2002) and simultaneous multiple slices (SMS) (Feinberg, Moeller et al. 2010, Setsompop, Gagoski et al. 2012) can change these tradeoffs by reducing the amount of data that need to be acquired and thereby increasing scan efficiency. SMS methods, for example, reduce the repetition time (TR) needed to acquire whole brain coverage by factors of 8 or more (e.g., (Chen, Vu et al. 2015)). The faster acquisition in turn allows more time frames to be collected, increasing the statistical power or enabling more complex temporal inferences, e.g., identifying dynamically changing brain repertoires in the temporal domain (Smith, Miller et al. 2012), but is often accompanied by reduced SNR. The higher scan efficiency afforded by acceleration can alternatively allow scan time to be reduced, thereby decreasing chances for head motion and increasing access for, and compliance by, populations such as children, older adults and patients.
Functional acquisition protocols include other sequences as well as the functional scan(s) themselves, and thus their scan times must also be considered when setting up a protocol to minimize the protocol duration. T2-weighted sequences (FSE or TSE) can be used to rapidly acquire high resolution slices with the same scan prescription as the functional scan. These are typically replaced or accompanied by T1 Inversion Recovery prepared whole brain acquisitions (3D MPRAGE or 3DFSPGR (Mugler and Brookeman 1990)) to enable normalization of subject data to a brain template in order to make group inferences (see Preprocessing below). In addition, many investigators collect diffusion tensor information in order to derive structural connectivity maps (“white matter tracks”) (Le Bihan, Breton et al. 1986), and to correlate structure with function (Werring, Clark et al. 1998, Zhu, Zhang et al. 2014). Infrequently, because of the added scan time and complexity, some studies also employ Arterial Spin Labeling (ASL) methods (Williams, Detre et al. 1992, Detre and Wang 2002, Borogovac and Asllani 2012), or hypercapnic challenges using CO2 breathing (Davis, Kwong et al. 1998, Kim, Rostrup et al. 1999) or breath holding (Kastrup, Kruger et al. 1999, Thomason, Foland et al. 2007, Chang, Thomason et al. 2008) to derive more quantitative measurements or correct for confounds such as inter-subject differences in vascular reactivity
As described previously, T2* weighting is usually employed for BOLD signal contrast, which requires long echo times (Bandettini, Wong et al. 1992), but which unfortunately also results in geometric distortion (Hutton, Bork et al. 2002) due to off-resonance near frontal-orbital and parietal regions, where the susceptibility difference between air and tissue generates substantial static magnetic field gradients. The distortion can be corrected using maps of magnetic field (see 4.1.4 distortion correction); therefore, field maps are often also acquired.
4. Analysis
4.1 Preprocessing
As fMRI detects neural activity indirectly via hemodynamic response to changes in metabolic consumption of oxygen, the collected time series are inevitably confounded by non-neurally related sources of variations, such as subjects’ head motion, physiological cycles, and magnetic field inhomogeneity. If not corrected, these unwanted fluctuations may obscure the intrinsic patterns of neural activity, reduce the detection power of further statistical analysis, or in worst cases, alter experimental conclusions by introducing structured noise that contaminates the real neurally-related results.
Several computational procedures, collectively termed as the preprocessing pipeline, have been proposed to remove the confounding sources of variations from the fMRI time series, and increase the functional signal to noise ratio (fSNR) before further analysis. The most frequently employed steps are detailed below.
4.1.1 Quality assurance
quality assurance (QA) testing is an indispensible but often ignored aspect of preprocessing in fMRI studies nowadays. The corruption of fMRI data may occur during data acquisition due to extreme scanner noise, e.g., “spike noise” or other scanner problems such as signal drift (even for the best-maintained scanner). If unnoticed, these corrupted datasets may propagate artifacts into final results of a study. To avoid frustration later with unusable data, the imaging data should be examined immediately post scan (e.g., check subjects’ motion parameters, physiological data, or view the stack of 3D brain images in a movie). In this way, it may be possible to prompt the subject to diminish excessive motion or observe and correct for instrument failures before continuing. QA procedures should also be employed throughout the preprocessing pipeline using visual inspection and simple tests (e.g., examining the mean intensity and standard derivation of slices, calculating the fSNR for each dataset (Murphy, Bodurka et al. 2007)) to guarantee the data quality prior to the next step of analysis.
4.1.2 Slice timing correction
the majority of fMRI studies use a two-dimensional pulse sequence that images one slice at a time, resulting in inconsistent acquisition time among different brain slices within one TR. Such slice-timing errors, if uncorrected, may pose severe inaccuracy in cases where the temporal information is critical, e.g., studies positing a causal relationship among different cortical regions or in rapid event-related experiments. A most common approach to correct for slice-timing errors is temporal interpolation, which estimates the signal amplitude of each slice/voxel at a reference time point by interpolating information from neighboring TRs. This method works most effectively when the single-slice sampling rate is much faster than the signal variability induced by the on-going experiment (Huettel 2008).
4.1.3 Head motion correction
subjects’ head motion is a prominent concern in most fMRI studies, particularly those involving hour-long scan duration (subjects may become increasingly drowsy and restless as time goes by), tasks requiring physical responses (subjects’ motion synchronizes with the on-going stimulus), or particular types of subjects (the young, the elderly and the diseased people).
Subjects’ head motion can affect the collected data quality in various ways. To list a few: motion mixes signals from neighboring voxels, yielding dramatic signal variability at the edge of distinct cortical regions; motion induces spurious distance-dependent variance (more similar between voxels nearby than far apart) that may alter the intrinsic correlation structure of the data; motion interplays with field inhomogeneity and slice excitation, bringing in more complicated noisy fluctuations (Huettel 2008, Van Dijk, Sabuncu et al. 2012, Power, Schlaggar et al. 2015).
Disruptive as it appears, motion can be considerably suppressed by strategies during or post acquisition. Head immobilization techniques, such as fixation devices (bite bars, masks, fixation pads- including inflatable air bags) and use of a mock scanner (training subject in a simulated environment) (Barnea-Goraly, Weinzimer et al. 2014), can diminish head movement during the scans. In addition, myriad retrospective methods have been proposed to correct for motion post acquisition (see (Power, Schlaggar et al. 2015) for a review of approaches and associated concerns). These approaches generally rely on motion parameters characterized by rigid body realignment, which assumes the brain to be a rigid object and estimates at each time point its displacement from a reference position (along three translational and three rotational axes). Motion induced signal variance can be mitigated by projecting the motion measures together with their higher order derivatives out of the data (Friston, Williams et al. 1996, Satterthwaite, Elliott et al. 2013, Yan, Cheung et al. 2013, Power, Mitra et al. 2014), or realigning the brain volume acquired at each time point to a fixed position using spatial interpolation. One may also identify those problematic time points by visually inspecting the time series of motion parameters, and apply censoring (excluding those volumes from further analysis, e.g., (Barch, Sabb et al. 1999, Lemieux, Salek-Haddadi et al. 2007, Kennedy and Courchesne 2008)), or temporal interpolation (extrapolating adjacent volumes, (Power, Mitra et al. 2014)) to suppress motion artifacts. Alternative to approaches that employ the estimated motion parameters, several other techniques attempt to extract motion-related fluctuations from the collected data itself based on its disparity with neural-related fluctuations in spatial distribution and temporal characteristics (Liao, Krolik et al. 2005, Behzadi, Restom et al. 2007, Kundu, Inati et al. 2012, Satterthwaite, Elliott et al. 2013, Patel, Kundu et al. 2014).
As an alternative to retrospective motion correction applied during preprocessing, prospective motion correction (“Promocor”) techniques can be employed during acquisition (see (Maclaren, Herbst et al. 2013) for a review). These methods utilize head position information to adjust the slice plane as the scan progresses so that the imaging plane orientation attempts to follow that of the head. One class of methods uses motion information acquired from the fMRI images or a navigator to alter the scan plane for the next TR. While this method requires no additional instrumentation, it is only applicable for motion that is slow compared to the slice collection time (TR/number_slices) because the correction always lags the motion by one TR. Another class of Promocor methods employs external instruments (visual (Forman, Aksoy et al. 2011) or electrical (Sengupta, Tadanki et al. 2014) to track head orientation in order to obtain truly real-time (<TR) orientation information. Such methods have improved correction but entail additional setup time for the tracking device.
4.1.4 Distortion correction
fMRI signals may suffer from geometric or intensity distortion due to inhomogeneity in the static/excitation fields. Field heterogeneity distorts the shape and location of tissue in the image because the reconstruction assumes a linear relationship between signal frequency and space. Hardware shimming embedded in the MR system can compensate for the magnetic field non-uniformity to a certain extent. In addition, techniques have been developed to correct for distortion in the reconstructed MR images by measuring the field heterogeneity with an additional acquisition of a magnetic field map and employing image or k-space interpolation during reconstruction (Jezzard and Balaban 1995, Hutton, Bork et al. 2002, Cusack, Brett et al. 2003, Sutton, Noll et al. 2003), or in cases when the field maps are not available (Sled, Zijdenbos et al. 1998, Arnold, Liow et al. 2001, Lewis and Fox 2004, Studholme, Cardenas et al. 2004, Vovk, Pernus et al. 2004).
4.1.5 Temporal filtering
in cases where the spectrum distributions of signal and noise components do not strictly overlap with each other, temporal filtering – which eliminates noisy frequencies but preserves signal frequency – can help enhance fSNR of the data. For instance, in studies employing block-design paradigm (the task-related signals reside in very narrow frequency bands), the detection power of the experiments can be effectively improved by suppressing the power of frequencies other than that of the task. Another common type of temporal filtering is to remove the slow fluctuations induced by scanner drift. Such high-pass filtering procedure is also referred to as detrending (Tanabe, Miller et al. 2002), and has been included as a routine step in most software packages. Besides improving fSNR of a time series, moderate temporal filtering can also reduce the bias in ensuing statistical analysis by obscuring the disparity between assumed and intrinsic models of the data (Friston, Josephs et al. 2000).
4.1.6 Spatial smoothing
Benefits from spatial smoothing are mainly threefold. First, spatial smoothing can improve the fSNR of the data. Due to functional similarity of adjacent brain areas and signal blurring caused by vascular origins, fMRI data are inherently spatially correlated as acquired. As a result, proper spatial smoothing, i.e., typically implemented by convolving the data with a Gaussian kernel that matches the inherent spatial correlation of fMRI data, could suppress noise sources uncorrelated among adjacent imaging voxels and increase the tSNR of the data (Lowe and Sorenson 1997, Skudlarski, Constable et al. 1999, Parrish, Gitelman et al. 2000, LaConte, Anderson et al. 2003). Second, spatial smoothing may also improve the validity of ensuing statistical analysis by mitigating the difference between inherent spatial structure of the data and the assumed model, e.g., increasing the Gaussianity of the data (a key assumption of the general linear model, and random field theory (Worsley, Cao et al. 1998)), or achieving valid estimation of the degrees of freedom in ensuing multiple comparisons (Worsley 2005). Lastly, proper spatial smoothing can also ameliorate the anatomical or functional variations among different subjects. Unfortunately, the optimum kernel sizes determined by different goals above are not consistent. For example, to maximize fSNR, the kernel size should match the spatial correlations of each region, while to approximate the assumed smooth Gaussian field, the ideal kernel size should be at least twice the size of a voxel (Worsley 2005). Meanwhile, several drawbacks of spatial smoothing should be considered as well. For instance, a larger kernel size will reduce the spatial resolution of acquired data, and may blur the functional boundaries or shift the activation loci of a task to an unacceptable level (Geissler, Lanzenberger et al. 2005, Sacchet and Knutson 2013). Therefore, there is inherent difficulty in choosing an appropriate kernel size (see (White, O'Leary et al. 2001, Worsley 2005, Scouten, Papademetris et al. 2006, Mikl, Marecek et al. 2008, Weibull, Gustavsson et al. 2008) for exploratory studies and detailed discussions). As oversimplified recommendations for conventional studies adopting fixed kernel size throughout the brain (in contrast to adaptive smoothing strategies, e.g., (Penny, Trujillo-Barreto et al. 2005, Yue, Loh et al. 2010)), a modest smoothing kernel size (~4mm) is suggested for single subject analysis, while a wider kernel size (6~8mm) can be applied for a group-level analysis. However, examining the results with no or modest kernel width is always recommended when a wide smoothing kernel is applied.
4.1.7 Physiological noise correction
as BOLD contrast originates from hemodynamically-driven changes in tissue and vessel oxygenation, it naturally contains non-neural fluctuations incurred by physiological processes, such as cardiac pulsatility and respiration (Birn 2012).
Briefly, the cardiac and respiratory-related physiological noise can be classified into two categories based on their spectral distributions. The first category refers to time-locked fluctuations directly synchronized with the cardiac (~0.8–1.3 Hz) and respiratory cycles (~0.1–0.3 Hz): cardiac pulsatility induces tissue movement and blood inflow that may cause signal fluctuations adjacent to large brain vessels (Dagli, Ingeholm et al. 1999); respiration engenders chest movement that can alter the magnetic susceptibility and MR signal intensity (Raj, Anderson et al. 2001, Brosch, Talavage et al. 2002). Such periodical noises have been demonstrated to be greater than system and thermal noise at 3T or higher magnetic field (Kruger and Glover 2001). With effectively faster acquisition (e.g., TR < 0.5s), the cyclic fluctuations can be resolved and temporally filtered out of the data (Biswal, DeYoe et al. 1996, Mitra and Pesaran 1999). However, despite the emergence of fast acquisition techniques, the majority of fMRI studies nowadays still use TR ≥ 2s for the whole brain acquisition, causing the cardiac noise to be aliased onto lower frequencies. To correct for the aliased physiological noise, several retrospective techniques have been proposed (Hu, Le et al. 1995, Le and Hu 1996, Glover, Li et al. 2000, Chuang and Chen 2001, Pfeuffer, Van De Moortele et al. 2002, Verstynen and Deshpande 2011). These approaches first characterize the physiological noise by either modeling them from external physiological recordings (e.g., photoplethysmography, respiratory belt and pulse oximetry (Verstynen and Deshpande 2011)), or estimating them directly from the acquired data, then extract the estimated noisy fluctuations out of the time course of each voxel.
A second category of physiological noise relates to variations of respiratory volume and heart rate. Variations of breathing depth and rate lead to altered levels of arterial CO2, a potent vasodilator modulating blood flow and consequently the amplitude of BOLD signals (Modarreszadeh and Bruce 1994, Van den Aardweg and Karemaker 2002, Wise, Ide et al. 2004, Birn, Diamond et al. 2006, Birn, Murphy et al. 2008, Chang and Glover 2009). The variability of heart rate possesses, which extends to broader spectrum compared to pulsalitity cycles, has been shown to account for considerable amounts of BOLD fluctuations in resting state (Shmueli, van Gelderen et al. 2007, Chang, Cunningham et al. 2009). Numerous studies have been proposed to model such noisy fluctuations from external recordings of physiological data (Birn, Smith et al. 2008, Chang, Cunningham et al. 2009, Verstynen and Deshpande 2011) or the data itself (in a manner similar to the removal of motion artifacts) (Beall and Lowe 2007, Behzadi, Restom et al. 2007, Perlbarg, Bellec et al. 2007, Weissenbacher, Kasess et al. 2009, Jo, Saad et al. 2010, Anderson, Druzgal et al. 2011).
4.1.8 Functional-structural co-registration
the collected 3D stack of anatomical and functional images generally do not match each other due to different MR contrasts and acquisitions (e.g., inconsistent slice orientation, voxel resolution and image distortion), causing problems in mapping activity (from functional data, e.g., the task-activation map) to the anatomical image. Computational procedures that map functional and structural images to each other are termed functional-structural co-registration. These procedures typically resample the anatomical data to the spatial resolution of functional data first, then employ a rigid body transformation where a cost function (e.g., mutual information) is minimized (see (Gholipour, Kehtarnavaz et al. 2007, Klein, Andersson et al. 2009) for reviews).
4.1.9 Spatial normalization
In most neuroscience studies, we may need to aggregate brain activities across multiple individuals. Given that the shape and size of brains are rather inconsistent across subjects, a standard approach is to normalize each individual’s brain to a template estimated locally from specific populations (Guimond, Meunier et al. 2000) or published ones (Talairach atlas (Talairach 1988) and MNI templates are most commonly used, see (Brett, Johnsrude et al. 2002, Devlin and Poldrack 2007, Lancaster, Tordesillas-Gutierrez et al. 2007) for differences and transformations between the two coordinate systems). Spatial normalization can be intensity, landmark, or surface based (see (Gholipour, Kehtarnavaz et al. 2007, Klein, Andersson et al. 2009) for reviews). It is typically implemented by either registering each individual’s functional images to a functional template directly, or in two steps: (1) co-registering functional and structural images; (2) registering the anatomical image to a high-resolution structural template. These two approaches each have their own advantages and shortcomings – the former approach avoids inconsistent geometric distortions induced by different imaging contrasts; while the latter approach appears more robust due to improved resolution and quality of the structural image – the employment of which depends on the particular scanning environment and imaging protocols.
For reference, Fig. 5 offers a summary framework for preprocessing of fMRI data. Notably, the determination of specific preprocessing pipeline interacts with numerous factors, e.g., types of stimulus, experimental hypothesis, and acquisition environment (Strother 2006, Huettel 2008). For instance, it is more proper to perform slice time correction prior to motion correction with interleaved slice acquisition, while the order should be switched in a sequential acquisition (green rectangle). Moreover, for processes that operate linearly on the datasets, switching orders would yield no differences in the final results (e.g., procedures in the pink rectangle). Furthermore, it can be questioned whether to normalize the functional images prior to or after statistical analysis. The former avoids extra smoothing, image distortions introduced by imperfect normalization in the ensuing analysis, whereas the latter makes statistical analysis demanding matched voxels from different subjects plausible (e.g., group independent component analysis (Calhoun, Adali et al. 2001), atlas-based graph analysis).
Figure 5.
The basic scheme of preprocessing pipelines.
The preprocessing steps listed above apply to different task paradigms as well as resting state scans. Compared to block trial type designs, event-related designs have relatively lower detection power and high demand on the temporal precision. Therefore, removal of various non-neural confounds and slice timing correction are essential and indispensible for event-related studies. In resting state studies, an additional procedure – global signal regression (GSR) is sometimes included in the preprocessing pipeline. GSR averages the time series across all brain voxels and projects the averaged global signal out of each voxel’s time series using linear regression, assuming that the averaged signal is dominated by non-neural fluctuations that affects brain’s time series globally. GSR has been shown to improve the specificity of functional connectivity, mitigate motion artifacts (Satterthwaite, Elliott et al. 2013, Yan, Cheung et al. 2013, Power, Mitra et al. 2014), and yield prominent anti-correlations in resting state studies (Fox, Snyder et al. 2005, Fox, Zhang et al. 2009). However, this procedure has been controversial in resting state studies, because global signal may also carry information related to neural activity (Scholvinck, Maier et al. 2010, Wong, Olafsson et al. 2013). Moreover, it has been shown both theoretically and practically that GSR shifts the center of correlation values (by reducing positive correlations and introducing artificial negative correlations) such that all the correlation values across the brain sum to a negative value (Murphy, Birn et al. 2009, Weissenbacher, Kasess et al. 2009, Saad, Gotts et al. 2012, Gotts, Saad et al. 2013). Therefore, the inclusion of GSR as a preprocessing procedure is advised with great caution. Generally, noise sources that affect large areas of the brain (e.g., physiological noise, motion) can be modeled by reasonable alternatives discussed earlier (see 4.1.3 head motion correction, 4.1.7 physiological noise correction above). However, there are a few situations where GSR can be considered. For instance, if the alternative methods are not accessible (e.g., without external recordings of cardiac or respiratory waveforms) or the data contain global confounds that cannot be effectively modeled by existing approaches, GSR could be tested, but it is highly recommended to reexamine the results without GSR. Besides, in studies that do not directly investigate the interaction patterns among different brain regions, e.g., using pattern recognition methods to classify two mental states, GSR could be employed as a common data manipulation procedure. One needs to be careful with the interpretation of results, because the inherent interaction structure of the brain has already been altered.
As introduced in the acquisition section, there has been growing interest in fMRI studies with faster sampling rates (from conventional seconds to sub-second scales). Faster acquisition promises higher temporal resolution, increased statistical power (more sampling points with a fixed scan duration), and the examination of neural information in higher frequency bands. The majority of existing studies with faster acquisition (Wu, Gu et al. 2008, Boubela, Kalcher et al. 2013, Boyacioglu, Beckmann et al. 2013, Lee, Zahneisen et al. 2013, Chen and Glover 2015, Gohel and Biswal 2015) still follow the routine preprocessing pipeline employed in conventional studies, which may lack rigorous validation. We mention a few concerns regarding this issue that warrant careful explorations in the future. First, as BOLD contrast results from an inherently slow hemodynamic process, the spectrum of observed neural information (<0.3 Hz according to the canonical HRF model in SPM8, Wellcome Trust Centre for Neuroimaging, University College London, UK) is less likely to accommodate the observation of functional connectivity at very high frequencies. It is therefore not yet clear whether one can apply similar de-noising procedures as used previously to the observed high-frequency (> 0.1 Hz) BOLD functional connectivity data (see (Chen and Glover 2015) for discussion of HRF in the resting state and tSNR vs. frequency).. Second, BOLD time series are inherently auto-correlated, suggesting that the effective number of degrees of freedoms will not scale linearly with the number of time frames collected at a fixed scan duration. Unfortunately, properly accounting for the degrees of freedom is ignored in many studies, leading to over-estimation of statistical significance. Third, the energy of BOLD time series is dominated by low-frequency signals (e.g. at resting state, power spectrums of spontaneous fluctuations mainly reside below 0.1 Hz). As a result, if we apply the conventional preprocessing pipeline to acquired full band time series, parameter fittings involved in different steps may be driven by the low-frequency data and cannot effectively de-noise the high-frequency band data. For instance, if we linearly project motion regressors (see 4.1.3 motion correction above) out of each voxel’s time series, the scaling parameter of each regressor would to a large extent depend on the low-frequency component (main fluctuation) of each time series. However, there has been no clear evidence that the relative relationships between signals and motion noises are consistent across different frequencies. Indeed, additional noise (high-frequency components of the motion regressors) may be introduced due to this preprocessing step and alter the structure of true neural-related high-frequency signals.
4.2 Analysis of task studies
After preprocessing, the next step is to examine the research hypothesis of the designed experiment. In this section, we take the two-condition blocked design (experimental vs. control), a widely adopted experimental paradigm in fMRI studies, as an example to illuminate several common methodologies of statistical analysis.
In this particular setting, we want to identify voxels actively involved in the imposed task, i.e., voxels whose temporal behaviors differ significantly between the experimental and control conditions. Specifically, we test the research hypothesis H1: experimental condition ≠ control condition, against its null hypothesis H0: experimental condition = control condition.
4.2.1 The t test
To introduce the concept of statistical inference in fMRI, the simplest procedure is to treat each time point as an independent sample (at least initially we assume so), and compare signal amplitudes under different conditions using a standard two-sample student’s t test. This procedure is repeated for each brain voxel. To quantify the statistical significance of the estimated t-value, a p value is defined – the chance of observing a statistic t-value or more extreme results under the null hypothesis. If a voxel’s p value is smaller than a user defined significance level α, we can hence reject the null hypothesis and classify the voxel as ‘active’.
Primary challenges of the t-test analysis are twofold. First, the fMRI time series is filtered by a sluggish hemodynamic process; as a result the actual response of an active voxel may lag ~5s of the condition boxcar, requiring that this delay be accounted for in assigning time points to one or the other state. More importantly, the transition between brain states cannot be represented as belonging uniquely to either state, which can represent a serious loss of statistical power. Therefore, straightforward as it appears, this t-test approach is rarely used directly with time series data in fMRI studies.
4.2.2 Correlation Analysis
A more elegant approach is to examine the temporal synchrony between each voxel’s time series and the predicted response of the experiment (Bandettini, Jesmanowicz et al. 1993) – derived by convolving the task-control boxcar waveform with a canonical HRF. The correlation coefficient (eqn. (1)) is the most frequently used metric to quantify the correspondence (or sometimes referred to as functional connectivity) between two time series x and y (equal length):
| eqn. (1) |
where n denotes the number of time points per signal, x̄/ȳ and σx/σy denote the means and standard deviations of x and y.
The correlation coefficient r ranges from −1 to 1, with 0 meaning no correlation (the null hypothesis), and ±1 meaning perfect positive/negative correlations. An r value can be converted to student’s t-value based on its degrees of freedom (unconstrained number of parameters), and we can therefore identify active voxels using similar ways introduced above (see the t test above). To allow for the variability of temporal delays in HRFs across different brain voxels, cross correlation – which estimates the correlation coefficient as a function of the temporal lag of one signal relative to the other, may be applied (Friston 1994).
Correlation analyses can only be applied when a single hypothesis is to be tested, i.e.,, to test a voxel’s time series against another time series, such as that generated as a model for activation as described above, or to test for similarity with the time series of another voxel in resting state studies. However, fMRI experiments frequently involve more than one manipulation or condition to characterize responses during different temporal events within the scan, i.e., there are multiple hypotheses to be tested. In this case, correlation analysis cannot be used, and a more general method must be employed, as described next.
4.2.3 The general linear model (GLM) analysis
As an alternative to or expansion upon correlation analysis, the GLM analysis has been widely employed in the fMRI community to examine the temporal synchrony between experimental observations and the predicted responses (see Fig. 6A) (Friston 1995). Briefly, the fMRI time series y, is modeled as a linear mixture of several model factors (e.g., predicted task response) and white Gaussian distributed additive noise term ε as below:
| eqn. (2) |
Figure 6.
The majority of model-based and model-free analyses in fMRI studies can be incorporated into a coherent scheme of matrix decomposition. Specifically, the 4-D fMRI dataset can be rearranged into a 2-D matrix by aligning all voxels of the same time point in a row; different approaches (e.g., GLM (A), MVPA (B), ICA/PCA (C)) attempt to decompose the 2-D matrix into sub-components by imposing various assumptions of the de-composed matrix structure (blue rectangles), then extract the spatial patterns (network patterns, pink rectangles) of neural-related contributions.
where xi denotes each model factor, and the parameter weight βi is the scaling term indicating the contribution of a model factor to the dependent variable y. When y refers to a large number of dependent variables, such as different time points across a scan in an fMRI study, eqn. (2) represents the GLM.
Statistical testing of the GLM estimates how well each voxel’s time series is fit by the linear combination of model factors. There exist routine ways (if the autocorrelation structure of ε is known) of converting the fitted results to t-, or F-statistics to assess the significance of each model factor’s contribution to y (assuming the null hypothesis: βi = 0, i > 0, i.e., no contribution from xi) (Friston, Holmes et al. 1995, Worsley and Friston 1995).
For instance, in the two-conditioned block task case, we can estimate each voxel’s task relevance by including a single model factor x1 (the predicted response), and testing the statistical significance of β1. This is equivalent to correlational analysis. Through versatile modifications of model factors, this approach allows more flexible shapes of the predicted response (originating from inconsistent temporal delays and variability of HRF shapes across different brain regions), details can be found in chapter 5 of (Poldrack 2011), and chapter 10 of (Huettel 2008).
Compared to correlation analysis, the GLM approach allows for more flexible experimental designs (e.g., experiments involving three or more cognitive conditions), and can include any known sources of variability as model factors, such as nuisance components (e.g., motion parameters, physiological fluctuations) and non-imaging information (e.g., subjects’ age and behavioral data, genotypical information, multi-site scanning environment). Other applications of GLM also include characterizing the impulse responses in event-related designs (Dale 1999, Glover 1999), studying the psychophysiological interactions (PPI) among different regions (Friston, Buechel et al. 1997), and investigating the coupling between fMRI and other imaging modalities (e.g., EEG recordings (Goldman, Stern et al. 2000, Laufs, Kleinschmidt et al. 2003)).
The major limitation of the GLM lies in the validity of model assumptions in specific fMRI applications (pertaining to relationships among model factors, between noise and model factors, and the assumed serial structure of the noise term, see (Monti 2011, Poline and Brett 2012) for reviews), which unfortunately, is rarely discussed in most cases. Of course, the GLM also presupposes linearity of BOLD responses (Boynton, Engel et al. 1996), which may not be valid. In such cases, higher order models must be utilized (Friston, Josephs et al. 1998).
4.2.4 Multivariate pattern analysis
Correlation analysis and the GLM introduced above treat each brain voxel independently and examines its intensity differences irrespective of all other voxels. However, such a univariate assumption may not hold rigorously, given that our brain is a complex system with tight interactions between different cortical regions. For instance, it is possible that an experimental condition modulates the activity pattern among multiple voxels without altering the averaged intensity levels of each voxel. In such cases and others, it may be statistically advantageous to examine groups of voxels simultaneously. Therefore, in contrast to univariate analyses focusing on the independent activity of individual voxels, a multivariate pattern analysis (MVPA) scheme, which integrates responses of multiple voxels/regions in an experiment, may be exploited (see Fig. 6B) (Haxby, Gobbini et al. 2001). Briefly, this set of approaches reference the concept of pattern classification, seeking to characterize the combination of activities among multiple voxels/regions to differentiate between experimental conditions (see (O'Toole, Jiang et al. 2007, Pereira, Mitchell et al. 2009, Haxby 2012, Mahmoudi, Takerkart et al. 2012) for reviews). Since 2001, MVPA has been actively applied in fMRI studies to investigate activity patterns in visual systems and various cognitive processes (see (Haynes and Rees 2006, Norman, Polyn et al. 2006, Tong and Pratte 2012) for reviews); several MVPA toolboxes are readily available for non-technical users (e.g., a matlab-based Princeton MVPA toolbox (http://code.google.com/p/princeton-mvpa-toolbox/), a Python-based PyMVPA toolbox (http://www.pymvpa.org)). Although MVPA is more sensitive and informative than univariate analysis, several technical challenges still exist, which potentially prevent it from being the dominant approach in revealing brain activity patterns (see (Norman, Polyn et al. 2006, Haxby 2012, Tong and Pratte 2012) for more complete discussions). First, it is hard to identify the neural representations of the learnt classification patterns, e.g., questions such as what information is encoded in the pattern, remain unclear. Second, technical factors (e.g., which voxels/regions should be covered, what spatial/temporal scales should be encoded) interplay with the performance of MVPA in very complicated manners, but are not easily determined. Finally, due to the high demand of fine-grained topographies in distinguishing subtle differences across states, MVPA are typically done in each individual’s native dataset, posing problems in characterizing activity patterns at a group level (see (Haxby, Guntupalli et al. 2011) for a plausible solution).
4.2.5 Correction for multiple comparisons
As mentioned above, the ‘p value’ is defined so to rigorously assess the statistical significance associated with each observed metric. If a voxel’s p value is smaller than a user defined significance level α, we reject the null hypothesis and classify the voxel as ‘active’.
However, in a standard fMRI analysis, we are faced with the challenge of multiple comparisons. For instance, if we attempt to identify ‘active’ voxels across 100,000 voxels under the significance level α = 0.05, ~ 5,000 voxels may be falsely classified as active by random chance under the null hypothesis. To correct for these false positive errors (voxels identified as ‘active’ but which are indeed not) induced by multiple comparisons, several approaches have been proposed (see (Nichols 2012) for a review). These approaches generally fall into two categories. A first category directly introduces more stringent voxel-wise significance level α (the threshold of p values) by assigning new error criteria, e.g., Familywise Error Rate (FWE, the chance of one or more false discoveries (Nichols and Hayasaka 2003)) and False Discovery Rate (FDR, the expected proportion of false positives among detected activation (Genovese, Lazar et al. 2002)). FWE is typically controlled by Bonferroni procedure and is effective in suppressing false positives, however, it has less power than FDR in general. A second category of methods controls the false positive probability of an entire cluster (contiguous voxels) instead of a single voxel. These methods first define clusters by retaining ‘active’ voxels above a primary p threshold, then evaluate the statistical significance of cluster activation by testing its size against the null hypothesis of no active voxels in that cluster. Popular approaches include random field theory (Worsley, Evans et al. 1992, Worsley, Taylor et al. 2004), Monte Carlo simulation (Forman, Cohen et al. 1995) and nonparametric permutation (Holmes, Blair et al. 1996, Nichols and Holmes 2002). Compared to the first category of approaches, these methods are advantageous in less stringent thresholds, high sensitivity, and incorporation of the spatial correlation, but have limited spatial specificity for large clusters (see (Woo, Krishnan et al. 2014) for detailed discussion).
4.2.6 Inter-subject analysis
The analyses discussed above have focused on identifying task activations in a single subject. However, an fMRI study typically recruits several or many subjects in order to probe biodiversity and generalize across a population or disease state. Therefore, we need to combine results across subjects to better test the experimental hypothesis. There exist two main approaches to make the group-level inference of task activation (Huettel 2008).
The first, and more straightforward way is to combine (through temporal concatenation or averaging) the time points of all the examined subjects in a single time series, and perform single-subject level analysis as introduced above. This approach relies on the key assumption that the experimental effect is fixed across all the recruited subjects, so it is termed a fixed-effect analysis. If this assumption (inter-subject variation = 0, only intra-subject variation exists) holds true, temporal combination of each subject’s data can improve the detection power by either increasing the degrees of freedom (concatenation) or reducing intra-subject variations (averaging). However, due to this assumption, the result of fixed-effect analysis is very sensitive to outliers within the recruited subjects (subjects with extreme task responses). Consequently, the conclusions are restricted to the specific subjects scanned within the study, and may not generalize to a larger population.
In contrast to fixed-effect analysis, a random-effect analysis is more commonly applied in fMRI studies nowadays. This analysis assumes that each subject is drawn from a large population of subjects, and that his response represents an independent sample from the overall distribution of task effects. The random-effect analysis is performed in two stages. In the first stage, summary statistics regarding task activation from each individual subject is analyzed independently. In the second stage, the distribution of summary statistics derived from the first stage is tested for significance. For instance, we can use a simple t-test to examine whether the summary statistics from all the subjects are drawn from a distribution with a mean of zero. If intra-subject variations in the first stage are carried up to the second stage, this analysis can also be referred to as mixed-effect analysis (Poldrack 2011). Because random-effect analysis permits the inferences of the entire population from which the subjects are drawn, it is preferable to fixed-effect analysis in most applications, and has been made available with various fMRI statistical toolboxes (see Table 2).
Table 2.
Several of the major fMRI toolboxes with flexible preprocessing pipelines and statistical analysis models
| Toolbox | website |
|---|---|
| AFNI | http://afni.nimh.nih.gov/afni/ |
| Brain Voyager | http://www.brainvoyager.com |
| FIASCO | http://www.stat.cmu.edu/~fiasco/ |
| FreeSurfer | http://surfer.nmr.mgh.harvard.edu |
| FSL | http://fsl.fmrib.ox.ac.uk/fsl/ |
| MEDx | http://ftp.medicalnumerics.com/products/medx/index.html |
| REST | http://restfmri.net/forum/index.php |
| SPM | http://www.fil.ion.ucl.ac.uk/spm/ |
| Stimulate | https://www.cmrr.umn.edu/stimulate/ |
4.3 Analysis of resting state data
In task-based experiments (blocked, event-related, or mixed designs), we can target brain regions/patterns associated with the on-going stimulus by examining each brain voxel’s temporal synchrony with the task waveform. By contrast, during task-free mental conditions (e.g., resting state, levels of consciousness, continuous hypercapnia/hypocapnia challenges), we do not have explicit timing information to model the temporal behavior of neural-related fluctuations inherent in each brain voxel. To reveal the patterns of functional connectivity governing a task-free condition, several schemes of analyses have been proposed in the past two decades.
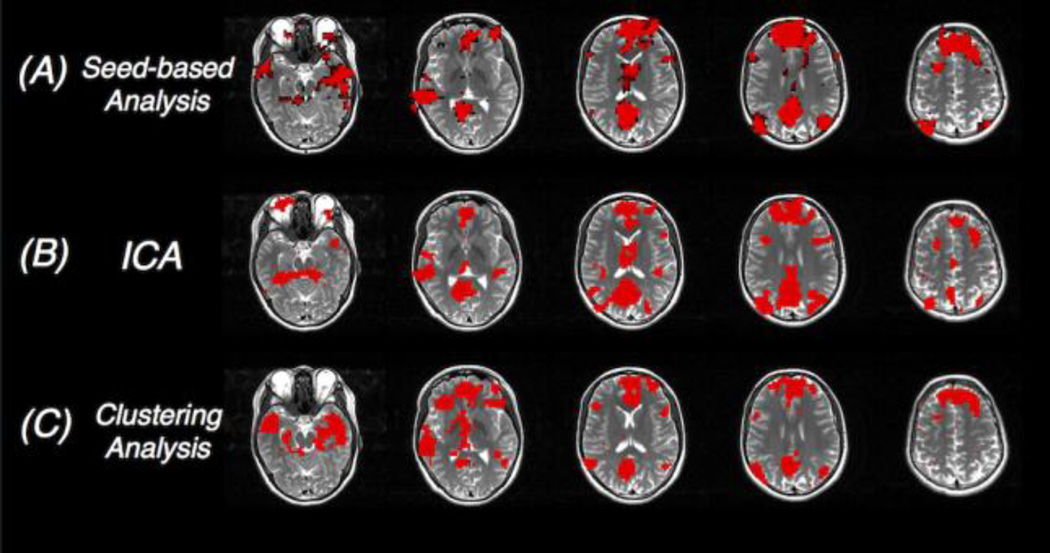
The first approach is seed-voxel based analysis (see Figs. 6A and 7A) (Biswal, Yetkin et al. 1995, Cordes, Haughton et al. 2000, Greicius, Krasnow et al. 2003, Hyde and Jesmanowicz 2012). This approach extracts the time series of a seed region, typically the activation/deactivation locus delineated from a prior task scan or a node within the network under investigation, and estimates its temporal synchrony with the rest brain voxels using GLM or correlation analysis introduced earlier. The topography of the network, i.e., regions significantly coupled with the seed voxel, informs the functional interaction patterns of the seed/network, and can be further compared under types of mental conditions or groups of subjects (healthy controls vs. clinical population) to examine its modulation by cognitive loads and neuropsychiatric disorders.
Figure 7.
Network patterns of the default mode network (a special resting state network) generated by different analyses approaches (red overlays). Results from the seed-based correlation analysis and ICA are thresholded for display.
A second type of analysis enables the exploration of whole-brain functional connectivity configuration without prior selections of network seeds using data-driven or model-free methods. Among all these model-free approaches, i.e., approaches without prior assumptions, independent component analysis (ICA) is the most frequently employed in task-free fMRI studies (see Figs. 6C and 7B) (McKeown and Sejnowski 1998, Calhoun, Adali et al. 2001, van de Ven, Formisano et al. 2004, Beckmann, DeLuca et al. 2005, Smith, Fox et al. 2009, Beckmann 2012). Very briefly, ICA separates the whole brain voxels into additive subcomponents by assuming that the subcomponents are non-Gaussian and they are statistically independent from each other. The spatial patterns of the obtained independent components (ICs) resemble those network patterns resolved by seed-based analysis (Greicius, Srivastava et al. 2004), and are consistent across different studies or subjects (Damoiseaux, Rombouts et al. 2006). Besides, ICA is able to identify certain non-neural sources of variability, such as motion or physiological noise, as separate subcomponents, and can therefore aid preprocessing (Liao, Krolik et al. 2005, Beckmann 2012). Major shortcomings of this approach include: (1) ICs correspond to more complicated representation of the raw fMRI data than seed-based functional connectivity maps, making it difficult to interpret group differences in clinical or neuropsychiatric applications; (2) the resolved ICs and their spatial patterns vary as a function of the number of subcomponents specified by the user; and (3) the classification of components into noise or signal is subject to user-induced interpretation bias.
In addition to ICA, several other model-free approaches have been proposed to characterize the functional connectivity patterns in task-free states. For instance, principal component analysis (PCA) projects the raw fMRI data into orthogonal spaces – principal components (PCs), and only focuses on the space spanned by the leading few PCs (i.e., PCs explaining the most variance of the original dataset) (Friston, Frith et al. 1993). A number of clustering techniques (Fig. 7C), such as hierarchical clustering (Cordes, Haughton et al. 2002), Normalized-cut (van den Heuvel, Mandl et al. 2008), Laplacian based clustering (Thirion, Dodel et al. 2006), fuzzy clustering (Chuang, Chiu et al. 1999), and spectral clustering (Craddock, James et al. 2012), have been applied to produce resting state networks as well. Clustering analysis attempts to parcellate the brain into distinct clusters such that intra-cluster similarity is higher than inter-cluster similarity. Naturally, voxels belonging to the same functional network (with strong temporal synchrony) will fall within the same cluster, if the cluster number is properly chosen.
A third type of analysis simplifies cortical regions as distributed functional nodes, and computes the pair-wise functional correlations of these nodes to achieve a global view of functional organization. The functional nodes can be derived by spatially parcellating the brain voxels into functionally homogeneous ROIs, or more conveniently, employing the recently reported functional atlas (Craddock, James et al. 2012, Shirer, Ryali et al. 2012). Obviously, this approach offers a more intuitive, comprehensive characterization of the connection patterns. However, as it assumes functional homogeneity within each functional ROI/node and is assumed to inherit the information carried at individual-voxel level, whether these atlas ROIs can be generalized to broader populations or mental disorders is questionable. For instance, it has been demonstrated that the functional connectivity map at rest may reorganize under different types of neuropsychological disease or age modulation (see (Fox and Greicius 2010, van den Heuvel and Hulshoff Pol 2010, Rosazza and Minati 2011, Lee, Smyser et al. 2013) for reviews). At the very least, given that alternations of atlas topography may be more or less reflected as changes in the node-level connectivity, we can still use this functional node level analysis as a preliminary step to target candidate brain regions, then employ approaches such as seed-based analysis to examine in detail the disruptive functional dissociations in more detailed manners.
4.4 Advanced analysis
4.4.1 From functional connectivity to effective connectivity
The vast majority of clinical inferences drawn from resting fMRI studies stem from quantifications of functional connectivity – the direct temporal synchrony among distributed cortical regions. Nonetheless, these metrics do not inform further the directional causal influence between neuronal systems that underlie the observed macroscopic correlation. Therefore, there have been growing efforts exploiting effective connectivity, the directed causal influence that one neuronal system exerts on another (see (Friston 2009, Friston 2011, Poldrack 2011, Valdes-Sosa, Roebroeck et al. 2011, Stephan and Roebroeck 2012) for reviews). Approaches estimating the effective connectivity generally start with sets of assumptions on the inherent data structure (time series, correlation matrix or higher-order statistics) or underlying biophysics to be modeled, then seek the optimum models using criteria such as maximum likelihoods or Bayesian inferences, and finally invoke the learned model parameters to conclude causality or conditional dependences. The most common approaches include dynamic causal modeling (DCM) (Friston, Harrison et al. 2003, Penny, Stephan et al. 2004, Lee, Friston et al. 2006, Stephan, Weiskopf et al. 2007, Marreiros, Kiebel et al. 2008, Stephan, Kasper et al. 2008, Penny, Stephan et al. 2010, Schuyler, Ollinger et al. 2010, Seghier, Zeidman et al. 2010, Stephan, Penny et al. 2010, Daunizeau, David et al. 2011, Friston, Li et al. 2011, Li, Daunizeau et al. 2011, Lohmann, Erfurth et al. 2012, Friston, Kahan et al. 2014), Granger causality analysis (Granger 1969, Goebel, Roebroeck et al. 2003, Harrison, Penny et al. 2003, Roebroeck, Formisano et al. 2005, Deshpande, LaConte et al. 2009), structural equation modeling (SEM) (McIntosh 1994, Buchel and Friston 1997, Horwitz, Tagamets et al. 1999, Bullmore, Horwitz et al. 2000), psychophysiological interaction (Friston, Buechel et al. 1997), graphical causal modeling (Pearl 2000, Spirtes 2000), dynamic Bayesian networks (Rajapakse and Zhou 2007), and switching linear dynamic system (Smith, Pillai et al. 2010); and have been actively employed in clinical studies to identify abnormal interactions in patients (e.g., Alzheimer’s disease (Agosta, Rocca et al. 2010, Rytsar, Fornari et al. 2011, Liu, Zhang et al. 2012, Neufang, Akhrif et al. 2014, Zhong, Huang et al. 2014), depression (Schlosser, Wagner et al. 2008, Almeida, Versace et al. 2009, Goulden, McKie et al. 2010, Moses-Kolko, Perlman et al. 2010, Hamilton, Chen et al. 2011, Iwabuchi, Peng et al. 2014, Liu, Wu et al. 2015), and schizophrenia (Schlosser, Gesierich et al. 2003, Kim, Burge et al. 2008, Benetti, Mechelli et al. 2009, Crossley, Mechelli et al. 2009, Dima, Roiser et al. 2009, Allen, Stephan et al. 2010, Diaconescu, Jensen et al. 2011, Deserno, Sterzer et al. 2012, Guller, Tononi et al. 2012, Mukherjee, Whalley et al. 2012, Birnbaum and Weinberger 2013, Zhang, Wei et al. 2013, de la Iglesia-Vaya, Escarti et al. 2014, Hutcheson, Sreenivasan et al. 2015)). As suggested by the way it is termed, effective connectivity opens a promising avenue to perceive the neural-related couplings of our brain systems. Nevertheless, such goals are challenging to achieve in real implementations due to the biophysics of fMRI and several technical limitations. First, fMRI by nature inevitably contain spatiotemporal variability from hemodynamic sources. Without rigorously correcting for hemodynamic variability, it may not be sensible to claim that the observed causal relationship reflects a neuronal origin (Chang, Thomason et al. 2008, David, Guillemain et al. 2008, Deshpande, Sathian et al. 2010, Roebroeck, Formisano et al. 2011, Webb, Ferguson et al. 2013). Second, to make integrative and precise inferences of information flow at the neuronal level, an ideal model should include the whole set of brain regions (even for cases assessing the effective connectivity between two regions) and superior to all alternative possible structures. The problem is thus problematic due to computational complexity (enormously large dimension expanded by the model space) and inadequate samples (the number of unknown free parameters is much larger than the number of time points per fMRI scan). To tackle the problem, we can enforce specific constraints on the model space by prior assumptions or briefly characterizing the causal structure of the data using approaches such as graphical causal modeling (Ramsey, Hanson et al. 2010). Apart from the two major concerns discussed here, effective connectivity faces other challenges, e.g., modeling causal structure across multiple subjects, see (Ramsey, Hanson et al. 2010, Poldrack 2011, Valdes-Sosa, Roebroeck et al. 2011) for discussions and plausible solutions.
4.4.2 From voxel/ROI-wise correlations to complex network behavior
As mentioned earlier (see analysis of resting state data above), a systematic view of the brain’s functional organization could be achieved by parcellating brain voxels into discrete functional nodes and examining the global interactions among these nodes. Indeed, such data manipulation and simplification also make it plausible to characterize the network-wise, or community-wise behavior of the data. Numerous quantitative metrics, originally proposed in graph theory, have been introduced to learn the complex network behavior of our brain’s functional structure, such as small-world topology (Watts and Strogatz 1998), scale-free network patterns (Barabasi and Albert 1999), rich club behavior (van den Heuvel and Sporns 2011), efficiency of global/local information flow (Latora and Marchiori 2001), hierarchies and modular structures (Meunier, Lambiotte et al. 2010), etc. (see (Bullmore and Sporns 2009, Meunier, Lambiotte et al. 2010, Rubinov and Sporns 2010, Bullmore and Sporns 2012, Sporns 2013) for reviews of complex network measures). These metrics, complimentary to conventional node-wise measures, have provided new opportunities to understand brain functions and neuropsychological diseases under the setting of a complex system (see (Bassett and Bullmore 2009, Stam and van Straaten 2012, Filippi, van den Heuvel et al. 2013, Hulshoff Pol and Bullmore 2013, Stam 2014) for reviews of clinical investigations). Despite the wide application of complex measures in clinical explorations, several potential challenges still lie ahead. For instance, the robustness of the estimated network-wise behavior is apparently vulnerable to choice of functional ROIs (Smith, Miller et al. 2011). If the ROIs employed to extract node time series for further network analysis do not match the actual functional boundaries of the data well, time series from different functional regions may mix with each other and obscure the actual community behavior of our brain. In this sense, functional, locally derived atlases are generally preferable to anatomical, standard atlases defined from large groups of subjects. Besides, these metrics from graph theory usually summarize the brain’s complex network-wise behavior in one single value. One can question whether the observed differences between groups of subjects (e.g., healthy controls vs. clinical populations) originate from neural sources, or from those confounds that affect brain voxels globally, e.g., motion and physiological processes (Smith 2012). Therefore, rigorous modeling and careful removal of potential noise confounds are essential for relevant studies. Examining the dependence of derived metrics to the processing pipeline (by including or excluding certain skeptical steps) can also enhance the reliability of the results. Furthermore, these complex network behaviors typically deviate substantially from the bottom-level temporal synchrony among functional nodes, making clinical interpretations on the results not easy.
4.4.3 From static functional connectivity to brain dynamics
Until a few years ago, studies investigating the RS functional connectivity have invoked the key assumption of temporal constancy, i.e., that interactions among different cortical regions remain unchanged during the entire scan. However, such assumptions are invalidated by recent observations that the network patterns may undergo substantial changes across a single RS scan (Chang and Glover 2010, Kiviniemi, Vire et al. 2011, Handwerker, Roopchansingh et al. 2012, Jones, Vemuri et al. 2012, Hutchison, Womelsdorf et al. 2013, Allen, Damaraju et al. 2014). In contrast to extracting the functional connectivity metrics by integrating time points over the whole scan as performed in conventional static studies, these dynamic analyses investigate the variability of brain connectivity metrics at sub time periods across the scan session (see (Hutchison, Womelsdorf et al. 2013, Calhoun, Miller et al. 2014) for reviews of methodology). The dynamic property of RS functional connectivity may carry information (at least) as important as those time-averaged metrics widely explored in neuroscience studies or clinical applications, e.g., it is entirely possible that clinical populations possess disrupted dynamics, which taken together with abnormal time-averaged metrics, may offer better understanding of the associated disorders. Preliminary applications include mental disorders such as schizophrenia (Sakoglu, Pearlson et al. 2010, Damaraju, Allen et al. 2014, Ma, Calhoun et al. 2014, Rashid, Damaraju et al. 2014, Shen, Li et al. 2014, Yu, Erhardt et al. 2015), major depression (Allen and Cohen 2010), Alzheimer’s disease (Jones, Vemuri et al. 2012), opioid analgesia (Robinson, Atlas et al. 2015), temporal lobe epilepsy (Morgan, Abou-Khalil et al. 2015) and childhood autism (Price, Wee et al. 2014). Of note, as studies of brain dynamic functional connectivity are at quite an exploratory stage, the associated interpretations of disrupted dynamics in disorders are still very cursory – it is yet hard to identify the true mechanism from candidates such as changes in autonomic processes, vigilance states, or behavioral origins (see (Hutchison, Womelsdorf et al. 2013) for a review). Hence, external measurements of physiological processes (e.g., galvanic skin response, respiratory and cardiac data) will be surely beneficial for confound reduction or identification of potential mechanisms.
It is important to highlight two technical challenges of dynamic studies. The first challenge lies in the inability of a standard fMRI scan (with minutes-long duration) to characterize the complete patterns of time-varying functional connectivity (an implicit premise for between group comparison). The total number of interaction patterns that different cortical regions may exhibit, although not quantified yet, is presumably huge. In contrast, the patterns that can be captured by ~10 minute long scanning snapshots are very limited. It is therefore speculative whether the altered patterns of brain dynamics in a clinical group (if they exist) indeed result from the associated disorder, or simply the stochastic nature of limited samples per scan. A second challenge relates to the number of time points (independent observations, if not considering the hemodynamic autocorrelation in time series) involved in the estimation of each connectivity pattern. Most current dynamic analyses implicitly assume that the time our brain spends in each network pattern is substantially shorter than the total scan duration. There is therefore a tradeoff between the statistical power of a limited snapshot of the brain’s behavior and the temporal resolution with which it is desired to test for dynamic changes. The consequent danger is that the number of time points collected when the brain is in a certain network pattern may not be adequate to yield statistically robust estimations. Collectively, at the current stage, longer scan durations (more network patterns) and faster acquisition rates (more samples) may improve the reliability of dynamic brain connectivity.
4.5 Analysis software
Many software packages have been developed for the analysis of fMRI data. These programs have flexible pipelines for preprocessing and multilevel task and resting state analysis (see Table 2 for some popular toolboxes).
Conclusions
Functional MRI has had a long history of development for, and application to, a large body of basic systems-level neuroscience investigation (see (Bandettini 2012) for a review). The original block design experiments have been augmented by many other types of experimental design, but remain a workhorse method for psychiatric studies. While most aspects of BOLD contrast are by now well understood, it remains an active area of research. Furthermore, acquisition methods are reasonably standardized by now with the use of single-shot EPI, although faster, more efficient methods such as SMS have been recently introduced. Both task-based and resting state experiments have shown great promise in understanding the brain’s behavior in healthy and diseased populations, and in guiding clinical therapy. Pattern classification and other forms of multivariate analyses are being employed to develop biomarkers of disease (Nash, Wiley et al. 2013), and with which to predict outcome for therapeutic intervention (Hoeft, McCandliss et al. 2011). Because BOLD fMRI relies on an indirect indicator of metabolic contrast, and because the signals are small, it is crucial to perform adequate compensation for confounds such as physiological noise, dynamic brain states, and subject motion, and to use great rigor in the acquisition and analysis pipelines to make sure the imaging data and derivative results are robust and reproducible across the duration of the study. With proper attention to all these factors, fMRI has become an invaluable tool for psychiatric investigations.
Acknowledgement
This work was supported by NIH P41 EB015891.
References
- Agosta F, Rocca MA, Pagani E, Absinta M, Magnani G, Marcone A, Falautano M, Comi G, Gorno-Tempini ML, Filippi M. Sensorimotor network rewiring in mild cognitive impairment and Alzheimer's disease. Hum Brain Mapp. 2010;31(4):515–525. doi: 10.1002/hbm.20883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex. 2014;24(3):663–676. doi: 10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JJ, Cohen MX. Deconstructing the "resting" state: exploring the temporal dynamics of frontal alpha asymmetry as an endophenotype for depression. Front Hum Neurosci. 2010;4:232. doi: 10.3389/fnhum.2010.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen P, Stephan KE, Mechelli A, Day F, Ward N, Dalton J, Williams SC, McGuire P. Cingulate activity and fronto-temporal connectivity in people with prodromal signs of psychosis. Neuroimage. 2010;49(1):947–955. doi: 10.1016/j.neuroimage.2009.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida JR, Versace A, Mechelli A, Hassel S, Quevedo K, Kupfer DJ, Phillips ML. Abnormal amygdala-prefrontal effective connectivity to happy faces differentiates bipolar from major depression. Biol Psychiatry. 2009;66(5):451–459. doi: 10.1016/j.biopsych.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Druzgal TJ, Lopez-Larson M, Jeong EK, Desai K, Yurgelun-Todd D. Network anticorrelations, global regression, and phase-shifted soft tissue correction. Hum Brain Mapp. 2011;32(6):919–934. doi: 10.1002/hbm.21079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JB, Liow JS, Schaper KA, Stern JJ, Sled JG, Shattuck DW, Worth AJ, Cohen MS, Leahy RM, Mazziotta JC, Rottenberg DA. Qualitative and quantitative evaluation of six algorithms for correcting intensity nonuniformity effects. Neuroimage. 2001;13(5):931–943. doi: 10.1006/nimg.2001.0756. [DOI] [PubMed] [Google Scholar]
- Bandettini PA. Twenty years of functional MRI: the science and the stories. Neuroimage. 2012;62(2):575–588. doi: 10.1016/j.neuroimage.2012.04.026. [DOI] [PubMed] [Google Scholar]
- Bandettini PA, Jesmanowicz A, Wong EC, Hyde JS. Processing strategies for time-course data sets in functional MRI of the human brain. Magn Reson Med. 1993;30(2):161–173. doi: 10.1002/mrm.1910300204. [DOI] [PubMed] [Google Scholar]
- Bandettini PA, Wong EC, Hinks RS, Tikofsky RS, Hyde JS. Time course EPI of human brain function during task activation. Magn Reson Med. 1992;25(2):390–397. doi: 10.1002/mrm.1910250220. [DOI] [PubMed] [Google Scholar]
- Bandettini PA, Wong EC, Jesmanowicz A, Hinks RS, Hyde JS. Spin Echo and gradient echo EPI of human brain activation using BOLD contrast: a comparative study at 1.5T. NMR in Biomed. 1994;7:12–20. doi: 10.1002/nbm.1940070104. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286(5439):509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Barch DM, Sabb FW, Carter CS, Braver TS, Noll DC, Cohen JD. Overt verbal responding during fMRI scanning: empirical investigations of problems and potential solutions. Neuroimage. 1999;10(6):642–657. doi: 10.1006/nimg.1999.0500. [DOI] [PubMed] [Google Scholar]
- Barnea-Goraly N, Weinzimer SA, Ruedy KJ, Mauras N, Beck RW, Marzelli MJ, Mazaika PK, Aye T, White NH, Tsalikian E, Fox L, Kollman C, Cheng P, Reiss AL N. Diabetes Research in Children. High success rates of sedation-free brain MRI scanning in young children using simple subject preparation protocols with and without a commercial mock scanner--the Diabetes Research in Children Network (DirecNet) experience. Pediatr Radiol. 2014;44(2):181–186. doi: 10.1007/s00247-013-2798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Bullmore ET. Human brain networks in health and disease. Curr Opin Neurol. 2009;22(4):340–347. doi: 10.1097/WCO.0b013e32832d93dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall EB, Lowe MJ. Isolating physiologic noise sources with independently determined spatial measures. Neuroimage. 2007;37(4):1286–1300. doi: 10.1016/j.neuroimage.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Beckmann CF. Modelling with independent components. Neuroimage. 2012;62(2):891–901. doi: 10.1016/j.neuroimage.2012.02.020. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360(1457):1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 2007;37(1):90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti S, Mechelli A, Picchioni M, Broome M, Williams S, McGuire P. Functional integration between the posterior hippocampus and prefrontal cortex is impaired in both first episode schizophrenia and the at risk mental state. Brain. 2009;132(Pt 9):2426–2436. doi: 10.1093/brain/awp098. [DOI] [PubMed] [Google Scholar]
- Bernstein MA, King KF, Zhou XJ. Handbook of MRI Pulse sequences. New York: Elsevier Press; 2004. [Google Scholar]
- Birn RM. The role of physiological noise in resting-state functional connectivity. Neuroimage. 2012;62(2):864–870. doi: 10.1016/j.neuroimage.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Birn RM, Cox RW, Bandettini PA. Detection versus estimation in event-related fMRI: choosing the optimal stimulus timing. Neuroimage. 2002;15(1):252–264. doi: 10.1006/nimg.2001.0964. [DOI] [PubMed] [Google Scholar]
- Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. Neuroimage. 2006;31(4):1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Birn RM, Murphy K, Bandettini PA. The effect of respiration variations on independent component analysis results of resting state functional connectivity. Hum Brain Mapp. 2008;29(7):740–750. doi: 10.1002/hbm.20577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Murphy K, Handwerker DA, Bandettini PA. fMRI in the presence of task-correlated breathing variations. Neuroimage. 2009;47(3):1092–1104. doi: 10.1016/j.neuroimage.2009.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Smith MA, Jones TB, Bandettini PA. The respiration response function: the temporal dynamics of fMRI signal fluctuations related to changes in respiration. Neuroimage. 2008;40(2):644–654. doi: 10.1016/j.neuroimage.2007.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum R, Weinberger DR. Functional neuroimaging and schizophrenia: a view towards effective connectivity modeling and polygenic risk. Dialogues Clin Neurosci. 2013;15(3):279–289. doi: 10.31887/DCNS.2013.15.3/rbirnbaum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, DeYoe EA, Hyde JS. Reduction of physiological fluctuations in fMRI using digital filters. Magn Reson Med. 1996;35(1):107–113. doi: 10.1002/mrm.1910350114. [DOI] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34(4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Borogovac A, Asllani I. Arterial Spin Labeling (ASL) fMRI: advantages, theoretical constrains, and experimental challenges in neurosciences. Int J Biomed Imaging. 2012;2012:818456. doi: 10.1155/2012/818456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubela RN, Kalcher K, Huf W, Kronnerwetter C, Filzmoser P, Moser E. Beyond noise: using temporal ICA to extract meaningful information from high-frequency fMRI signal fluctuations during rest. Frontiers in Human Neuroscience. 2013;7 doi: 10.3389/fnhum.2013.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyacioglu R, Beckmann CF, Barth M. An Investigation of RSN Frequency Spectra Using Ultra-Fast Generalized Inverse Imaging. Front Hum Neurosci. 2013;7:156. doi: 10.3389/fnhum.2013.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton GM, Engel SA, Glover GH, Heeger DJ. Linear systems analysis of functional magnetic resonance imaging in human V1. J Neurosci. 1996;16(13):4207–4221. doi: 10.1523/JNEUROSCI.16-13-04207.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett M, Johnsrude IS, Owen AM. The problem of functional localization in the human brain. Nat Rev Neurosci. 2002;3(3):243–249. doi: 10.1038/nrn756. [DOI] [PubMed] [Google Scholar]
- Brosch JR, Talavage TM, Ulmer JL, Nyenhuis JA. Simulation of human respiration in fMRI with a mechanical model. IEEE Trans Biomed Eng. 2002;49(7):700–707. doi: 10.1109/TBME.2002.1010854. [DOI] [PubMed] [Google Scholar]
- Buchel C, Friston KJ. Modulation of connectivity in visual pathways by attention: cortical interactions evaluated with structural equation modelling and fMRI. Cereb Cortex. 1997;7(8):768–778. doi: 10.1093/cercor/7.8.768. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. The brain's default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10(3):186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. The economy of brain network organization. Nat Rev Neurosci. 2012;13(5):336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- Bullmore ET, Horwitz B, Honey G, Brammer M, Williams S, Sharma T. How good is good enough in path analysis of fMRI data? Neuroimage. 2000;11(4):289–301. doi: 10.1006/nimg.2000.0544. [DOI] [PubMed] [Google Scholar]
- Buxton RB, Wong EC, Frank LR. Dynamics of blood flow and oxygenation changes during brain activation: The balloon model. Magnetic Resonance in Medicine. 1998;39(6):855–864. doi: 10.1002/mrm.1910390602. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14(3):140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Miller R, Pearlson G, Adali T. The Chronnectome: Time-Varying Connectivity Networks as the Next Frontier in fMRI Data Discovery. Neuron. 2014;84(2):262–274. doi: 10.1016/j.neuron.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Cunningham JP, Glover GH. Influence of heart rate on the BOLD signal: the cardiac response function. Neuroimage. 2009;44(3):857–869. doi: 10.1016/j.neuroimage.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Relationship between respiration, end-tidal CO(2), and BOLD signals in resting-state fMRI. Neuroimage. 2009;47:1381–1393. doi: 10.1016/j.neuroimage.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage. 2010;50(1):81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Metzger CD, Glover GH, Duyn JH, Heinze HJ, Walter M. Association between heart rate variability and fluctuations in resting-state functional connectivity. Neuroimage. 2013;68:93–104. doi: 10.1016/j.neuroimage.2012.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Thomason ME, Glover GH. Mapping and correction of vascular hemodynamic latency in the BOLD signal. Neuroimage. 2008;43(1):90–102. doi: 10.1016/j.neuroimage.2008.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla D, Rees G, Friston KJ. The physiological basis of attentional modulation in extrastriate visual areas. Nat Neurosci. 1999;2(7):671–676. doi: 10.1038/10230. [DOI] [PubMed] [Google Scholar]
- Chen JYE, Glover GH. BOLD fractional contribution to resting-state functional connectivity above 0.1 Hz. Neuroimage. 2015;107:207–218. doi: 10.1016/j.neuroimage.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Vu AT, Xu J, Moeller S, Ugurbil K, Yacoub E, Feinberg DA. Evaluation of highly accelerated simultaneous multi-slice EPI for fMRI. Neuroimage. 2015;104:452–459. doi: 10.1016/j.neuroimage.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang KH, Chen JH. IMPACT: image-based physiological artifacts estimation and correction technique for functional MRI. Magn Reson Med. 2001;46(2):344–353. doi: 10.1002/mrm.1197. [DOI] [PubMed] [Google Scholar]
- Chuang KH, Chiu MJ, Lin CC, Chen JH. Model-free functional MRI analysis using Kohonen clustering neural network and fuzzy C-means. IEEE Trans Med Imaging. 1999;18(12):1117–1128. doi: 10.1109/42.819322. [DOI] [PubMed] [Google Scholar]
- Cordes D, Haughton V, Carew JD, Arfanakis K, Maravilla K. Hierarchical clustering to measure connectivity in fMRI resting-state data. Magn Reson Imaging. 2002;20(4):305–317. doi: 10.1016/s0730-725x(02)00503-9. [DOI] [PubMed] [Google Scholar]
- Cordes D, Haughton VM, Arfanakis K, Wendt GJ, Turski PA, Moritz CH, Quigley MA, Meyerand ME. Mapping functionally related regions of brain with functional connectivity MR imaging. AJNR Am J Neuroradiol. 2000;21(9):1636–1644. [PMC free article] [PubMed] [Google Scholar]
- Craddock RC, James GA, Holtzheimer PE, 3rd, Hu XP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp. 2012;33(8):1914–1928. doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley NA, Mechelli A, Fusar-Poli P, Broome MR, Matthiasson P, Johns LC, Bramon E, Valmaggia L, Williams SC, McGuire PK. Superior temporal lobe dysfunction and frontotemporal dysconnectivity in subjects at risk of psychosis and in first-episode psychosis. Hum Brain Mapp. 2009;30(12):4129–4137. doi: 10.1002/hbm.20834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack R, Brett M, Osswald K. An evaluation of the use of magnetic field maps to undistort echo-planar images. Neuroimage. 2003;18(1):127–142. doi: 10.1006/nimg.2002.1281. [DOI] [PubMed] [Google Scholar]
- Dagli MS, Ingeholm JE, Haxby JV. Localization of cardiac-induced signal change in fMRI. Neuroimage. 1999;9(4):407–415. doi: 10.1006/nimg.1998.0424. [DOI] [PubMed] [Google Scholar]
- Dale AM. Optimal experimental design for event-related fMRI. Hum Brain Mapp. 1999;8(2–3):109–114. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<109::AID-HBM7>3.0.CO;2-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Buckner RL. Selective averaging of rapidly presented individual trials using fMRI. Hum Brain Mapp. 1997;5(5):329–340. doi: 10.1002/(SICI)1097-0193(1997)5:5<329::AID-HBM1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Damaraju E, Allen EA, Belger A, Ford JM, McEwen S, Mathalon DH, Mueller BA, Pearlson GD, Potkin SG, Preda A, Turner JA, Vaidya JG, van Erp TG, Calhoun VD. Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. Neuroimage Clin. 2014;5:298–308. doi: 10.1016/j.nicl.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci U S A. 2006;103(37):13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daunizeau J, David O, Stephan KE. Dynamic causal modelling: A critical review of the biophysical and statistical foundations. Neuroimage. 2011;58(2):312–322. doi: 10.1016/j.neuroimage.2009.11.062. [DOI] [PubMed] [Google Scholar]
- David O, Guillemain I, Saillet S, Reyt S, Deransart C, Segebarth C, Depaulis A. Identifying neural drivers with functional MRI: an electrophysiological validation. PLoS Biol. 2008;6(12):2683–2697. doi: 10.1371/journal.pbio.0060315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Kwong KK, Weisskoff RM, Rosen BR. Calibrated functional MRI: Mapping the dynamics of oxidative metabolism. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(4):1834–1839. doi: 10.1073/pnas.95.4.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Iglesia-Vaya M, Escarti MJ, Molina-Mateo J, Marti-Bonmati L, Gadea M, Castellanos FX, Aguilar Garcia-Iturrospe EJ, Robles M, Biswal BB, Sanjuan J. Abnormal synchrony and effective connectivity in patients with schizophrenia and auditory hallucinations. Neuroimage Clin. 2014;6:171–179. doi: 10.1016/j.nicl.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deserno L, Sterzer P, Wustenberg T, Heinz A, Schlagenhauf F. Reduced prefrontal-parietal effective connectivity and working memory deficits in schizophrenia. J Neurosci. 2012;32(1):12–20. doi: 10.1523/JNEUROSCI.3405-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, LaConte S, James GA, Peltier S, Hu XP. Multivariate Granger Causality Analysis of fMRI Data. Human Brain Mapping. 2009;30(4):1361–1373. doi: 10.1002/hbm.20606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Sathian K, Hu X. Effect of hemodynamic variability on Granger causality analysis of fMRI. Neuroimage. 2010;52(3):884–896. doi: 10.1016/j.neuroimage.2009.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detre JA, Wang J. Technical aspects and utility of fMRI using BOLD and ASL. Clin Neurophysiol. 2002;113(5):621–634. doi: 10.1016/s1388-2457(02)00038-x. [DOI] [PubMed] [Google Scholar]
- Devlin JT, Poldrack RA. In praise of tedious anatomy. Neuroimage. 2007;37(4):1033–1041. doi: 10.1016/j.neuroimage.2006.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaconescu AO, Jensen J, Wang H, Willeit M, Menon M, Kapur S, McIntosh AR. Aberrant Effective Connectivity in Schizophrenia Patients during Appetitive Conditioning. Front Hum Neurosci. 2011;4:239. doi: 10.3389/fnhum.2010.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dima D, Roiser JP, Dietrich DE, Bonnemann C, Lanfermann H, Emrich HM, Dillo W. Understanding why patients with schizophrenia do not perceive the hollow-mask illusion using dynamic causal modelling. Neuroimage. 2009;46(4):1180–1186. doi: 10.1016/j.neuroimage.2009.03.033. [DOI] [PubMed] [Google Scholar]
- Edelstein WA, Glover GH, Hardy CJ, Redington RW. The intrinsic signal-to-noise ratio in NMR imaging. Magn Reson Med. 1986;3(4):604–618. doi: 10.1002/mrm.1910030413. [DOI] [PubMed] [Google Scholar]
- Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Glasser MF, Miller KL, Ugurbil K, Yacoub E. Multiplexed Echo Planar Imaging for Sub-Second Whole Brain FMRI and Fast Diffusion Imaging. Plos One. 2010;5(12) doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari M, Giannini I, Sideri G, Zanette E. Continuous Non Invasive Monitoring of Human-Brain by near-Infrared Spectroscopy. Advances in Experimental Medicine and Biology. 1985;191:873–882. doi: 10.1007/978-1-4684-3291-6_88. [DOI] [PubMed] [Google Scholar]
- Filippi M, van den Heuvel MP, Fornito A, He Y, Hulshoff Pol HE, Agosta F, Comi G, Rocca MA. Assessment of system dysfunction in the brain through MRI-based connectomics. Lancet Neurol. 2013;12(12):1189–1199. doi: 10.1016/S1474-4422(13)70144-3. [DOI] [PubMed] [Google Scholar]
- Forman C, Aksoy M, Hornegger J, Bammer R. Self-encoded marker for optical prospective head motion correction in MRI. Med Image Anal. 2011;15(5):708–719. doi: 10.1016/j.media.2011.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman SD, Cohen JD, Fitzgerald M, Eddy WF, Mintun MA, Noll DC. Improved assessment of significant activation in functional magnetic resonance imaging (fMRI): use of a cluster-size threshold. Magn Reson Med. 1995;33(5):636–647. doi: 10.1002/mrm.1910330508. [DOI] [PubMed] [Google Scholar]
- Fox MD, Greicius M. Clinical applications of resting state functional connectivity. Front Syst Neurosci. 2010;4:19. doi: 10.3389/fnsys.2010.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(27):9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Zhang DY, Snyder AZ, Raichle ME. The Global Signal and Observed Anticorrelated Resting State Brain Networks. Journal of Neurophysiology. 2009;101(6):3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PT, Raichle ME. Focal Physiological Uncoupling of Cerebral Blood-Flow and Oxidative-Metabolism during Somatosensory Stimulation in Human-Subjects. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(4):1140–1144. doi: 10.1073/pnas.83.4.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PT, Raichle ME, Mintun MA, Dence C. Nonoxidative glucose consumption during focal physiologic neural activity. Science. 1988;241(4864):462–464. doi: 10.1126/science.3260686. [DOI] [PubMed] [Google Scholar]
- Frahm J, Merboldt KD, Hanicke W, Kleinschmidt A, Boecker H. Brain or vein--oxygenation or flow? On signal physiology in functional MRI of human brain activation. NMR Biomed. 1994;7(1–2):45–53. doi: 10.1002/nbm.1940070108. [DOI] [PubMed] [Google Scholar]
- Friston KHAP, Worsley KJ, Poline JP, Frith CD, Frackowiak RSJ. Statistical parametric maps in functional imaging: a general linear approach. Human Brain Mapping. 1995;2:22. [Google Scholar]
- Friston KJ. Causal Modelling and Brain Connectivity in Functional Magnetic Resonance Imaging. Plos Biology. 2009;7(2):220–225. doi: 10.1371/journal.pbio.1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ. Dynamic causal modeling and Granger causality Comments on: The identification of interacting networks in the brain using fMRI: Model selection, causality and deconvolution. Neuroimage. 2011;58(2):303–305. doi: 10.1016/j.neuroimage.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ. Functional and effective connectivity: a review. Brain Connect. 2011;1(1):13–36. doi: 10.1089/brain.2011.0008. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Buechel C, Fink GR, Morris J, Rolls E, Dolan RJ. Psychophysiological and modulatory interactions in neuroimaging. Neuroimage. 1997;6(3):218–229. doi: 10.1006/nimg.1997.0291. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Frith CD, Liddle PF, Frackowiak RS. Functional connectivity: the principal-component analysis of large (PET) data sets. J Cereb Blood Flow Metab. 1993;13(1):5–14. doi: 10.1038/jcbfm.1993.4. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Harrison L, Penny W. Dynamic causal modelling. Neuroimage. 2003;19(4):1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Holmes AP, Poline JB, Grasby PJ, Williams SC, Frackowiak RS, Turner R. Analysis of fMRI time-series revisited. Neuroimage. 1995;2(1):45–53. doi: 10.1006/nimg.1995.1007. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Josephs O, Rees G, Turner R. Nonlinear event-related responses in fMRI. Magn Reson Med. 1998;39(1):41–52. doi: 10.1002/mrm.1910390109. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Josephs O, Rees G, Turner R. Nonlinear event-related responses in fMRI. Magnetic Resonance in Medicine. 1998;39(1):41–52. doi: 10.1002/mrm.1910390109. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Josephs O, Zarahn E, Holmes AP, Rouquette S, Poline J. To smooth or not to smooth? Bias and efficiency in fMRI time-series analysis. Neuroimage. 2000;12(2):196–208. doi: 10.1006/nimg.2000.0609. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Kahan J, Biswal B, Razi A. A DCM for resting state fMRI. Neuroimage. 2014;94:396–407. doi: 10.1016/j.neuroimage.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Li BJ, Daunizeau J, Stephan KE. Network discovery with DCM. Neuroimage. 2011;56(3):1202–1221. doi: 10.1016/j.neuroimage.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJP, Turner R. Analysis of Functional MRI Time-Series. Human Brain Mapping. 1994;1:19. [Google Scholar]
- Friston KJ, Williams S, Howard R, Frackowiak RS, Turner R. Movement-related effects in fMRI time-series. Magn Reson Med. 1996;35(3):346–355. doi: 10.1002/mrm.1910350312. [DOI] [PubMed] [Google Scholar]
- Geissler A, Fischmeister FPS, Grabner G, Wurnig M, Rath J, Foki T, Matt E, Trattnig S, Beisteiner R, Robinson SD. Comparing the microvascular specificity of the 3-and 7-T BOLD response using ICA and susceptibility-weighted imaging. Frontiers in Human Neuroscience. 2013;7 doi: 10.3389/fnhum.2013.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler A, Lanzenberger R, Barth M, Tahamtan AR, Milakara D, Gartus A, Beisteiner R. Influence of fMRI smoothing procedures on replicability of fine scale motor localization. Neuroimage. 2005;24(2):323–331. doi: 10.1016/j.neuroimage.2004.08.042. [DOI] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15(4):870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Gholipour A, Kehtarnavaz N, Briggs R, Devous M, Gopinath K. Brain functional localization: a survey of image registration techniques. IEEE Trans Med Imaging. 2007;26(4):427–451. doi: 10.1109/TMI.2007.892508. [DOI] [PubMed] [Google Scholar]
- Glover GH. Deconvolution of impulse response in event-related BOLD fMRI. Neuroimage. 1999;9(4):416–429. doi: 10.1006/nimg.1998.0419. [DOI] [PubMed] [Google Scholar]
- Glover GH, Law CS. Spiral-in/out BOLD fMRI for increased SNR and reduced susceptibility artifacts. Magnetic Resonance in Medicine. 2001;46(3):515–522. doi: 10.1002/mrm.1222. [DOI] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med. 2000;44(1):162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Goebel R, Roebroeck A, Kim DS, Formisano E. Investigating directed cortical interactions in time-resolved fMRI data using vector autoregressive modeling and Granger causality mapping. Magn Reson Imaging. 2003;21(10):1251–1261. doi: 10.1016/j.mri.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Gohel SR, Biswal BB. Functional integration between brain regions at rest occurs in multiple-frequency bands. Brain Connect. 2015;5(1):23–34. doi: 10.1089/brain.2013.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RI, Stern JM, Engel J, Jr, Cohen MS. Acquiring simultaneous EEG and functional MRI. Clin Neurophysiol. 2000;111(11):1974–1980. doi: 10.1016/s1388-2457(00)00456-9. [DOI] [PubMed] [Google Scholar]
- Gotts SJ, Saad ZS, Jo HJ, Wallace GL, Cox RW, Martin A. The perils of global signal regression for group comparisons: a case study of Autism Spectrum Disorders. Front Hum Neurosci. 2013;7:356. doi: 10.3389/fnhum.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulden N, McKie S, Suckling J, Williams SR, Anderson IM, Deakin JF, Elliott R. A comparison of permutation and parametric testing for between group effective connectivity differences using DCM. Neuroimage. 2010;50(2):509–515. doi: 10.1016/j.neuroimage.2009.11.059. [DOI] [PubMed] [Google Scholar]
- Granger C. Investigating causal relations by econometric models and cross-spectral methods. Econometrica. 1969;37:15. [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A. 2003;100(1):253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci U S A. 2004;101(13):4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang JM, Kiefer B, Haase A. Generalized Autocalibrating Partially Parallel Acquisitions (GRAPPA) Magnetic Resonance in Medicine. 2002;47(6):1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- Guimond A, Meunier J, Thirion JP. Average brain models: A convergence study. Computer Vision and Image Understanding. 2000;77(2):192–210. [Google Scholar]
- Guller Y, Tononi G, Postle BR. Conserved functional connectivity but impaired effective connectivity of thalamocortical circuitry in schizophrenia. Brain Connect. 2012;2(6):311–319. doi: 10.1089/brain.2012.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton JP, Chen G, Thomason ME, Schwartz ME, Gotlib IH. Investigating neural primacy in Major Depressive Disorder: multivariate Granger causality analysis of resting-state fMRI time-series data. Mol Psychiatry. 2011;16(7):763–772. doi: 10.1038/mp.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerker DA, Roopchansingh V, Gonzalez-Castillo J, Bandettini PA. Periodic changes in fMRI connectivity. Neuroimage. 2012;63(3):1712–1719. doi: 10.1016/j.neuroimage.2012.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison L, Penny WD, Friston KJ. Multivariate autoregressive modeling of fMRI time series. Neuroimage. 2003;19(4):1477–1491. doi: 10.1016/s1053-8119(03)00160-5. [DOI] [PubMed] [Google Scholar]
- Haxby JV. Multivariate pattern analysis of fMRI: the early beginnings. Neuroimage. 2012;62(2):852–855. doi: 10.1016/j.neuroimage.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxby JV, Gobbini MI, Furey ML, Ishai A, Schouten JL, Pietrini P. Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science. 2001;293(5539):2425–2430. doi: 10.1126/science.1063736. [DOI] [PubMed] [Google Scholar]
- Haxby JV, Guntupalli JS, Connolly AC, Halchenko YO, Conroy BR, Gobbini MI, Hanke M, Ramadge PJ. A common, high-dimensional model of the representational space in human ventral temporal cortex. Neuron. 2011;72(2):404–416. doi: 10.1016/j.neuron.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes JD, Rees G. Decoding mental states from brain activity in humans. Nat Rev Neurosci. 2006;7(7):523–534. doi: 10.1038/nrn1931. [DOI] [PubMed] [Google Scholar]
- Hoeft F, McCandliss BD, Black JM, Gantman A, Zakerani N, Hulme C, Lyytinen H, Whitfield-Gabrieli S, Glover GH, Reiss AL, Gabrieli JD. Neural systems predicting long-term outcome in dyslexia. Proc Natl Acad Sci U S A. 2011;108(1):361–366. doi: 10.1073/pnas.1008950108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AP, Blair RC, Watson JD, Ford I. Nonparametric analysis of statistic images from functional mapping experiments. J Cereb Blood Flow Metab. 1996;16(1):7–22. doi: 10.1097/00004647-199601000-00002. [DOI] [PubMed] [Google Scholar]
- Horwitz B, Tagamets MA, McIntosh AR. Neural modeling, functional brain imaging, and cognition. Trends in Cognitive Sciences. 1999;3(3):91–98. doi: 10.1016/s1364-6613(99)01282-6. [DOI] [PubMed] [Google Scholar]
- Hu X, Le TH, Parrish T, Erhard P. Retrospective estimation and correction of physiological fluctuation in functional MRI. Magn Reson Med. 1995;34(2):201–212. doi: 10.1002/mrm.1910340211. [DOI] [PubMed] [Google Scholar]
- Huettel SASAW, McCarthy G. Functional Magnetic Resonance Imaging, 2nd Edition. Sunderland: Sinauer Associates, Inc.; 2008. [Google Scholar]
- Hulshoff Pol H, Bullmore E. Neural networks in psychiatry. Eur Neuropsychopharmacol. 2013;23(1):1–6. doi: 10.1016/j.euroneuro.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Hutcheson NL, Sreenivasan KR, Deshpande G, Reid MA, Hadley J, White DM, Ver Hoef L, Lahti AC. Effective connectivity during episodic memory retrieval in schizophrenia participants before and after antipsychotic medication. Hum Brain Mapp. 2015;36(4):1442–1457. doi: 10.1002/hbm.22714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, Della Penna S, Duyn JH, Glover GH, Gonzalez-Castillo J, Handwerker DA, Keilholz S, Kiviniemi V, Leopold DA, de Pasquale F, Sporns O, Walter M, Chang C. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage. 2013;80:360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton C, Bork A, Josephs O, Deichmann R, Ashburner J, Turner R. Image distortion correction in fMRI: A quantitative evaluation. Neuroimage. 2002;16(1):217–240. doi: 10.1006/nimg.2001.1054. [DOI] [PubMed] [Google Scholar]
- Hyde JS, Jesmanowicz A. Cross-correlation: an fMRI signal-processing strategy. Neuroimage. 2012;62(2):848–851. doi: 10.1016/j.neuroimage.2011.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi SJ, Peng D, Fang Y, Jiang K, Liddle EB, Liddle PF, Palaniyappan L. Alterations in effective connectivity anchored on the insula in major depressive disorder. Eur Neuropsychopharmacol. 2014;24(11):1784–1792. doi: 10.1016/j.euroneuro.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Jezzard P, Balaban RS. Correction for geometric distortion in echo planar images from B0 field variations. Magn Reson Med. 1995;34(1):65–73. doi: 10.1002/mrm.1910340111. [DOI] [PubMed] [Google Scholar]
- Jo HJ, Saad ZS, Simmons WK, Milbury LA, Cox RW. Mapping sources of correlation in resting state FMRI, with artifact detection and removal. Neuroimage. 2010;52(2):571–582. doi: 10.1016/j.neuroimage.2010.04.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Vemuri P, Murphy MC, Gunter JL, Senjem ML, Machulda MM, Przybelski SA, Gregg BE, Kantarci K, Knopman DS, Boeve BF, Petersen RC, Jack CR., Jr Non-stationarity in the "resting brain's" modular architecture. PLoS One. 2012;7(6):e39731. doi: 10.1371/journal.pone.0039731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastrup A, Kruger G, Glover GH, Moseley ME. Assessment of cerebral oxidative metabolism with breath holding and fMRI. Magnetic Resonance in Medicine. 1999;42(3):608–611. doi: 10.1002/(sici)1522-2594(199909)42:3<608::aid-mrm26>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Kennedy DP, Courchesne E. The intrinsic functional organization of the brain is altered in autism. Neuroimage. 2008;39(4):1877–1885. doi: 10.1016/j.neuroimage.2007.10.052. [DOI] [PubMed] [Google Scholar]
- Kim D, Burge J, Lane T, Pearlson GD, Kiehl KA, Calhoun VD. Hybrid ICA-Bayesian network approach reveals distinct effective connectivity differences in schizophrenia. Neuroimage. 2008;42(4):1560–1568. doi: 10.1016/j.neuroimage.2008.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Rostrup E, Larsson HB, Ogawa S, Paulson OB. Determination of relative CMRO2 from CBF and BOLD changes: significant increase of oxygen consumption rate during visual stimulation. Magn Reson Med. 1999;41(6):1152–1161. doi: 10.1002/(sici)1522-2594(199906)41:6<1152::aid-mrm11>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Kiviniemi V, Vire T, Remes J, Elseoud AA, Starck T, Tervonen O, Nikkinen J. A sliding time-window ICA reveals spatial variability of the default mode network in time. Brain Connect. 2011;1(4):339–347. doi: 10.1089/brain.2011.0036. [DOI] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46(3):786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger G, Glover GH. Physiological noise in oxygenation-sensitive magnetic resonance imaging. Magn Reson Med. 2001;46(4):631–637. doi: 10.1002/mrm.1240. [DOI] [PubMed] [Google Scholar]
- Kruger G, Kastrup A, Glover GH. Neuroimaging at 1.5 T and 3.0 T: comparison of oxygenation-sensitive magnetic resonance imaging. Magn Reson Med. 2001;45(4):595–604. doi: 10.1002/mrm.1081. [DOI] [PubMed] [Google Scholar]
- Kundu P, Inati SJ, Evans JW, Luh WM, Bandettini PA. Differentiating BOLD and non-BOLD signals in fMRI time series using multi-echo EPI. Neuroimage. 2012;60(3):1759–1770. doi: 10.1016/j.neuroimage.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong KK, Belliveau JW, Chesler DA, Goldberg IE, Weisskoff RM, Poncelet BP, Kennedy DN, Hoppel BE, Cohen MS, Turner and R, et al. Dynamic magnetic resonance imaging of human brain activity during primary sensory stimulation. Proc Natl Acad Sci U S A. 1992;89(12):5675–5679. doi: 10.1073/pnas.89.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaConte S, Anderson J, Muley S, Ashe J, Frutiger S, Rehm K, Hansen LK, Yacoub E, Hu XP, Rottenberg D, Strother S. The evaluation of preprocessing choices in single-subject BOLD fMRI using NPAIRS performance metrics. Neuroimage. 2003;18(1):10–27. doi: 10.1006/nimg.2002.1300. [DOI] [PubMed] [Google Scholar]
- Lancaster JL, Tordesillas-Gutierrez D, Martinez M, Salinas F, Evans A, ZilleS K, Mazziotta JC, Fox PT. Bias between MNI and talairach coordinates analyzed using the ICBM-152 brain template. Human Brain Mapping. 2007;28(11):1194–1205. doi: 10.1002/hbm.20345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latora V, Marchiori M. Efficient behavior of small-world networks. Phys Rev Lett. 2001;87(19):198701. doi: 10.1103/PhysRevLett.87.198701. [DOI] [PubMed] [Google Scholar]
- Laufs H, Kleinschmidt A, Beyerle A, Eger E, Salek-Haddadi A, Preibisch C, Krakow K. EEG-correlated fMRI of human alpha activity. Neuroimage. 2003;19(4):1463–1476. doi: 10.1016/s1053-8119(03)00286-6. [DOI] [PubMed] [Google Scholar]
- Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, Laval-Jeantet M. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology. 1986;161(2):401–407. doi: 10.1148/radiology.161.2.3763909. [DOI] [PubMed] [Google Scholar]
- Le TH, Hu X. Retrospective estimation and correction of physiological artifacts in fMRI by direct extraction of physiological activity from MR data. Magn Reson Med. 1996;35(3):290–298. doi: 10.1002/mrm.1910350305. [DOI] [PubMed] [Google Scholar]
- Lee HL, Zahneisen B, Hugger T, Levan P, Hennig J. Tracking dynamic resting-state networks at higher frequencies using MR-encephalography. Neuroimage. 2013;65:216–222. doi: 10.1016/j.neuroimage.2012.10.015. [DOI] [PubMed] [Google Scholar]
- Lee L, Friston K, Horwitz B. Large-scale neural models and dynamic causal modelling. Neuroimage. 2006;30(4):1243–1254. doi: 10.1016/j.neuroimage.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Lee MH, Smyser CD, Shimony JS. Resting-state fMRI: a review of methods and clinical applications. AJNR Am J Neuroradiol. 2013;34(10):1866–1872. doi: 10.3174/ajnr.A3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux L, Salek-Haddadi A, Lund TE, Laufs H, Carmichael D. Modelling large motion events in fMRI studies of patients with epilepsy. Magn Reson Imaging. 2007;25(6):894–901. doi: 10.1016/j.mri.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Lewis EB, Fox NC. Correction of differential intensity inhomogeneity in longitudinal MR images. Neuroimage. 2004;23(1):75–83. doi: 10.1016/j.neuroimage.2004.04.030. [DOI] [PubMed] [Google Scholar]
- Li BJ, Daunizeau J, Stephan KE, Penny W, Hu DW, Friston KJ. Generalised filtering and stochastic DCM for fMRI. Neuroimage. 2011;58(2):442–457. doi: 10.1016/j.neuroimage.2011.01.085. [DOI] [PubMed] [Google Scholar]
- Liao R, Krolik JL, McKeown MJ. An information-theoretic criterion for intrasubject alignment of FMRI time series: motion corrected independent component analysis. IEEE Trans Med Imaging. 2005;24(1):29–44. doi: 10.1109/tmi.2004.837791. [DOI] [PubMed] [Google Scholar]
- Liu TT. The development of event-related fMRI designs. Neuroimage. 2012;62(2):1157–1162. doi: 10.1016/j.neuroimage.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TT, Behzadi Y, Restom K, Uludag K, Lu K, Buracas GT, Dubowitz DJ, Buxton RB. Caffeine alters the temporal dynamics of the visual BOLD response. Neuroimage. 2004;23(4):1402–1413. doi: 10.1016/j.neuroimage.2004.07.061. [DOI] [PubMed] [Google Scholar]
- Liu TT, Frank LR, Wong EC, Buxton RB. Detection power, estimation efficiency, and predictability in event-related fMRI. Neuroimage. 2001;13(4):759–773. doi: 10.1006/nimg.2000.0728. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wu X, Zhang J, Guo X, Long Z, Yao L. Altered effective connectivity model in the default mode network between bipolar and unipolar depression based on resting-state fMRI. J Affect Disord. 2015;182:8–17. doi: 10.1016/j.jad.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhang Y, Bai L, Yan H, Dai R, Zhong C, Wang H, Wei W, Xue T, Feng Y, You Y, Tian J. Investigation of the effective connectivity of resting state networks in Alzheimer's disease: a functional MRI study combining independent components analysis and multivariate Granger causality analysis. NMR Biomed. 2012;25(12):1311–1320. doi: 10.1002/nbm.2803. [DOI] [PubMed] [Google Scholar]
- Lohmann G, Erfurth K, Muller K, Turner R. Critical comments on dynamic causal modelling. Neuroimage. 2012;59(3):2322–2329. doi: 10.1016/j.neuroimage.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Lowe MJ, Sorenson JA. Spatially filtering functional magnetic resonance imaging data. Magnetic Resonance in Medicine. 1997;37(5):723–729. doi: 10.1002/mrm.1910370514. [DOI] [PubMed] [Google Scholar]
- Ma S, Calhoun VD, Phlypo R, Adali T. Dynamic changes of spatial functional network connectivity in healthy individuals and schizophrenia patients using independent vector analysis. Neuroimage. 2014;90:196–206. doi: 10.1016/j.neuroimage.2013.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclaren J, Herbst M, Speck O, Zaitsev M. Prospective motion correction in brain imaging: a review. Magn Reson Med. 2013;69(3):621–636. doi: 10.1002/mrm.24314. [DOI] [PubMed] [Google Scholar]
- Mahmoudi A, Takerkart S, Regragui F, Boussaoud D, Brovelli A. Multivoxel pattern analysis for FMRI data: a review. Comput Math Methods Med. 2012;2012:961257. doi: 10.1155/2012/961257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malonek D, Grinvald A. Interactions between electrical activity and cortical microcirculation revealed by imaging spectroscopy: Implications for functional brain mapping. Science. 1996;272(5261):551–554. doi: 10.1126/science.272.5261.551. [DOI] [PubMed] [Google Scholar]
- Mansfield P. Multi-Planar Image-Formation Using Nmr Spin Echoes. Journal of Physics C-Solid State Physics. 1977;10(3):L55–L58. [Google Scholar]
- Marreiros AC, Kiebel SJ, Friston KJ. Dynamic causal modelling for fMRI: A two-state model. Neuroimage. 2008;39(1):269–278. doi: 10.1016/j.neuroimage.2007.08.019. [DOI] [PubMed] [Google Scholar]
- McIntosh AR, F G-L. Structural equation modelling and its application to network analysis in functional brain imaging. Human Brain Mapping. 1994;2:21. [Google Scholar]
- McKeown MJ, Sejnowski TJ. Independent component analysis of fMRI data: examining the assumptions. Hum Brain Mapp. 1998;6(5–6):368–372. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<368::AID-HBM7>3.0.CO;2-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier D, Lambiotte R, Bullmore ET. Modular and hierarchically modular organization of brain networks. Front Neurosci. 2010;4:200. doi: 10.3389/fnins.2010.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miezin FM, Maccotta L, Ollinger JM, Petersen SE, Buckner RL. Characterizing the hemodynamic response: effects of presentation rate, sampling procedure, and the possibility of ordering brain activity based on relative timing. Neuroimage. 2000;11(6 Pt 1):735–759. doi: 10.1006/nimg.2000.0568. [DOI] [PubMed] [Google Scholar]
- Mikl M, Marecek R, Hlustik P, Pavlicova M, Drastich A, Chlebus P, Brazdil M, Krupa P. Effects of spatial smoothing on fMRI group inferences. Magn Reson Imaging. 2008;26(4):490–503. doi: 10.1016/j.mri.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Mitra PP, Pesaran B. Analysis of dynamic brain imaging data. Biophys J. 1999;76(2):691–708. doi: 10.1016/S0006-3495(99)77236-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarreszadeh M, Bruce EN. Ventilatory variability induced by spontaneous variations of PaCO2 in humans. J Appl Physiol 1985. 1994;76(6):2765–2775. doi: 10.1152/jappl.1994.76.6.2765. [DOI] [PubMed] [Google Scholar]
- Monti MM. Statistical Analysis of fMRI Time-Series: A Critical Review of the GLM Approach. Front Hum Neurosci. 2011;5:28. doi: 10.3389/fnhum.2011.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan VL, Abou-Khalil B, Rogers BP. Evolution of functional connectivity of brain networks and their dynamic interaction in temporal lobe epilepsy. Brain Connect. 2015;5(1):35–44. doi: 10.1089/brain.2014.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses-Kolko EL, Perlman SB, Wisner KL, James J, Saul AT, Phillips ML. Abnormally reduced dorsomedial prefrontal cortical activity and effective connectivity with amygdala in response to negative emotional faces in postpartum depression. Am J Psychiatry. 2010;167(11):1373–1380. doi: 10.1176/appi.ajp.2010.09081235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugler JP, 3rd, Brookeman JR. Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE) Magn Reson Med. 1990;15(1):152–157. doi: 10.1002/mrm.1910150117. [DOI] [PubMed] [Google Scholar]
- Mukherjee P, Whalley HC, McKirdy JW, McIntosh AM, Johnstone EC, Lawrie SM, Hall J. Lower effective connectivity between amygdala and parietal regions in response to fearful faces in schizophrenia. Schizophr Res. 2012;134(2–3):118–124. doi: 10.1016/j.schres.2011.09.033. [DOI] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage. 2009;44(3):893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Bodurka J, Bandettini PA. How long to scan? The relationship between fMRI temporal signal to noise ratio and necessary scan duration. Neuroimage. 2007;34(2):565–574. doi: 10.1016/j.neuroimage.2006.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P, Wiley K, Brown J, Shinaman R, Ludlow D, Sawyer AM, Glover G, Mackey S. Functional magnetic resonance imaging identifies somatotopic organization of nociception in the human spinal cord. Pain. 2013;154(6):776–781. doi: 10.1016/j.pain.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Neufang S, Akhrif A, Riedl V, Forstl H, Kurz A, Zimmer C, Sorg C, Wohlschlager AM. Predicting effective connectivity from resting-state networks in healthy elderly and patients with prodromal Alzheimer's disease. Hum Brain Mapp. 2014;35(3):954–963. doi: 10.1002/hbm.22226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols T, Hayasaka S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Stat Methods Med Res. 2003;12(5):419–446. doi: 10.1191/0962280203sm341ra. [DOI] [PubMed] [Google Scholar]
- Nichols TE. Multiple testing corrections, nonparametric methods, and random field theory. Neuroimage. 2012;62(2):811–815. doi: 10.1016/j.neuroimage.2012.04.014. [DOI] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15(1):1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman KA, Polyn SM, Detre GJ, Haxby JV. Beyond mind-reading: multi-voxel pattern analysis of fMRI data. Trends Cogn Sci. 2006;10(9):424–430. doi: 10.1016/j.tics.2006.07.005. [DOI] [PubMed] [Google Scholar]
- O'Toole AJ, Jiang F, Abdi H, Penard N, Dunlop JP, Parent MA. Theoretical, statistical, and practical perspectives on pattern-based classification approaches to the analysis of functional neuroimaging data. J Cogn Neurosci. 2007;19(11):1735–1752. doi: 10.1162/jocn.2007.19.11.1735. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Lee TM, Nayak AS, Glynn P. Oxygenation-sensitive contrast in magnetic resonance image of rodent brain at high magnetic fields. Magn Reson Med. 1990;14(1):68–78. doi: 10.1002/mrm.1910140108. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Tank DW, Menon R, Ellermann JM, Kim SG, Merkle H, Ugurbil K. Intrinsic signal changes accompanying sensory stimulation: functional brain mapping with magnetic resonance imaging. Proc Natl Acad Sci U S A. 1992;89(13):5951–5955. doi: 10.1073/pnas.89.13.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollinger JM, Corbetta M, Shulman GL. Separating processes within a trial in event-related functional MRI - II. Analysis. Neuroimage. 2001;13(1):218–229. doi: 10.1006/nimg.2000.0711. [DOI] [PubMed] [Google Scholar]
- Parrish TB, Gitelman DR, LaBar KS, Mesulam MM. Impact of signal-to-noise on functional MRI. Magnetic Resonance in Medicine. 2000;44(6):925–932. doi: 10.1002/1522-2594(200012)44:6<925::aid-mrm14>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Patel AX, Kundu P, Rubinov M, Jones PS, Vertes PE, Ersche KD, Suckling J, Bullmore ET. A wavelet method for modeling and despiking motion artifacts from resting-state fMRI time series. Neuroimage. 2014;95:287–304. doi: 10.1016/j.neuroimage.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriat R, Molloy EK, Meier TB, Kirk GR, Nair VA, Meyerand ME, Prabhakaran V, Birn RM. The effect of resting condition on resting-state fMRI reliability and consistency: A comparison between resting with eyes open, closed, and fixated. Neuroimage. 2013;78:463–473. doi: 10.1016/j.neuroimage.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl J. Causality: Models, Reasoning and Inference. Cambridge University Press; 2000. [Google Scholar]
- Penny WD, Stephan KE, Daunizeau J, Rosa MJ, Friston KJ, Schofield TM, Leff AP. Comparing Families of Dynamic Causal Models. Plos Computational Biology. 2010;6(3) doi: 10.1371/journal.pcbi.1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny WD, Stephan KE, Mechelli A, Friston KJ. Comparing dynamic causal models. Neuroimage. 2004;22(3):1157–1172. doi: 10.1016/j.neuroimage.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Penny WD, Trujillo-Barreto NJ, Friston KJ. Bayesian fMRI time series analysis with spatial priors. Neuroimage. 2005;24(2):350–362. doi: 10.1016/j.neuroimage.2004.08.034. [DOI] [PubMed] [Google Scholar]
- Pereira F, Mitchell T, Botvinick M. Machine learning classifiers and fMRI: a tutorial overview. Neuroimage. 2009;45(1 Suppl):S199–S209. doi: 10.1016/j.neuroimage.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlbarg V, Bellec P, Anton JL, Pelegrini-Issac M, Doyon J, Benali H. CORSICA: correction of structured noise in fMRI by automatic identification of ICA components. Magn Reson Imaging. 2007;25(1):35–46. doi: 10.1016/j.mri.2006.09.042. [DOI] [PubMed] [Google Scholar]
- Petersen SE, Dubis JW. The mixed block/event-related design. Neuroimage. 2012;62(2):1177–1184. doi: 10.1016/j.neuroimage.2011.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeuffer J, Van De Moortele PF, Ugurbil K, Hu X, Glover GH. Correction of physiologically induced global off-resonance effects in dynamic echo-planar and spiral functional imaging. Magn Reson Med. 2002;47(2):344–353. doi: 10.1002/mrm.10065. [DOI] [PubMed] [Google Scholar]
- Poldrack RAMJA, Nichols TE. Handbook of functional MRI data analysis. New York: Cambridge University Press; 2011. [Google Scholar]
- Poline JB, Brett M. The general linear model and fMRI: does love last forever? Neuroimage. 2012;62(2):871–880. doi: 10.1016/j.neuroimage.2012.01.133. [DOI] [PubMed] [Google Scholar]
- Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, Petersen SE. Recent progress and outstanding issues in motion correction in resting state fMRI. Neuroimage. 2015;105:536–551. doi: 10.1016/j.neuroimage.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price T, Wee CY, Gao W, Shen D. Multiple-network classification of childhood autism using functional connectivity dynamics. Med Image Comput Comput Assist Interv. 2014;17(Pt 3):177–184. doi: 10.1007/978-3-319-10443-0_23. [DOI] [PubMed] [Google Scholar]
- Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: Sensitivity encoding for fast MRI. Magnetic Resonance in Medicine. 1999;42(5):952–962. [PubMed] [Google Scholar]
- Raichle ME. Developing a functional anatomy of the human brain with positron emission tomography. Current Neurology. 1989;9:8. [Google Scholar]
- Raichle ME, Grubb RL, Gado MH, Eichling JO, Terpogossian MM. Correlation between Regional Cerebral Blood-Flow and Oxidative-Metabolism. Archives of Neurology. 1976;33(8):523–526. doi: 10.1001/archneur.1976.00500080001001. [DOI] [PubMed] [Google Scholar]
- Raj D, Anderson AW, Gore JC. Respiratory effects in human functional magnetic resonance imaging due to bulk susceptibility changes. Phys Med Biol. 2001;46(12):3331–3340. doi: 10.1088/0031-9155/46/12/318. [DOI] [PubMed] [Google Scholar]
- Rajapakse JC, Zhou J. Learning effective brain connectivity with dynamic Bayesian networks. Neuroimage. 2007;37(3):749–760. doi: 10.1016/j.neuroimage.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Ramsey JD, Hanson SJ, Hanson C, Halchenko YO, Poldrack RA, Glymour C. Six problems for causal inference from fMRI. Neuroimage. 2010;49(2):1545–1558. doi: 10.1016/j.neuroimage.2009.08.065. [DOI] [PubMed] [Google Scholar]
- Rashid B, Damaraju E, Pearlson GD, Calhoun VD. Dynamic connectivity states estimated from resting fMRI Identify differences among Schizophrenia, bipolar disorder, and healthy control subjects. Front Hum Neurosci. 2014;8:897. doi: 10.3389/fnhum.2014.00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson LF, Atlas LY, Wager TD. Dynamic functional connectivity using state-based dynamic community structure: method and application to opioid analgesia. Neuroimage. 2015;108:274–291. doi: 10.1016/j.neuroimage.2014.12.034. [DOI] [PubMed] [Google Scholar]
- Roebroeck A, Formisano E, Goebel R. Mapping directed influence over the brain using Granger causality and fMRI. Neuroimage. 2005;25(1):230–242. doi: 10.1016/j.neuroimage.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Roebroeck A, Formisano E, Goebel R. The identification of interacting networks in the brain using fMRI: Model selection, causality and deconvolution. Neuroimage. 2011;58(2):296–302. doi: 10.1016/j.neuroimage.2009.09.036. [DOI] [PubMed] [Google Scholar]
- Roland E, Larsen B. Focal increase of cerebral blood flow during stereognostic testing in man. Arch Neurol. 1976;33(8):551–558. doi: 10.1001/archneur.1976.00500080029005. [DOI] [PubMed] [Google Scholar]
- Rosazza C, Minati L. Resting-state brain networks: literature review and clinical applications. Neurol Sci. 2011;32(5):773–785. doi: 10.1007/s10072-011-0636-y. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52(3):1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Rytsar R, Fornari E, Frackowiak RS, Ghika JA, Knyazeva MG. Inhibition in early Alzheimer's disease: an fMRI-based study of effective connectivity. Neuroimage. 2011;57(3):1131–1139. doi: 10.1016/j.neuroimage.2011.05.029. [DOI] [PubMed] [Google Scholar]
- Saad ZS, Gotts SJ, Murphy K, Chen G, Jo HJ, Martin A, Cox RW. Trouble at rest: how correlation patterns and group differences become distorted after global signal regression. Brain Connect. 2012;2(1):25–32. doi: 10.1089/brain.2012.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchet MD, Knutson B. Spatial smoothing systematically biases the localization of reward-related brain activity. Neuroimage. 2013;66:270–277. doi: 10.1016/j.neuroimage.2012.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoglu U, Pearlson GD, Kiehl KA, Wang YM, Michael AM, Calhoun VD. A method for evaluating dynamic functional network connectivity and task-modulation: application to schizophrenia. MAGMA. 2010;23(5–6):351–366. doi: 10.1007/s10334-010-0197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Elliott MA, Gerraty RT, Ruparel K, Loughead J, Calkins ME, Eickhoff SB, Hakonarson H, Gur RC, Gur RE, Wolf DH. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage. 2013;64:240–256. doi: 10.1016/j.neuroimage.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser R, Gesierich T, Kaufmann B, Vucurevic G, Hunsche S, Gawehn J, Stoeter P. Altered effective connectivity during working memory performance in schizophrenia: a study with fMRI and structural equation modeling. Neuroimage. 2003;19(3):751–763. doi: 10.1016/s1053-8119(03)00106-x. [DOI] [PubMed] [Google Scholar]
- Schlosser RG, Wagner G, Koch K, Dahnke R, Reichenbach JR, Sauer H. Fronto-cingulate effective connectivity in major depression: a study with fMRI and dynamic causal modeling. Neuroimage. 2008;43(3):645–655. doi: 10.1016/j.neuroimage.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Scholvinck ML, Maier A, Ye FQ, Duyn JH, Leopold DA. Neural basis of global resting-state fMRI activity. Proc Natl Acad Sci U S A. 2010;107(22):10238–10243. doi: 10.1073/pnas.0913110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler B, Ollinger JM, Oakes TR, Johnstone T, Davidson RJ. Dynamic Causal Modeling applied to fMRI data shows high reliability. Neuroimage. 2010;49(1):603–611. doi: 10.1016/j.neuroimage.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scouten A, Papademetris X, Constable RT. Spatial resolution, signal-to-noise ratio, and smoothing in multi-subject functional MRI studies. Neuroimage. 2006;30(3):787–793. doi: 10.1016/j.neuroimage.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Seghier ML, Zeidman P, Neufeld NH, Leff AP, Price CJ. Identifying abnormal connectivity in patients using dynamic causal modeling of FMRI responses. Front Syst Neurosci. 2010;4 doi: 10.3389/fnsys.2010.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Tadanki S, Gore JC, Welch EB. Prospective Real-Time Head Motion Correction Using Inductively Coupled Wireless NMR Probes. Magnetic Resonance in Medicine. 2014;72(4):971–985. doi: 10.1002/mrm.25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magnetic Resonance in Medicine. 2012;67(5):1210–1224. doi: 10.1002/mrm.23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Li Z, Zeng LL, Yuan L, Chen F, Liu Z, Hu D. Internetwork dynamic connectivity effectively differentiates schizophrenic patients from healthy controls. Neuroreport. 2014;25(17):1344–1349. doi: 10.1097/WNR.0000000000000267. [DOI] [PubMed] [Google Scholar]
- Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD. Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cereb Cortex. 2012;22(1):158–165. doi: 10.1093/cercor/bhr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmueli K, van Gelderen P, de Zwart JA, Horovitz SG, Fukunaga M, Jansma JM, Duyn JH. Low-frequency fluctuations in the cardiac rate as a source of variance in the resting-state fMRI BOLD signal. Neuroimage. 2007;38(2):306–320. doi: 10.1016/j.neuroimage.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skudlarski P, Constable RT, Gore JC. ROC analysis of statistical methods used in functional MRI: Individual subjects. Neuroimage. 1999;9(3):311–329. doi: 10.1006/nimg.1999.0402. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Smith JF, Pillai A, Chen K, Horwitz B. Identification and validation of effective connectivity networks in functional magnetic resonance imaging using switching linear dynamic systems. Neuroimage. 2010;52(3):1027–1040. doi: 10.1016/j.neuroimage.2009.11.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM. The future of FMRI connectivity. Neuroimage. 2012;62(2):1257–1266. doi: 10.1016/j.neuroimage.2012.01.022. [DOI] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci U S A. 2009;106(31):13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Moeller S, Xu J, Auerbach EJ, Woolrich MW, Beckmann CF, Jenkinson M, Andersson J, Glasser MF, Van Essen DC, Feinberg DA, Yacoub ES, Ugurbil K. Temporally-independent functional modes of spontaneous brain activity. Proc Natl Acad Sci U S A. 2012;109(8):3131–3136. doi: 10.1073/pnas.1121329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Ramsey JD, Woolrich MW. Network modelling methods for FMRI. Neuroimage. 2011;54(2):875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- Sodickson DK, Manning WJ. Simultaneous acquisition of spatial harmonics (SMASH): Fast imaging with radiofrequency coil arrays. Magnetic Resonance in Medicine. 1997;38(4):591–603. doi: 10.1002/mrm.1910380414. [DOI] [PubMed] [Google Scholar]
- Sokoloff L, Reivich M, Kennedy C, Des Rosiers MH, Patlak CS, Pettigrew KD, Sakurada O, Shinohara M. The [14C]deoxyglucose method for the measurement of local cerebral glucose utilization: theory, procedure, and normal values in the conscious and anesthetized albino rat. J Neurochem. 1977;28(5):897–916. doi: 10.1111/j.1471-4159.1977.tb10649.x. [DOI] [PubMed] [Google Scholar]
- Spirtes PGC, Scheines R. Causation, Prediction and Search. 2nd Edition. Cambridge, MA: MIT Press; 2000. [Google Scholar]
- Sporns O. Structure and function of complex brain networks. Dialogues Clin Neurosci. 2013;15(3):247–262. doi: 10.31887/DCNS.2013.15.3/osporns. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam CJ. Modern network science of neurological disorders. Nat Rev Neurosci. 2014;15(10):683–695. doi: 10.1038/nrn3801. [DOI] [PubMed] [Google Scholar]
- Stam CJ, van Straaten EC. The organization of physiological brain networks. Clin Neurophysiol. 2012;123(6):1067–1087. doi: 10.1016/j.clinph.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Stephan KE, Kasper L, Harrison LM, Daunizeau J, den Ouden HEM, Breakspear M, Friston KJ. Nonlinear dynamic causal models for fMRI. Neuroimage. 2008;42(2):649–662. doi: 10.1016/j.neuroimage.2008.04.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Penny WD, Moran RJ, den Ouden HEM, Daunizeau J, Friston KJ. Ten simple rules for dynamic causal modeling. Neuroimage. 2010;49(4):3099–3109. doi: 10.1016/j.neuroimage.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Roebroeck A. A short history of causal modeling of fMRI data. Neuroimage. 2012;62(2):856–863. doi: 10.1016/j.neuroimage.2012.01.034. [DOI] [PubMed] [Google Scholar]
- Stephan KE, Weiskopf N, Drysdale PM, Robinson PA, Friston KJ. Comparing hemodynamic models with DCM. Neuroimage. 2007;38(3):387–401. doi: 10.1016/j.neuroimage.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strother SC. Evaluating fMRI preprocessing pipelines. IEEE Eng Med Biol Mag. 2006;25(2):27–41. doi: 10.1109/memb.2006.1607667. [DOI] [PubMed] [Google Scholar]
- Studholme C, Cardenas V, Song E, Ezekiel F, Maudsley A, Weiner M. Accurate template-based correction of brain MRI intensity distortion with application to dementia and aging. IEEE Trans Med Imaging. 2004;23(1):99–110. doi: 10.1109/TMI.2003.820029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton BP, Noll DC, Fessler JA. Fast, iterative image reconstruction for MRI in the presence of field inhomogeneities. IEEE Trans Med Imaging. 2003;22(2):178–188. doi: 10.1109/tmi.2002.808360. [DOI] [PubMed] [Google Scholar]
- Talairach JTP. Co-planar stereotaxic atlas of the human brain. Stuttgart, Germany, Georg: Theme Verlag; 1988. [Google Scholar]
- Tanabe J, Miller D, Tregellas J, Freedman R, Meyer FG. Comparison of detrending methods for optimal fMRI preprocessing. Neuroimage. 2002;15(4):902–907. doi: 10.1006/nimg.2002.1053. [DOI] [PubMed] [Google Scholar]
- Teplan Fundamentals of EEG measurement. Measurement science review. 2002;2:11. [Google Scholar]
- Thirion B, Dodel S, Poline JB. Detection of signal synchronizations in resting-state fMRI datasets. Neuroimage. 2006;29(1):321–327. doi: 10.1016/j.neuroimage.2005.06.054. [DOI] [PubMed] [Google Scholar]
- Thomason ME, Foland LC, Glover GH. Calibration of BOLD fMRI using breath holding reduces group variance during a cognitive task. Human Brain Mapping. 2007;28(1):59–68. doi: 10.1002/hbm.20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulborn KR, Waterton JC, Matthews PM, Radda GK. Oxygenation dependence of the transverse relaxation time of water protons in whole blood at high field. Biochim Biophys Acta. 1982;714(2):265–270. doi: 10.1016/0304-4165(82)90333-6. [DOI] [PubMed] [Google Scholar]
- Tong F, Pratte MS. Decoding patterns of human brain activity. Annu Rev Psychol. 2012;63:483–509. doi: 10.1146/annurev-psych-120710-100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes-Sosa PA, Roebroeck A, Daunizeau J, Friston K. Effective connectivity: influence, causality and biophysical modeling. Neuroimage. 2011;58(2):339–361. doi: 10.1016/j.neuroimage.2011.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Ven VG, Formisano E, Prvulovic D, Roeder CH, Linden DE. Functional connectivity as revealed by spatial independent component analysis of fMRI measurements during rest. Hum Brain Mapp. 2004;22(3):165–178. doi: 10.1002/hbm.20022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Aardweg JG, Karemaker JM. Influence of chemoreflexes on respiratory variability in healthy subjects. Am J Respir Crit Care Med. 2002;165(8):1041–1047. doi: 10.1164/ajrccm.165.8.2104100. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Mandl R, Hulshoff Pol H. Normalized cut group clustering of resting-state FMRI data. PLoS One. 2008;3(4):e2001. doi: 10.1371/journal.pone.0002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Hulshoff Pol HE. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol. 2010;20(8):519–534. doi: 10.1016/j.euroneuro.2010.03.008. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. Rich-club organization of the human connectome. J Neurosci. 2011;31(44):15775–15786. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KR, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59(1):431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstynen TD, Deshpande V. Using pulse oximetry to account for high and low frequency physiological artifacts in the BOLD signal. Neuroimage. 2011;55(4):1633–1644. doi: 10.1016/j.neuroimage.2010.11.090. [DOI] [PubMed] [Google Scholar]
- Visscher KM, Miezin FM, Kelly JE, Buckner RL, Donaldson DI, McAvoy MP, Bhalodia VM, Petersen SE. Mixed blocked/event-related designs separate transient and sustained activity in fMRI. Neuroimage. 2003;19(4):1694–1708. doi: 10.1016/s1053-8119(03)00178-2. [DOI] [PubMed] [Google Scholar]
- Vovk U, Pernus F, Likar B. MRI intensity inhomogeneity correction by combining intensity and spatial information. Phys Med Biol. 2004;49(17):4119–4133. doi: 10.1088/0031-9155/49/17/020. [DOI] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of 'small-world' networks. Nature. 1998;393(6684):440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Webb JT, Ferguson MA, Nielsen JA, Anderson JS. BOLD Granger Causality Reflects Vascular Anatomy. Plos One. 2013;8(12) doi: 10.1371/journal.pone.0084279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibull A, Gustavsson H, Mattsson S, Svensson J. Investigation of spatial resolution, partial volume effects and smoothing in functional MRI using artificial 3D time series. Neuroimage. 2008;41(2):346–353. doi: 10.1016/j.neuroimage.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Weissenbacher A, Kasess C, Gerstl F, Lanzenberger R, Moser E, Windischberger C. Correlations and anticorrelations in resting-state functional connectivity MRI: a quantitative comparison of preprocessing strategies. Neuroimage. 2009;47(4):1408–1416. doi: 10.1016/j.neuroimage.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Weisskoff RM, Zuo CS, Boxerman JL, Rosen BR. Microscopic susceptibility variation and transverse relaxation: theory and experiment. Magn Reson Med. 1994;31(6):601–610. doi: 10.1002/mrm.1910310605. [DOI] [PubMed] [Google Scholar]
- Werring DJ, Clark CA, Barker GJ, Miller DH, Parker GJ, Brammer MJ, Bullmore ET, Giampietro VP, Thompson AJ. The structural and functional mechanisms of motor recovery: complementary use of diffusion tensor and functional magnetic resonance imaging in a traumatic injury of the internal capsule. J Neurol Neurosurg Psychiatry. 1998;65(6):863–869. doi: 10.1136/jnnp.65.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, O'Leary D, Magnotta V, Arndt S, Flaum M, Andreasen NC. Anatomic and functional variability: the effects of filter size in group fMRI data analysis. Neuroimage. 2001;13(4):577–588. doi: 10.1006/nimg.2000.0716. [DOI] [PubMed] [Google Scholar]
- Williams DS, Detre JA, Leigh JS, Koretsky AP. Magnetic-Resonance-Imaging of Perfusion Using Spin Inversion of Arterial Water. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(1):212–216. doi: 10.1073/pnas.89.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RG, Ide K, Poulin MJ, Tracey I. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage. 2004;21(4):1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Wong CW, Olafsson V, Tal O, Liu TT. The amplitude of the resting-state fMRI global signal is related to EEG vigilance measures. Neuroimage. 2013;83:983–990. doi: 10.1016/j.neuroimage.2013.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CW, Krishnan A, Wager TD. Cluster-extent based thresholding in fMRI analyses: pitfalls and recommendations. Neuroimage. 2014;91:412–419. doi: 10.1016/j.neuroimage.2013.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsley KJ. Spatial smoothing of autocorrelations to control the degrees of freedom in fMRI analysis. Neuroimage. 2005;26(2):635–641. doi: 10.1016/j.neuroimage.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Cao J, Paus T, Petrides M, Evans AC. Applications of random field theory to functional connectivity. Hum Brain Mapp. 1998;6(5–6):364–367. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<364::AID-HBM6>3.0.CO;2-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsley KJ, Evans AC, Marrett S, Neelin P. A three-dimensional statistical analysis for CBF activation studies in human brain. J Cereb Blood Flow Metab. 1992;12(6):900–918. doi: 10.1038/jcbfm.1992.127. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Friston KJ. Analysis of fMRI time-series revisited--again. Neuroimage. 1995;2(3):173–181. doi: 10.1006/nimg.1995.1023. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Taylor JE, Tomaiuolo F, Lerch J. Unified univariate and multivariate random field theory. Neuroimage. 2004;23(Suppl 1):S189–S195. doi: 10.1016/j.neuroimage.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Wu CWW, Gu H, Lu HB, Stein EA, Chen JH, Yang YH. Frequency specificity of functional connectivity in brain networks. Neuroimage. 2008;42(3):1047–1055. doi: 10.1016/j.neuroimage.2008.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoub E, Shmuel A, Pfeuffer J, Van de Moortele PF, Adriany G, Andersen P, Vaughan JT, Merkle H, Ugurbil K, Hu XP. Imaging brain function in humans at 7 Tesla. Magnetic Resonance in Medicine. 2001;45(4):588–594. doi: 10.1002/mrm.1080. [DOI] [PubMed] [Google Scholar]
- Yan CG, Cheung B, Kelly C, Colcombe S, Craddock RC, Di Martino A, Li Q, Zuo XN, Castellanos FX, Milham MP. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. Neuroimage. 2013;76:183–201. doi: 10.1016/j.neuroimage.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Erhardt EB, Sui J, Du Y, He H, Hjelm D, Cetin MS, Rachakonda S, Miller RL, Pearlson G, Calhoun VD. Assessing dynamic brain graphs of time-varying connectivity in fMRI data: application to healthy controls and patients with schizophrenia. Neuroimage. 2015;107:345–355. doi: 10.1016/j.neuroimage.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Loh JM, Lindquist MA. Adaptive spatial smoothing of fMRI images. Statistics and Its Interface. 2010;3(1):3–13. [Google Scholar]
- Zhang H, Wei X, Tao H, Mwansisya TE, Pu W, He Z, Hu A, Xu L, Liu Z, Shan B, Xue Z. Opposite effective connectivity in the posterior cingulate and medial prefrontal cortex between first-episode schizophrenic patients with suicide risk and healthy controls. PLoS One. 2013;8(5):e63477. doi: 10.1371/journal.pone.0063477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Huang L, Cai S, Zhang Y, von Deneen KM, Ren A, Ren J I. Alzheimer's Disease Neuroimaging. Altered effective connectivity patterns of the default mode network in Alzheimer's disease: an fMRI study. Neurosci Lett. 2014;578:171–175. doi: 10.1016/j.neulet.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Zhang T, Jiang X, Hu X, Chen H, Yang N, Lv J, Han J, Guo L, Liu T. Fusing DTI and fMRI data: a survey of methods and applications. Neuroimage. 2014;102(Pt 1):184–191. doi: 10.1016/j.neuroimage.2013.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]