Abstract
The role of Notch signaling in osteoclast differentiation is controversial with conflicting experimental evidence indicating both stimulatory and inhibitory roles. Differences in experimental protocols and in vivo versus in vitro models may explain the discrepancies between studies. In this study, we investigated cell autonomous roles of Notch signaling in osteoclast differentiation and function by altering Notch signaling during osteoclast differentiation using stimulation with immobilized ligands Jagged1 or Delta-like1 or by suppression with γ-secretase inhibitor DAPT or transcriptional inhibitor SAHM1. Stimulation of Notch signaling in committed osteoclast precursors resulted in larger osteoclasts with a greater number of nuclei and resorptive activity whereas suppression resulted in smaller osteoclasts with fewer nuclei and suppressed resorptive activity. Conversely, stimulation of Notch signaling in osteoclast precursors prior to induction of osteoclastogenesis resulted in fewer osteoclasts. Our data support a mechanism of context-specific Notch signaling effects wherein Notch stimulation inhibits commitment to osteoclast differentiation, but enhances the maturation and function of committed precursors.
Keywords: Osteoclasts, Notch signaling, Bone resorption, Osteoclast fusion, Osteoclast commitment
Molecular signaling for osteoclast differentiation is initiated by monocyte/macrophage colony-stimulating factor (MCSF) and Receptor Activator of NF-κB Ligand (RANKL)[Feng, 2005; Teitelbaum, 2007]. While indispensable for proper osteoclast formation, these pathways are subject to modulation[Hong et al., 2013; Izawa et al., 2012; Jules et al., 2012; McCoy et al., 2013]. Signaling pathways regulating osteoclast precursor proliferation, osteoclast fusion, function, and survival potentially impact the number and activity of osteoclasts differentiated under MCSF/RANKL stimulation[Feng, 2005; Li et al., 2011; Maeda et al., 2012; Park et al., 2013]. Notch signaling is an important, yet incompletely understood, pathway that has been suggested to have multiple roles in the differentiation of osteoclasts[Duan et al., 2008]. Understanding the seemingly variegated roles of Notch signaling in osteoclasts is critical to the rational design of bone-preserving therapies targeting this pathway.
Signaling from the mammalian homologues of the Drosophila Notch receptor is an essential component of embryonic development, tissue patterning, and differentiation of stem cell populations[Artavanis-Tsakonas et al., 1999]. In mammals, there are four Notch receptors (Notch1, 2, 3, and 4), and multiple ligands of the Delta-like (Delta-like1, Delta-like3, and Delta-like4) and Jagged (Jagged1 and Jagged2) families[Chen et al., 2014]. Notch signaling has two distinguishing characteristics. First, Notch signaling can only be properly initiated in a target cell via receptor binding by a ligand on the plasma membrane of another cell (trans-activation); interactions between ligand and receptor on the same cell suppress Notch signaling (cis-inhibition) [Cordle et al., 2008; Wang et al., 2011]. In addition, activation of Notch signaling requires tensile force between receptor and ligand, and, because of this, Notch signaling can only be efficiently activated in culture through the use of immobilized ligands [Kopan and Ilagan, 2009; Wang and Ha, 2013]. Second, upon ligand stimulation, proteolytic cleavage events catalyzed by Tumor Necrosis Factor α-converting enzyme (TACE) and γ-secretase release the intracellular domain of Notch (NICD) which translocates to the nucleus where it forms a complex with mastermind-like (MAML), CSL, and general transcription factors to activate expression of target genes[Groot et al., 2014]. Notch signaling can be inhibited with γ-secretase inhibitors, such as N-[N-(3,5-difluorophenacetyl-L-alanyl)]-(S)-phenylglycine t-butyl ester (DAPT). SAHM1, a peptide mimetic of a dominant negative form of MAML, inhibits canonical Notch transcription complex formation [Jones, 2009; Kornilova et al., 2003; Moellering et al., 2009; Morohashi et al., 2006].
The controversial findings of previous studies regarding Notch signaling in osteoclasts may be summarized as follows: (1) Notch signaling suppresses osteoclast differentiation, (2) Notch signaling enhances osteoclast differentiation, and (3) Notch1 signaling stimulated by Jagged1 (JAG1) suppresses osteoclast differentiation and Notch2 signaling stimulated by Delta-like1 (DLL1) enhances osteoclast differentiation [Bai et al., 2008; Fukushima et al., 2008; Schwarzer et al., 2008; Sekine et al., 2012; Sethi et al., 2011; Yamada et al., 2003; Zhou et al., 2014; Zhou et al., 2015]. The disparate findings of these studies likely stem from differences in experimental approach and may be evidence for context-dependent roles for Notch signaling in osteoclasts. Resolution of these conflicting findings is critical for informed design of mechanistic studies of Notch signaling in osteoclast function.
This study investigates osteoclast differentiation and function in the context of Notch stimulation and suppression in a cell-autonomous fashion using cultured osteoclast precursors, Notch ligands, and Notch signaling inhibitors. This study utilizes multiple specific methodologies in order to isolate Notch signaling effects at distinct points during differentiation and clarifies the context-dependent roles of Notch signaling in osteoclastogenesis.
Materials and Methods
Animal care and use
Mice were maintained according to Institutional Animal Care and Use Committee guidelines of the University of Pennsylvania.
Cell culture reagents
Osteoclast/macrophage medium (α-10) was prepared by dissolving one vial of Eagle’s minimum essential medium powder (Sigma-Aldrich, M0894-10X1L) and 2g sodium bicarbonate (Fisher Scientific, BP328-500) in 890mL deionized water and adding 100mL heat-inactivated fetal bovine serum, 10mL 10000u/mL penicillin/10000μg/mL streptomycin (Life Technologies, 15140122), and 10mL 200mM L-glutamine (Life Technologies, 25030-081). Medium was sterile filtered using two 500mL 0.22μm filter bottles (Corning, 431097) and stored at 4°C. 10% L929 cell-conditioned medium was added to α-10 as a source of monocyte/macrophage colony-stimulating factor (MCSF)
Recombinant mouse Receptor Activator of NF-κB Ligand (RANKL) (Shenandoah Biotechnology, 200-04) was reconstituted in sterile phosphate buffered saline (PBS) (CellGro, 21-031-CV) at 100μg/mL and stored at −80°C. Recombinant human Jagged1-Fc (R&D Systems, 1277-JG-050) and Delta-like1-Fc (Adipogen, AG-40A-0116Y-C050) were reconstituted at 200μg/mL in sterile PBS and stored at −80°C. Fc fragment-specific goat anti-human IgG (Jackson ImmunoResearch, 109-001-008) was reconstituted at 200μg/mL in sterile water and stored at −80°C. DAPT (Sigma Aldrich, D5942-5MG) and SAHM1 (EMD Millipore, 491002-1MG) were dissolved in DMSO (Fisher Scientific, BP231-100) at 10mM and stored at −80°C.
Modulation of Notch signaling with immobilized ligands and soluble inhibitors
Notch signaling was induced by seeding cells onto IgG- (control), JAG1-, or DLL1-coated surfaces. Notch ligand-coated plates were prepared as previously described[Zhu et al., 2013]. 250μL (for 24-well plates) of 10μg/mL goat anti-human IgG in PBS were added to each well to be coated and incubated for 1 hour at room temperature. Wells were washed 3x with PBS, and 250μL of PBS (IgG wells), 10μg/mL Jagged1-Fc, or 10μg/mL Delta-like1-Fc were added and incubated for 2 hours at room temperature. Wells were washed 3x with PBS. Wells were kept filled with PBS until cell seeding to prevent drying. Notch signaling was inhibited by culturing cells in medium containing either 10μM DAPT, which inhibits γ-secretase, or 10μM SAHM1, which inhibits NICD-MAML-CSL transcription complex formation. Figure 1 summarizes the general mechanism of Notch signaling and points of experimental manipulation.
Figure 1. Summary of experimental manipulation of Notch signaling.
Notch signaling was stimulated by plating cells on antibody-immobilized Fc-fusion proteins of either Jagged1 or Delta-like1; antibody only was used the control for ligand-stimulated cells. Notch signaling was inhibited with either the γ-secretase inhibitor, DAPT, which inhibits Notch signaling at the point of NICD release, or the dominant-negative MAML peptide mimetic, SAHM1, which inhibits Notch signaling at the transcriptional level; DMSO, the vehicle for both inhibitors, was used as the control for Notch inhibition.
Osteoclast precursor culture and differentiation
Osteoclast precursors were obtained and differentiated according to an established protocol[Ashley et al., 2011]. Mice were sacrificed by CO2 inhalation followed by cervical dislocation. Tibiae and femura from each mouse were dissected and each was flushed with 2.5mL α-10 via a 10mL syringe (BD 14-823-2A) with a 25-gauge needle (BD, 305125) into a single 15mL conical tube which was mixed by inversion. Tissue debris were allowed to settle and the cell suspension was transferred to a new 15mL tube and pelleted at 300×g for 5 min. Pellets were resuspended in 1mL ACK lysing buffer (Life Technologies, A10492-01) and incubated at 37°C for 2 min to lyse red blood cells. 8mL PBS was then added to the tube and mixed by inversion. Cells were pelleted at 300×g for 5 min and resuspended in α-10 without MCSF, plated in a 100mm tissue culture treated plate (Corning, 430167), and incubated in a humidified 37°C, 5% CO2 incubator overnight. The next morning, non-adherent cells were collected for differentiation into osteoclast using one of thee protocols.
RANK signaling stimulation prior to Notch signaling stimulation [Ashley et al., 2011]. Non-adherent bone marrow cells were evenly distributed between 5 60mm non tissue culture-treated suspension plates (Corning, 08-772-31) in α-10 with MCSF. Three days after seeding, adherent macrophages were washed once with PBS and incubated with 0.5mL 5mM EDTA (Fisher Scientific, S312-500) in PBS per plate for 3–5 min at 37°C. 1.5mL α-10 was added to each plate and lifted cells were pooled in a single 50mL conical tube and counted using a hemocytometer. Cells were diluted to 2 × 105 cells/mL in α-10 with MCSF, and 250μL of the cell suspension was added to each well of a 24-well plate to achieve 5 × 104 cells/well (2.6 × 104 cells/cm2). To each well, 250μL α-10 with MCSF and double concentrations of each well’s particular treatment (e.g. 200ng/mL RANKL, 20μM DAPT) was added to achieve correct final concentrations. 48 hours after seeding, media and treatments in wells were refreshed. For wells treated with SAHM1, additional SAHM1 to a final concentration of 10μM was added to the wells daily. 72 hours after seeding, cells were washed with PBS and either fixed and stained for TRAP expression using a leukocyte acid phosphatase staining kit (Sigma-Aldrich, 387A) or lysed in 1mL TRIzol (Life Technologies, 15596-026) for RNA extraction.
Near-simultaneous stimulation of Notch and RANK signaling [Bradley and Oursler, 2008]. Non-adherent bone marrow cells were transferred to a conical tube and counted. Variable numbers of non-adherent cells (2 – 10 × 105) were seeded into a 24-well plate with MCSF and 100ng/mL RANKL. Differentiation medium was refreshed on day 3 and day 4 of the differentiation. Cells were TRAP stained after 5 days of differentiation.
RANK signaling stimulation after Notch signaling stimulation [Tevlin et al., 2014]. Non-adherent bone marrow cells were transferred to a conical tube and counted. Variable numbers of non-adherent cells (1 – 8 × 105) were seeded into a 24-well plate with MCSF and cultured for 3 days. On the third day of culture medium was replaced with α-10 containing MCSF and 100ng/mL RANKL. Medium was refreshed daily, and differentiated cells were TRAP stained 3 days later.
Quantification of stained area, osteoclast number, osteoclast size, and nuclear number
Following TRAP staining, wells were dried and plates were scanned at 4800DPI using a flatbed scanner. Using ImageJ (http://rsb.info.nih.gov/ij/), well images were converted to grayscale and black/white thresholded at brightness 0–110 of a 0–255 histogram. Using equal sized ellipse selections, the stained area fraction of each well was measured to generate stained area values.
Two fields (at 20X magnification) per well showing the highest density of osteoclasts were imaged in color. Prior to analysis, images were automatically white-balanced using the adjustment layer>levels function of Adobe Photoshop. Osteoclasts with more than 3 nuclei were counted manually. Using ImageJ, images were converted to grayscale, automatically contrast enhanced, and black/white thresholded at brightness 0–190 of a 0–255 histogram. Using the Analyze Particles function, the median size of osteoclasts were determined in pixels and converted to μm2 using a microscope calibration standard. After all measurements were complete median osteoclast sizes for each group were averaged.
To quantify nuclear number, TRAP-stained wells were observed at 40X magnification. Nuclei within the 20 largest osteoclasts per well were manually counted.
RNA isolation and quantitative RT-PCR
To isolate RNA from TRIzol lysates, 200μL chloroform (Fisher Scientific, BP1145-1) was added to each sample and shaken for 15 seconds. Samples were incubated at room temperature for 5 minutes and centrifuged at 12000g at 4°C for 15 min. 500μL of the top, aqueous layer was transferred to a new microcentrifuge tube and mixed with 500μL 100% ethanol. This mixture was applied to the columns of an RNeasy mini kit (Qiagen, 74106) and the remainder of the isolation was carried out according to the manufacturer’s protocol. RNA concentration was determined using a NanoDrop spectrophotometer (Thermo Scientific).
To generate cDNA, 2μg or as much as possible RNA was used as template in a reverse transcriptase reaction using the SuperScript VILO cDNA synthesis kit (Life Technologies, 11754-050). The resulting cDNA solutions were diluted to an effective starting RNA mass of 100ng (20-fold dilution when starting with 2μg).
Primers for real time PCR listed in Table 1 were designed using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) and synthesized by Integrated DNA Technologies (Table 1). For each reaction primer mix tubes and cDNA mix tubes were prepared. Primer tubes contained 5μL 2X SYBR Select master mix (Life Technologies, 4472908), 2μL 2μM primer mix, and 3μL deionized water per reaction. Complementary DNA mixtures contained 5μL 2X SYBR Select master mix, 2μL cDNA, and 3μL deionized water per reaction. 10μL each of primer mix and cDNA mix were added to the wells of a MicroAmp Fast Optical 96-well reaction plate (Life Technologies, 4346907). Plates were sealed with optical adhesive film (Life Technologies, 4311971) and run in an ABI 7500 real time PCR system using the following program: (1) 50°C for 2 min, (2) 95°C for 2 min, (3) 95°C for 15 sec, (4) 60°C for 1 min, (5) repeat 3–4 40X. Single products were confirmed by determining melting curves at the conclusion of the reaction. Relative expression was calculated using the 2−ΔΔCt method normalized to 18S (endogenous loading control).
Table 1.
| Primer Name | Sequence (5′ to 3′) | Measured Transcript |
|---|---|---|
| 18S_RTQ_F | GGTAACCCGTTGAACCCCAT | 18S - Loading Control |
| 18S_RTQ_R | CAACGCAAGCTTATGACCCG | |
| Notch1_RTQ_F | GCTCCGAGGAGATCAACGAG | Notch1 |
| Notch1_RTQ_R | TTGACATCACCCTCACACCG | |
| Notch2_RTQ_F | ATATCGACGACTGCCCCAAC | Notch2 |
| Notch2_RTQ_R | CCATAGCCTCCGTTTCGGTT | |
| Notch3_RTQ_F | ATGTGGCCGTGGCTATACTG | Notch3 |
| Notch3_RTQ_R | AGTGCTTGCACACTCATCCA | |
| Notch4_RTQ_F | GTCCTGAGGGCTATTCCTGC | Notch4 |
| Notch4_RTQ_R | CTTGGCAGGTTGCCTTGTTC | |
| DLL1_RTQ_F | CAATCTGTCTGCCAGGGTGT | Delta-like1 |
| DLL1_RTQ_R | CGGATGCACTCATCGCAGTA | |
| DLL4_RTQ_F | GATGAGGGATGGGGAGGTCT | Delta-like4 |
| DLL4_RTQ_R | CGCGCAGGTCAAGGTACTAT | |
| JAG1_RTQ_F | GCAACGACCGTAATCGCATC | Jagged1 |
| JAG1_RTQ_R | CCATTGCCGGCTAGGGTTTA | |
| JAG2_RTQ_F | GGGTGGCAACTCCTTCTACC | Jagged2 |
| JAG2_RTQ_R | GTCATTGTCCCAGTCCCAGG | |
| HES1_RTQ_F | GAGGCTGCCAAGGTTTTTGG | HES1 |
| HES1_RTQ_R | ACTTTACGGGTAGCAGTGGC | |
| HEY1_RTQ_F | GCTCACCCAGACTACAGCTC | HEY1 |
| HEY1_RTQ_R | CGCTTCTCGATGATGCCTCT | |
| ACP5_RTQ_F | CAGCTCAGTTGGGTAGCACA | TRAP |
| ACP5_RTQ_R | AGCCACAAATCTCAGGGTGG | |
| MMP9_RTQ_F | CGCTCATGTACCCGCTGTAT | MMP9 |
| MMP9_RTQ_R | CCGTGGGAGGTATAGTGGGA | |
| CTSK_RTQ_F | CAGTGTTGGTGGTGGGCTAT | Cathepsin K |
| CTSK_RTQ_R | CATGTTGGTAATGCCGCAGG | |
| CTR_RTQ_F | GGTGCGGCGGGATCCTATAA | Calcitonin Receptor |
| CTR_RTQ_R | CACGAGTGATGGCGTGGATA | |
| CD200_RTQ_F | GGGGTGAATCATCACAGGGG | CD200 |
| CD200_RTQ_R | CAAATCCCTCACAGGCTCGT | |
| DCSTAMP_RTQ_F | CATGTGGGTGCTGTTTGCC | DC-STAMP |
| DCSTAMP_RTQ_R | GACTCCTTGGGTTCCTTGCTT | |
| CD9_RTQ_F | TTCTGTCCCAGTCGTTCGTG | CD9 |
| CD9_RTQ_R | CTGAGAGTCGAATCGGAGCC | |
| CD81_RTQ_F | GAACTGGGAAACAAACCGGC | CD81 |
| CD81_RTQ_R | ATAGCACCCCAGGAAGCCTA | |
| SIRPA_RTQ_F | GAAACCATACCGTGCTGGGA | Signal-regulatory protein alpha |
| SIRPA_RTQ_R | CTGGGTTATTTCCCTGGCGT | |
| ATP6V0D2_RTQ_F | ATGCAAAGCCAGCCTCCTAA | v-ATPase V0 subunit D2 |
| ATP6V0D2_RTQ_R | TTGCCATAGTCCGTGGTCTG |
In vitro resorption
Osteoclasts were differentiated from adherent bone marrow macrophages according to protocol (1) for 3 days in 24-well Osteo Assay plates (Corning, 3987) which contain a layer of mineral hydroxyapatite. Medium was refreshed every other day and osteoclasts were cultured for an additional 1 or 3 days to allow resorption to occur. Osteoclasts were removed from the plate at the conclusion of the resorption period by washing wells once with PBS and incubating for 5 min with 10% bleach followed by 2 washes with deionized water. To enhance contrast between non-resorbed and resorbed areas, wells were stained using a modified von Kossa protocol. 300μL 5% silver nitrate (Midland Scientific Inc., 6828-16) was added to each well an incubated for 30 min at room temperature in the dark. Plates were then soaked in deionized water for 5 min. Stain was developed by adding 300μL 5% sodium carbonate (Fisher Scientific, S263-500) in formalin and incubating at room temperature for 4 min. Sodium carbonate solution was discarded and plates were dried at 50°C for 1 hour.
Plates were scanned at 600DPI using a flatbed scanner. Using ImageJ, well images were converted to grayscale and black/white thresholded at brightness 0–110 of a 0–255 histogram. Using equal sized ellipse selections, the stained area fraction of each well was measured to generate stained area values. Resorbed area was calculated as 100% - % stained area.
Statistical analysis
Statistical significance was evaluated using 1-tailed student’s T tests for real time RT-PCR data and 2-tailed student’s T tests for all other data. P-values less than 0.05 were considered statistically significant.
Results
Notch receptors and ligands are expressed by osteoclast precursors and osteoclasts
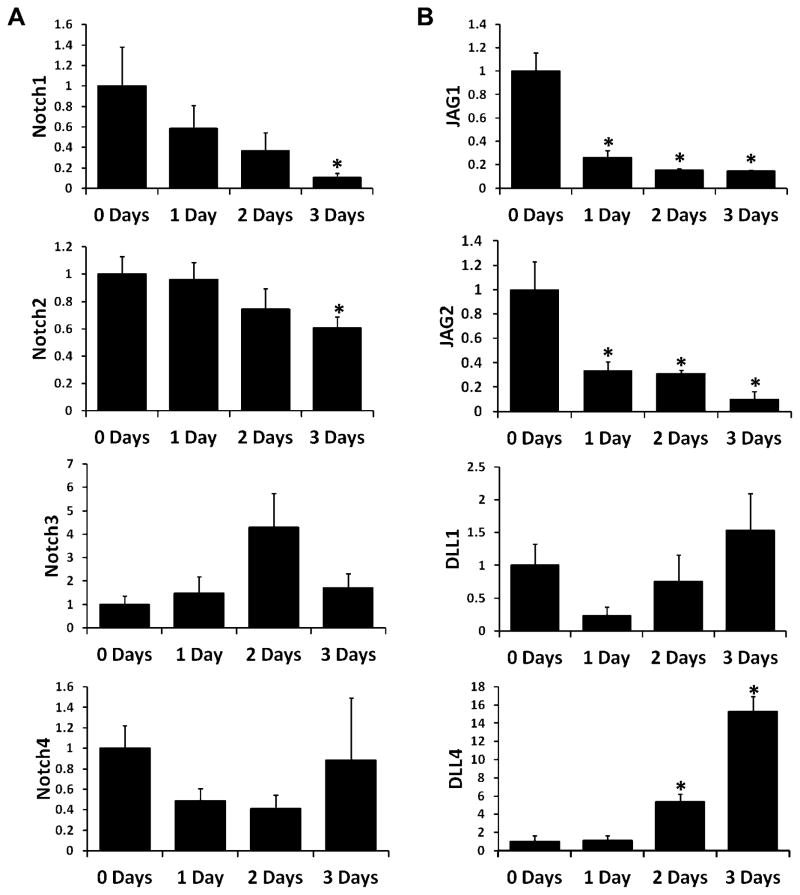
Notch receptor (Fig. 2A) and ligand (Fig. 2B) gene expression at baseline and during osteoclastogenesis were measured via quantitative RT-PCR. Osteoclasts were differentiated from adherent bone marrow-derived macrophages (BMMs) with MCSF and RANKL, and RNA was collected after 1, 2, and 3 days of treatment. RNA samples from cells treated with MCSF alone for 3 days (0 days RANKL treatment) were also collected. Expression of all four Notch receptors was detectible throughout differentiation though expression of both Notch1 and Notch2 had decreased significantly by the end of the treatment (Fig. 2A). Among Notch ligands, expression of both Jagged1 (JAG1) and Jagged2 (JAG2) significantly decreased after 1 day of RANKL treatment and remained low for the remainder of the differentiation period (Fig. 2B). By contrast, Delta-like1 (DLL1) expression remained constant and Delta-like4 (DLL4) expression significantly increased after 2 and 3 days of RANKL treatment. Delta-like3 (DLL3) expression could not be detected at any time point (data not shown).
Figure 2. Notch receptor and ligand expression during osteoclastogenesis.
RNA was extracted from osteoclast precursors cultured with MCSF alone (0 Days) or with MCSF and 100ng/mL RANKL for 1, 2, or 3 days. Expression was measured via real-time RT-PCR. (A) Expression of Notch receptors (Notch1, Notch2, Notch3, and Notch4) relative to expression at 0 Days. (B) Expression of Notch ligands (Jagged1, Jagged2, Delta-like1, and Delta-like4) relative to expression at 0 Days. *, p<0.05 vs. 0 Days. Data are derived from three biological replicates.
Notch signaling manipulation alters expression of osteoclast function-related genes
mRNA levels of Notch-responsive genes and osteoclast functional were quantified via real-time quantitative RT-PCR. The Notch-responsive gene Hes1, but not Hey1, was upregulated by stimulation with JAG1, but not DLL1, in osteoclast precursors (Fig. 3A). Both JAG1 and DLL1 stimulation enhanced TRAP expression even in the absence of RANKL (Fig. 3B). JAG1 also significantly increased cathepsin K and MMP9 expression, where DLL1 increased only cathepsin K. With the exception of cathepsin K, which was significantly reduced by DAPT, there was no significant change in osteoclast functional genes under Notch signaling inhibition. No treatment altered expression of calcitonin receptor.
Figure 3. Expression of Notch target and osteoclast genes under Notch signaling manipulation.
RNA was extracted from osteoclast precursors cultured under Notch stimulation or inhibition with either MCSF only or MCSF and 100ng/mL RANKL for 3 days. (A) Expression of Notch target genes (HES1 and HEY1). (B) Expression of osteoclast functional genes (Tartrate-resistant acid phosphatase, TRAP; Calcitonin receptor, CTR; Cathepsin K, CTSK; Matrix metalloproteinase 9, MMP9). *, p<0.05 vs IgG; †, p<0.05 vs. DMSO. Data are derived from three biological replicates. N.D., not detected.
Notch signaling enhances osteoclastogenesis of osteoclast precursors pre-stimulated with RANKL
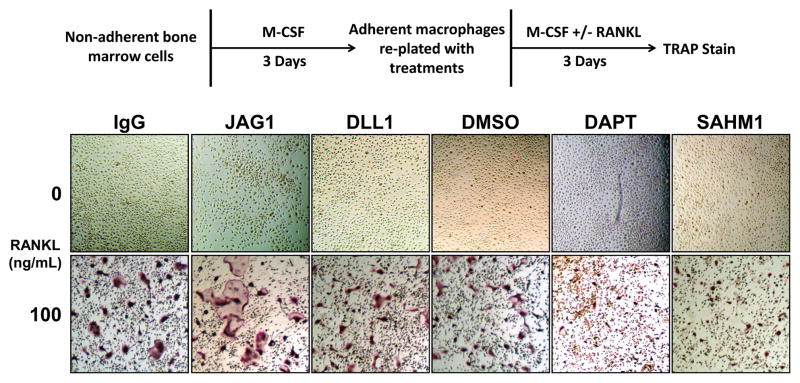
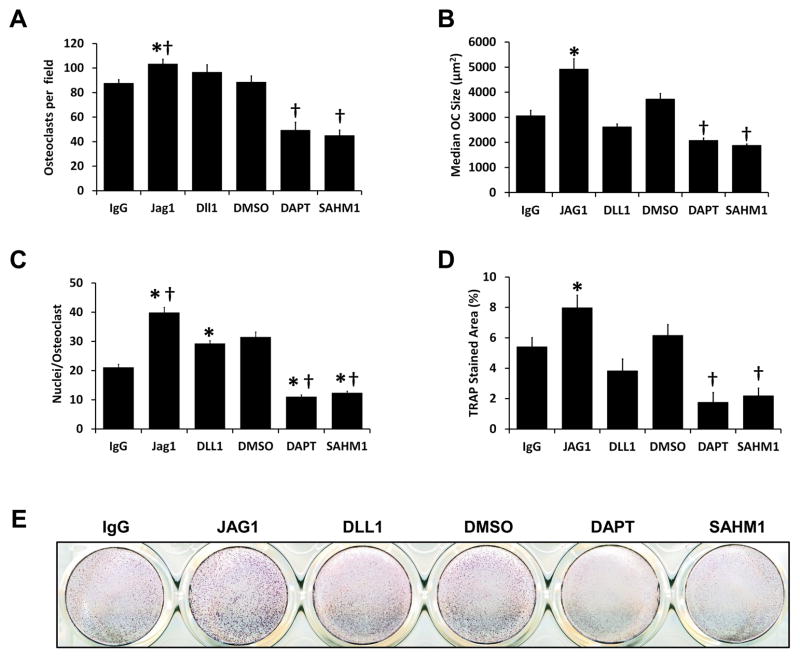
Adherent BMMs were differentiated to osteoclasts by seeding with MCSF and RANKL either onto JAG1- or DLL1-coated wells (control: IgG-coated wells) or with DAPT or SAHM1 (control: DMSO) (Fig. 4). Culture of cells on JAG1, but not DLL1, resulted in increased numbers of multinuclear osteoclasts; in contrast, Notch signaling inhibition with either DAPT or SAHM1 significantly reduced osteoclast numbers (Figs. 4 and 5A). JAG1 stimulation resulted in larger osteoclasts, where Notch inhibition by both DAPT and SAHM1 resulted in smaller osteoclasts (Figs. 4 and 5B). In addition, both JAG1 and DLL1-stimulated cells had significantly increased numbers of nuclei where nuclear numbers were significantly reduced in DAPT and SAHM1 treated cells (Fig. 5C). Alterations in osteoclast size and number were reflected in measurements of TRAP-stained areas; JAG1-stimulated cells demonstrated significantly higher and DAPT- and SAHM1-treated cells demonstrated significantly lower stained areas compared to their respective controls (Fig. 5D & E). These results suggest that activation of Notch signaling in RANKL-stimulated osteoclast precursors enhances osteoclast differentiation.
Figure 4. Osteoclastogenic differentiation of adherent BMMs under Notch stimulation and inhibition.
Non-adherent bone marrow cells were cultured with M-CSF for 3 days to generate adherent bone marrow-derived macropahges, which were lifted and re-plated into 24-well plates with indicated treatments. Osteoclasts precursors were cultured in either MCSF alone or MCSF and 100ng/mL RANKL for 3 days under either Notch ligand stimulation or Notch signaling inhibition. At the conclusion of differentiation, cells were TRAP stained. Red/purple staining indicates TRAP activity.
Figure 5. Quantification of in vitro osteoclastogenesis parameters.
(A) Mean number of osteoclasts per microscopic field. (B) Average of median osteoclast size in each visual field. (C) Mean number of nuclei per osteoclast. (D) Mean TRAP-stained areas. *, p<0.05 vs. IgG; †, p<0.05 vs. DMSO. Data are aggregate of three independent experiments. (E) Representative image of TRAP-stained wells.
Notch signaling inhibits osteoclastogenesis of non-committed osteoclast precursors
To investigate context-dependent effects of Notch signaling on osteoclastogenesis, osteoclast precursors were differentiated under two additional conditions (Fig. 6). First, varying numbers of non-adherent bone marrow cells were seeded with MCSF and RANKL into IgG- (control) or JAG1-coated wells. At the lowest density (1 × 105 cells), there was no significant difference in TRAP-stained areas between precursors cultured in IgG- or JAG1-coated wells (Fig. 6A). However, at intermediate densities (4 and 8 × 105 cells) osteoclast differentiation was significantly higher in IgG-coated wells. At the highest density (10 × 105 cells), there were similar levels of osteoclastogenesis in IgG- and JAG1-coated wells.
Figure 6. Differentiation of osteoclasts from non-adherent bone marrow cells.
(A) Osteoclasts were generated from varying seeding densities of non-adherent marrow cells on IgG- or JAG1-coated culture surfaces by culturing for 5 days with MCSF and RANKL. Following differentiation, cells were TRAP stained and TRAP-stained area was quantified. *, p < 0.05 vs. IgG. (B) Osteoclasts were generated from varying seeding densities of non-adherent marrow cells on IgG- or JAG1-coated culture surfaces by first culturing 3 days with MCSF only followed by 3 days of MCSF and RANKL. Following differentiation, cells were TRAP stained and TRAP-stained area was quantified. *, p < 0.05 vs. IgG. Each treatment was performed in duplicate. Images are representative and data are aggregate of 2 independent experiments.
Second, varying numbers of non-adherent bone marrow cells were seeded into IgG- or JAG1-coated wells with MCSF only and allowed to adhere and proliferate for 3 days prior to RANKL stimulation. Under this method, cells in IgG-coated wells demonstrated a greater amount of osteoclastogenesis regardless of seeding cell density (Fig. 6B). These results suggest that early activation of Notch signaling in osteoclast precursors suppresses osteoclastogenesis.
Notch signaling enhances osteoclastic resorption
Osteoclastic resorption of mineral surfaces was assessed under Notch signaling stimulation and suppression to determine whether alterations in osteoclast maturation translate to altered function. Osteoclast precursors were cultured with and without RANKL on mineral-coated OsteoAssay surfaces under Notch stimulation with immobilized JAG1 or DLL1 or Notch inhibition with DAPT or SAHM1 for 4 (Fig 7A) or 6 (Fig. 7B) days. After 4 days of culture, significant increases in resorption were evident in both JAG1 and DLL1-stimulated groups compared to IgG-coated wells, but there was not yet sufficient resorption in controls to assess effects of Notch inhibition (Fig. 7A). After 6 days of culture, resorption remained significantly higher in JAG1- and DLL1-coated wells compared to IgG control, and resorption under Notch-inhibition with both DAPT and SAHM1 was significantly reduced compared to DMSO control wells (Fig. 7B).
Figure 7. Osteoclastic hydroxyapatite resorption under Notch signaling manipulation.
Osteoclast precursors were cultured under Notch stimulation or inhibition with either MCSF only or MCSF and 100ng/mL RANKL for either 4 or 6 days. At the conclusion of the culture period, cells were removed and remaining mineral was darkened via von Kossa stain. (A) Representative von Kossa-stained plate following 4 days of culture. (B) Quantification of hydroxyapatite resorption area following 4 days of culture. *, p<0.05 vs. IgG. (C) Representative von Kossa-stained plate following 6 days of culture. (D) Quantification of hydroxyapatite resorption area following 6 days of culture. *, p<0.05 vs. IgG or DMSO, respectively. Images are representative and data are aggregate of two independent experiments.
Notch signaling manipulation alters expression of osteoclast fusion genes
The increases and decreases in nuclear number seen under Notch stimulation and inhibition, respectively, suggest that Notch signaling may contribute to the fusion of osteoclast precursors. To investigate whether Notch signaling might regulate fusion at the mRNA level, expression of established osteoclast fusion genes were measured in the context of Notch signaling stimulation (JAG1- and DLL1-coated wells; IgG-coated wells, control) and inhibition (DAPT and SAHM1 treatment; DMSO, control) (Fig. 8). Both CD200 and DC-STAMP were significantly upregulated by both JAG1 (64% and 41% increases over IgG) and DLL1 (85% and 13% increases over IgG) stimulation; similarly, Notch inhibition with DAPT significantly decreased expression of these genes where SAHM1 decreased only CD200 expression. v-ATPase V0 subunit d2 (ATP6V0D2), CD9, CD81, and signal regulatory protein α (SIRPA) expression were not altered with Notch signaling manipulation.
Figure 8. Expression of osteoclast fusion genes under Notch signaling manipulation.
RNA was extracted from osteoclast precursors cultured under Notch stimulation or inhibition with either MCSF only or MCSF and 100ng/mL RANKL for 3 days. Expression of osteoclast fusion genes CD200, DC-STAMP, v-ATPase V0 subunit d2 (ATP6V0D2), CD9, CD81, Signal-regulatory protein alpha (SIRPA) were measured via qRT-PCR. *, p<0.05 vs IgG; †, p<0.05 vs. DMSO. Data are derived from three biological replicates.
Discussion
The role of Notch signaling in osteoclastogenesis has been controversial with evidence supporting both inhibitory and stimulatory roles. The findings of this study suggest that Notch signaling is necessary for the maturation of osteoclasts, but it suppresses the differentiation of macrophages that have not yet been stimulated with RANKL.
Genetic studies by Bai et al. suggested an inhibitory role for Notch signaling in osteoclastogenesis as compound knockout of Notch1, 2, and 3 resulted in enhanced osteoclastogenesis due to increased osteoclast precursor proliferation and enhanced response to RANKL[Bai et al., 2008]. In the present study, however, we have found that osteoclasts and their precursors also express Notch4; thus, the enhanced osteoclastogenesis seen in this model may be attributable to signaling through Notch4 (Fig 2A). Yamada et al. similarly suggest an inhibitory role for Notch signaling in osteoclastogenesis in their experiments with immobilized DLL1 which demonstrated an inhibition of osteoclast formation and osteoclast precursor proliferation [Yamada et al., 2003]. However, the inhibitory effect of DLL1 has been challenged by the work of Sekine et al. who, using immobilized antibodies against specific Notch receptors and blocking antibodies against specific Notch ligands, demonstrated that Notch signaling initiated by DLL1 and Notch2 promote osteoclastogenesis and Notch signaling initiated by JAG1 and Notch1 suppress osteoclastogenesis [Sekine et al., 2012]. Our data agrees with that of Sekine et al. in that DLL1 increased osteoclast nuclear number and resporptive activity (Figs. 5C and 7).
In the above-described studies, while the exact details of the osteoclastogenesis protocols are not provided, a common aspect is that the osteoclast differentiation processes were concluded after six days of culture. In the present study, Notch signaling stimulation via immobilized JAG1 was carried out under three different osteoclastogenisis protocols: (1) seeding of adherent bone marrow-derived macrophages with MCSF and RANKL into coated plates wherein RANKL in the media stimualted RANK signaling prior to attachment to the ligand-coated surface (RANK first, Notch second), (2) seeding of non-adherent bone marrow cells with MCSF and RANKL into coated plates wherein cells become responsive to RANK and Notch signaling after they have differentiated into adherent macrophages (near-simultaneous stimulation of RANK and Notch), and (3) seeding of non-adherent bone marrow cells with MCSF only into coated plates followed by three days of culture prior to the addition of RANKL (Notch first, RANK second). Under the first protocol, Notch stimulation resulted in a clear enhancement of osteoclastogenesis evidenced not only by increased number of nuclei per osteoclast and increased osteoclast size (in the case of JAG1) (Figs. 4 and 5), but also by increased mineral resorption (Fig. 7). The importance of Notch signaling in osteoclast maturation is further evidenced by supression of osteoclast fusion and resorption by Notch inhibition with either DAPT or SAHM1.
Data from both Bai and Yamada suggest that the inhibitory effect of Notch signaling is due to suppression of osteoclast precursor proliferation[Bai et al., 2008; Yamada et al., 2003]. This is likely to be the case, as, when osteoclasts were differentiated according to the second protocol, JAG1 appeared to decrease osteoclastogenesis, but this defect was rescued by seeding a higher density of cells (Fig. 6A). By contrast, JAG1 inhibited osteoclastogenesis when osteoclast precursors were cultued on coated surfaces for three days prior to RANKL stimulation regardless of cell density (Fig. 6B). These data are not only consistent with findings by others, but also provide evidence for divergent roles for Notch signaling in osteoclastogenesis that are governed by whether osteoclast precursors are exposed to RANKL before or after Notch activation. Such effects are not unusual as TNF exerts a similar behavior wherin it suppresses osteoclast differentiation from uncommitted osteoclast precursors, but enhances osteoclastogenesis if precursors are pre-stimulated with RANKL [Jules et al., 2010].
While there were significant alterations in the expression of some osteoclast genes under Notch signaling manipulation (Fig. 3), the magnitude of gene expression alteration was not great relative to the difference between cells treated with MCSF alone and those treated with both MCSF and RANKL. Furthermore, while osteoclasts were smaller and had fewer nuclei under Notch signaling inhibition, they were, nevertheless, TRAP-positive (Figs. 3 and 4). These findings suggest that Notch signaling does not largely contribute to the initial differentiation of osteoclasts, but rather osteoclast maturation. A role for Notch signaling in osteoclast maturation is is further supported by increased expression of osteoclast fusion genes CD200 and DC-STAMP under Notch stimulation, and decreased expression of these genes under Notch inhibition (Fig. 8). As a bridge to the clinical side, inhibition of osteoclast maturation without inhibition of initial osteoclast differentiation could be a useful and powerful approach to restore bone mass in osteoporosis or other bone loss conditions, as small osteoclasts are deficient in resorption, but are nevertheless capable of stimulating osteogenesis[Henriksen et al., 2014].
Our final observation, which then leads to some interesting future mechanistic investigative directions, stems from the distinct effects of DAPT and SHMI. DAPT blocks Notch cleavage and liberation of NICD, and SAHMI interferes with canonical transcription complex formation [Kornilova et al., 2003; Moellering et al., 2009]. In the case of osteoclast genes, fusion genes, and hydroxyapatite resorption, DAPT demonstrated a more robust inhibitory effect than SAHM1 (Figs. 5, 7, & 8). The differences in intensity suggest that there are (1) targets of DAPT other than Notch signaling, such as ErbB4, involved in osteoclastogenesis and/or (2) elements of Notch signaling that are MAML-independent involved in osteoclastogenesis [Besse et al., 2007; Sardi et al., 2006]. It has been demonstrated that NICD1 and NICD2 interact with Calcium/calmodulin-dependent protein kinase IV and NF-κB subunit p65 respectively, and these interactions support osteoclastogenesis, and these may be examples of MAML-independent Notch signaling that cannot be observed with Notch receptor knockout [Choi et al., 2013; Fukushima et al., 2008]. Future studies are necessary to understand these pontentially important additional pathways that may interact with Notch signaling and what roles they play in osteoclast differentiation and function.
Acknowledgments
This work was supported by the Univeristy of Pennsylvania Center for Musculoskeletal Disorders (5 P30 AR050950-09) and the Philadelphia VA Medical Center Translational Musculoskeltal Research Center (JA, KDH) and an intramural orthopaedic departmental reseach development fund (JA). JWA is supported by the University of Pennsylvania Postdoctoral Opportunities in Research and Teaching (PENN-PORT) fellowship funded by the National Institute of General Medical Sciences Institutional Research and Career Development Award (IRACDA; 5 K12 GM081259-08).
Footnotes
Author contributions: Ashley JW – designed and performed experiments, analyzed and interpreted data, and drafted manuscript; Ahn J – designed experiments, interpreted data, and edited manuscript; Hankenson KD – designed experiments, interpreted data, and edited manuscript.
Disclosures
Ashley JW: Nothing to disclose
Ahn J: Unrelated research funding from Synthes. Consultant for Synthes and Merck. Part owner of Skelegen.
Hankenson KD: Co-founder of Skelegen
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Ashley JW, Shi Z, Zhao H, Li X, Kesterson RA, Feng X. Genetic ablation of CD68 results in mice with increased bone and dysfunctional osteoclasts. PLoS One. 2011;6:e25838. doi: 10.1371/journal.pone.0025838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S, Kopan R, Zou W, Hilton MJ, Ong CT, Long F, Ross FP, Teitelbaum SL. NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J Biol Chem. 2008;283:6509–18. doi: 10.1074/jbc.M707000200. [DOI] [PubMed] [Google Scholar]
- Besse A, Lamothe B, Campos AD, Webster WK, Maddineni U, Lin SC, Wu H, Darnay BG. TAK1-dependent signaling requires functional interaction with TAB2/TAB3. J Biol Chem. 2007;282:3918–28. doi: 10.1074/jbc.M608867200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley EW, Oursler MJ. Osteoclast culture and resorption assays. Methods Mol Biol. 2008;455:19–35. doi: 10.1007/978-1-59745-104-8_2. [DOI] [PubMed] [Google Scholar]
- Chen S, Lee BH, Bae Y. Notch signaling in skeletal stem cells. Calcif Tissue Int. 2014;94:68–77. doi: 10.1007/s00223-013-9773-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YH, Ann EJ, Yoon JH, Mo JS, Kim MY, Park HS. Calcium/calmodulin-dependent protein kinase IV (CaMKIV) enhances osteoclast differentiation via the up-regulation of Notch1 protein stability. Biochim Biophys Acta. 2013;1833:69–79. doi: 10.1016/j.bbamcr.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Cordle J, Johnson S, Tay JZ, Roversi P, Wilkin MB, de Madrid BH, Shimizu H, Jensen S, Whiteman P, Jin B, Redfield C, Baron M, Lea SM, Handford PA. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol. 2008;15:849–57. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L, de Vos P, Fan M, Ren Y. Notch is activated in RANKL-induced osteoclast differentiation and resorption. Front Biosci. 2008;13:7064–71. doi: 10.2741/3210. [DOI] [PubMed] [Google Scholar]
- Feng X. RANKing intracellular signaling in osteoclasts. IUBMB Life. 2005;57:389–95. doi: 10.1080/15216540500137669. [DOI] [PubMed] [Google Scholar]
- Fukushima H, Nakao A, Okamoto F, Shin M, Kajiya H, Sakano S, Bigas A, Jimi E, Okabe K. The association of Notch2 and NF-kappaB accelerates RANKL-induced osteoclastogenesis. Mol Cell Biol. 2008;28:6402–12. doi: 10.1128/MCB.00299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot AJ, Habets R, Yahyanejad S, Hodin CM, Reiss K, Saftig P, Theys J, Vooijs M. Regulated proteolysis of NOTCH2 and NOTCH3 receptors by ADAM10 and presenilins. Mol Cell Biol. 2014;34:2822–32. doi: 10.1128/MCB.00206-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen K, Karsdal MA, Martin TJ. Osteoclast-derived coupling factors in bone remodeling. Calcif Tissue Int. 2014;94:88–97. doi: 10.1007/s00223-013-9741-7. [DOI] [PubMed] [Google Scholar]
- Hong H, Shi Z, Qiao P, Li H, McCoy EM, Mao P, Xu H, Feng X, Wang S. Interleukin-3 plays dual roles in osteoclastogenesis by promoting the development of osteoclast progenitors but inhibiting the osteoclastogenic process. Biochem Biophys Res Commun. 2013;440:545–50. doi: 10.1016/j.bbrc.2013.09.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Zou W, Chappel JC, Ashley JW, Feng X, Teitelbaum SL. c-Src links a RANK/alphavbeta3 integrin complex to the osteoclast cytoskeleton. Mol Cell Biol. 2012;32:2943–53. doi: 10.1128/MCB.00077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA. Outsmarting a mastermind. Dev Cell. 2009;17:750–2. doi: 10.1016/j.devcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jules J, Shi Z, Liu J, Xu D, Wang S, Feng X. Receptor activator of NF-{kappa}B (RANK) cytoplasmic IVVY535-538 motif plays an essential role in tumor necrosis factor-{alpha} (TNF)-mediated osteoclastogenesis. J Biol Chem. 2010;285:37427–35. doi: 10.1074/jbc.M110.149484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jules J, Zhang P, Ashley JW, Wei S, Shi Z, Liu J, Michalek SM, Feng X. Molecular basis of requirement of receptor activator of nuclear factor kappaB signaling for interleukin 1-mediated osteoclastogenesis. J Biol Chem. 2012;287:15728–38. doi: 10.1074/jbc.M111.296228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornilova AY, Das C, Wolfe MS. Differential effects of inhibitors on the gamma-secretase complex. Mechanistic implications. J Biol Chem. 2003;278:16470–3. doi: 10.1074/jbc.C300019200. [DOI] [PubMed] [Google Scholar]
- Li C, Yang Z, Li Z, Ma Y, Zhang L, Zheng C, Qiu W, Wu X, Wang X, Li H, Tang J, Qian M, Li D, Wang P, Luo J, Liu M. Maslinic acid suppresses osteoclastogenesis and prevents ovariectomy-induced bone loss by regulating RANKL-mediated NF-kappaB and MAPK signaling pathways. J Bone Miner Res. 2011;26:644–56. doi: 10.1002/jbmr.242. [DOI] [PubMed] [Google Scholar]
- Maeda K, Kobayashi Y, Udagawa N, Uehara S, Ishihara A, Mizoguchi T, Kikuchi Y, Takada I, Kato S, Kani S, Nishita M, Marumo K, Martin TJ, Minami Y, Takahashi N. Wnt5a-Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nat Med. 2012;18:405–12. doi: 10.1038/nm.2653. [DOI] [PubMed] [Google Scholar]
- McCoy EM, Hong H, Pruitt HC, Feng X. IL-11 produced by breast cancer cells augments osteoclastogenesis by sustaining the pool of osteoclast progenitor cells. BMC Cancer. 2013;13:16. doi: 10.1186/1471-2407-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–8. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi Y, Kan T, Tominari Y, Fuwa H, Okamura Y, Watanabe N, Sato C, Natsugari H, Fukuyama T, Iwatsubo T, Tomita T. C-terminal fragment of presenilin is the molecular target of a dipeptidic gamma-secretase-specific inhibitor DAPT (N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester) J Biol Chem. 2006;281:14670–6. doi: 10.1074/jbc.M513012200. [DOI] [PubMed] [Google Scholar]
- Park KH, Park B, Yoon DS, Kwon SH, Shin DM, Lee JW, Lee HG, Shim JH, Park JH, Lee JM. Zinc inhibits osteoclast differentiation by suppression of Ca2+-Calcineurin-NFATc1 signaling pathway. Cell Commun Signal. 2013;11:74. doi: 10.1186/1478-811X-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell. 2006;127:185–97. doi: 10.1016/j.cell.2006.07.037. [DOI] [PubMed] [Google Scholar]
- Schwarzer R, Kaiser M, Acikgoez O, Heider U, Mathas S, Preissner R, Sezer O, Doerken B, Jundt F. Notch inhibition blocks multiple myeloma cell-induced osteoclast activation. Leukemia. 2008;22:2273–7. doi: 10.1038/leu.2008.138. [DOI] [PubMed] [Google Scholar]
- Sekine C, Koyanagi A, Koyama N, Hozumi K, Chiba S, Yagita H. Differential regulation of osteoclastogenesis by Notch2/Delta-like 1 and Notch1/Jagged1 axes. Arthritis Res Ther. 2012;14:R45. doi: 10.1186/ar3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19:192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum SL. Osteoclasts: what do they do and how do they do it? Am J Pathol. 2007;170:427–35. doi: 10.2353/ajpath.2007.060834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevlin R, McArdle A, Chan CK, Pluvinage J, Walmsley GG, Wearda T, Marecic O, Hu MS, Paik KJ, Senarath-Yapa K, Atashroo DA, Zielins ER, Wan DC, Weissman IL, Longaker MT. Osteoclast derivation from mouse bone marrow 2014. 2014 Nov 6;(93):e52056. doi: 10.3791/52056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Liu K, Chen L, Aihara K. Neural fate decisions mediated by trans-activation and cis-inhibition in Notch signaling. Bioinformatics. 2011;27:3158–65. doi: 10.1093/bioinformatics/btr551. [DOI] [PubMed] [Google Scholar]
- Wang X, Ha T. Defining single molecular forces required to activate integrin and notch signaling. Science. 2013;340:991–4. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Yamazaki H, Yamane T, Yoshino M, Okuyama H, Tsuneto M, Kurino T, Hayashi S, Sakano S. Regulation of osteoclast development by Notch signaling directed to osteoclast precursors and through stromal cells. Blood. 2003;101:2227–34. doi: 10.1182/blood-2002-06-1740. [DOI] [PubMed] [Google Scholar]
- Zhou J, Fujiwara T, Ye S, Li X, Zhao H. Downregulation of Notch modulators, tetraspanin 5 and 10, inhibits osteoclastogenesis in vitro. Calcif Tissue Int. 2014;95:209–17. doi: 10.1007/s00223-014-9883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Fujiwara T, Ye S, Li X, Zhao H. Ubiquitin E3 Ligase LNX2 is Critical for Osteoclastogenesis In Vitro by Regulating M-CSF/RANKL Signaling and Notch2. Calcif Tissue Int. 2015 doi: 10.1007/s00223-015-9967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Sweetwyne MT, Hankenson KD. PKCdelta is required for Jagged-1 induction of human mesenchymal stem cell osteogenic differentiation. Stem Cells. 2013;31:1181–92. doi: 10.1002/stem.1353. [DOI] [PubMed] [Google Scholar]