Summary
In recent years, tetracyclines, such as doxycycline, have become broadly used to control gene expression by virtue of the Tet-On/Tet-Off systems. The wide range of direct effects of tetracycline use has, however, not been fully appreciated. We show here that these antibiotics induce a mitonuclear protein imbalance through their effects on mitochondrial translation, an effect that likely reflects the evolutionary relationship between mitochondria and proteobacteria. Tetracyclines, Even at low concentrations, tetracyclines induce mitochondrial proteotoxic stress, leading to changes in nuclear gene expression and altered mitochondrial dynamics and function in commonly used cell types, as well as worms, flies, mice, and plants. Since tetracyclines are so widely applied in research, scientists should be aware of their potentially confounding effects on experimental results. Furthermore, these results caution against extensive use of tetracyclines in livestock due to potential downstream impacts on the environment and human health.
Introduction
Advances in the mechanistic understanding of gene function are often based on the characterization of gain-of-function (GOF) and loss-of-function (LOF) mutations in cells and model organisms. Constitutive GOF and LOF studies in cell and animal models have now become an essential part of the post-genomic biomedical toolkit (Argmann et al., 2005; Branda and Dymecki, 2004). Since many genes are essential for cellular function and/or animal development (i.e. they are lethal if knocked out from the embryonic state), conditional systems have been developed in which gene expression can be spatially or temporally controlled. In mammalian systems, cell-specific promoters are often used in genetic strategies to spatially control GOF and LOF. For example, tissue- or cell type-specific expression of the Cre recombinase is commonly employed to restrict recombination at LoxP sites, introduced at specific locations in the genomic DNA, to a given cell-type and/or tissue (Utomo et al., 1999). Temporal control often requires responsiveness to an exogenously added inducer. Two prototypical examples of such temporal control are the use of chimeric Cre recombinase proteins (Utomo et al., 1999) and the Tet-On/Tet-Off system (Gossen and Bujard, 1992) (reviewed in (Argmann et al., 2005; Ryding et al., 2001)). The best characterized chimeric Cre recombinase is the Cre-ERT2 protein, where recombinase activity is gated by a mutated version of the ligand binding domain of the estrogen receptor (ER), modified to be only responsive to the synthetic ER antagonist tamoxifen, which does not occur naturally (Feil et al., 1996). Similar chimeric Cre proteins have been developed using the affinity of the progesterone or ecdysone receptor ligand binding domains for RU-486 or ecdysone, respectively (Minamino et al., 2001; No et al., 1996). Long-lasting side effects of the use of these nuclear receptor ligands have been described (Lelliott et al., 2005; Lopez et al., 2006), which have to be factored in as potential confounders in functional genomic studies. The Tet-On/Tet-Off system employs a tetracycline, doxycycline, to activate or inactivate the tetracycline-responsive promoter (Gossen and Bujard, 1992). In Tet-On systems, doxycycline binds the tetracycline transactivator protein and thereby allows binding to a tetracycline response element and transcriptional activation to occur (Gossen et al., 1995). In Tet-Off systems, doxycycline binding to a slightly modified tetracycline transactivator protein impairs its ability to activate the responsive promoter, thus preventing transcriptional activation (Gossen and Bujard, 1992). Although the Tet-On/Tet-Off system provides exquisite flexibility to study gene function, few researchers consider the potential detrimental effects of the use of tetracyclines themselves, although prolonged antibiotic use is known to cause adverse effects in the clinic (Brummett and Fox, 1989; Mingeot-Leclercq and Tulkens, 1999; Selimoglu, 2007).
Work in the 1960’s described that tetracyclines, as well as chloramphenicol, inhibit translation of proteins encoded by mitochondrial DNA (mtDNA), but not by nuclear DNA (nDNA) (Clark-Walker and Linnane, 1966). We recently showed that this selective inhibition of mitochondrial protein translation by both types of antibiotics leads to a state of so-called “mitonuclear protein imbalance”, which disturbs mitochondrial proteostasis (Houtkooper et al., 2013). Mitonuclear protein imbalance ensues when protein synthesis from mtDNA is not matched by protein synthesis from nDNA. This abnormal mitochondrial proteostasis robustly induces the mitochondrial unfolded protein response (UPRmt), leading to a pronounced lifespan extension in the worm and marked metabolic and molecular changes in cells and mice (Houtkooper et al., 2013). Since tetracyclines are widely applied to control gene expression in cells and a large panel of model systems —we found over 18,000 hits in a Google Scholar search (using Tet-On OR Tet-Off OR “tet inducible” OR “tet-induced” OR “doxycycline inducible” OR “tetracycline inducible”)—we explored here in detail whether these antibiotics also interfere with mitochondrial function in these model organisms.
Results
Disturbed mitochondrial proteostasis and function after doxycycline treatment
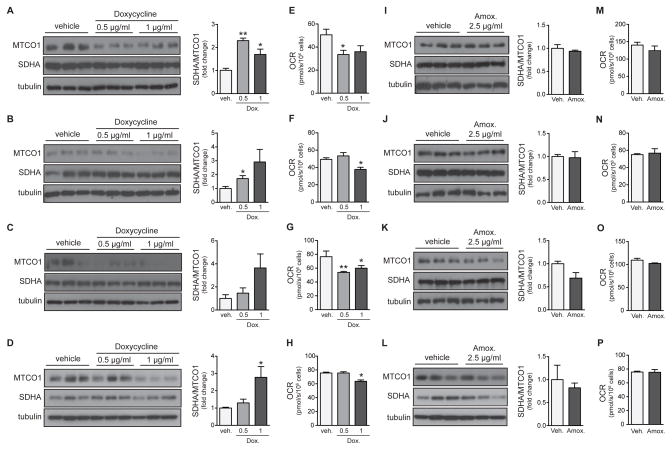
We first treated four commonly used cell lines originating from distinct tissues with increasing concentrations of doxycycline: (1) the human embryonic kidney-derived HEK293 cell line, (2) the human cervical carcinoma HeLa cell line, (3) the mouse hepatoma Hepa 1-6 cell line, and (4) the mouse hypothalamus cell line GT1-7 (Figure 1). We determined mitochondrial proteostasis or mitonuclear protein imbalance (Houtkooper et al., 2013) by analyzing the ratio between a mtDNA-encoded OXPHOS subunit (cytochrome c oxidase subunit I or MTCO1) and a nDNA-encoded OXPHOS subunit (succinate dehydrogenase A or SDHA). Indeed, in all four cell lines, doxycycline dose-dependently induced mitonuclear protein imbalance (Figure 1A–D). These effects occurred at doses that are typically used in Tet-On/Tet-Off experiments, i.e. in the low μg/mL range. In order to demonstrate that these concentrations are indeed relevant, we exposed the Tet-on cell line A549 to increasing concentration of doxycycline. This cell line expresses Tet-controlled luciferase, which was activated at 5–10 μg/ml after 24 hours treatment (Figure S1A). At the same concentrations, these cells displayed marked mitonuclear protein imbalance, comparing the expression of mtDNA-encoded (MTCO1 and MTCO2) and nDNA-encoded (SDHA and SDHB) OXPHOS subunits (Figure S1B–C). Furthermore when we extended the time of treatment to 48 hours, we observed the same mitonuclear protein imbalance at a concentration of 1 μg/ml (Figure S1D–E).
Figure 1. Doxycycline inhibits mitochondrial function in cells.
Effects of doxycycline and amoxicillin were tested in human HEK293 cells (A, E, I, M), human HeLa cells (B, F, J, N), mouse Hepa 1-6 cells (C, G, K, O), and mouse hypothalamus cell line GT1-7 (D, H, L, P).
(A–D) Doxycycline dose-dependently induces mitonuclear protein imbalance, as shown by the reduced ratio between mtDNA-encoded cytochrome c oxidase subunit 1 (MTCO1) and nDNA-encoded succinate dehydrogenase A (SDHA). Western blots are quantified in the right panels.
(E–H) Doxycycline dose-dependently impairs mitochondrial oxygen consumption rate (OCR).
(I–L) Amoxicillin, which does not interfere with bacterial/mitochondrial translation, does not change the balance between MTCO1 and SDHA protein levels. Western blots are quantified in the right panels.
(M–P) Amoxicillin treatment also does not change OCR in the different cell lines.
Bar graphs are expressed as mean+SEM, * p≤0.05; ** p≤0.01.
The mitonuclear protein imbalance in all these cellular systems was accompanied by a strong decrease in cellular respiration, indicative for severely impaired mitochondrial activity (Figure 1E–H). Even though the inhibition of respiration was dose-dependent with an EC50 at 6.3 μg/ml (Figure S1F), it was reversible when doxycycline was removed from the medium (Figure S1G). As the effects of doxycycline could also represent a general effect of antibiotic exposure, we tested the same parameters in the presence of the antibiotic amoxicillin, which is from the penicillin family and disrupts bacterial cell wall synthesis rather than translation (Cole et al., 1972). Consistent with its mode of action in bacteria, amoxicillin did not affect the mitonuclear protein imbalance (Figure 1I–L) and had no impact on mitochondrial function as reflected by the unchanged cellular respiration (Figure 1M–P).
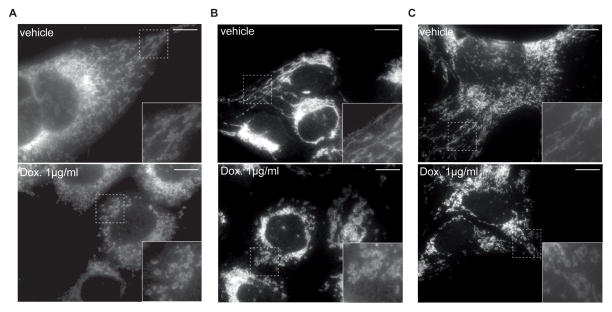
One key factor in the regulation of mitochondrial function is mitochondrial dynamics (Gao et al., 2014). Healthy mitochondrial dynamics consist of repeated cycles of mitochondrial fusion and fission, and is coupled to mitochondrial breakdown through mitophagy (Andreux et al., 2013; Gao et al., 2014; Liesa and Shirihai, 2013; Youle and van der Bliek, 2012). We previously showed that inhibiting mitochondrial translation promotes mitochondrial fission in worms (Houtkooper et al., 2013). Similarly, short-term exposure of Hepa 1-6, HeLa or GT1-7 cell lines to doxycycline induced a striking balance towards mitochondrial fission and a more fragmented mitochondrial appearance as reflected by TOM40 staining (Figure 2A–C).
Figure 2. Impact of doxycycline on mitochondrial morphology in cultured cells.
Mitochondria in Hepa 1-6 (A), HeLa (B), and GT1-7 (C) cells appear more fragmented after treatment with doxycycline at the indicated concentration.
Mitochondria were stained with TOM40 antibody. Insets showed higher magnification of the image, marked by the rectangle. Scale bar represents 10μM.
Doxycycline disrupts global cellular transcriptional profiles
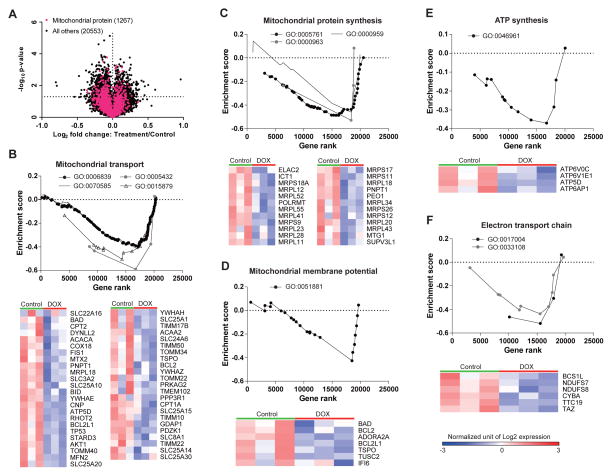
To examine how doxycycline impairs mitochondrial function and ignites global adaptive responses, we analyzed gene expression datasets generated following treatment of the human bladder cancer cell line RT112 with 1 μg/mL of doxycycline (Du et al., 2012). Overall, doxycycline treatment in the RT112 cell line altered the expression of 2181 genes (9.5% of total genes), including 142 mitochondrial protein-coding genes (8.9% of mitochondrial genes under GO accession: 0005739), of which 65.5% were significantly downregulated by doxycycline treatment (Figure 3A). We next performed a targeted gene set enrichment analysis (GSEA) focused on mitochondrial gene sets, to explore the influence of doxycycline on mitochondrial pathways (Figures 3B, S2). Pathways that were robustly downregulated included mitochondrial transport, mitochondrial protein synthesis, mitochondrial membrane potential, ATP synthesis, and electron transport chain (Figure 3B–F, S2). Of note, from the 124 mitochondria related genesets in analysis, many more mitochondrial gene sets were downregulated (55/124) than upregulated (0/124) in these human RT112 cells (Figure S2). Altogether, these results indicate that doxycycline leads to global repression of mitochondrial protein synthesis and function.
Figure 3. Impact of doxycycline on gene expression in the human bladder cancer cell line RT112.
(A) Volcano plot of microarray dataset that was reanalyzed from the original data initially included in Du et al. (Du et al., 2012). The mitochondrial geneset (GO:0005739) is represented in pink. The significance threshold is marked by the horizontal dashed line (p<0.05). 9.5% of total genes and 8.9% of genes in mitochondrial geneset show differential expression under doxycycline treatment.
(B–F) Mitochondrial GSEA and heatmaps of genes involved in mitochondrial transport (B), mitochondrial protein synthesis (C), mitochondrial membrane potential (D), ATP synthesis (E), and electron transport chain (F). Genesets were determined according to Gene Ontology annotation (www.geneontology.org). The heatmap represents only the genes in Core Enrichment, which is the subset of genes that contributes most to the enrichment result. Low expression is shown in blue, while high expression is in red.
Doxycycline impairs development and mitochondrial function in worms and flies
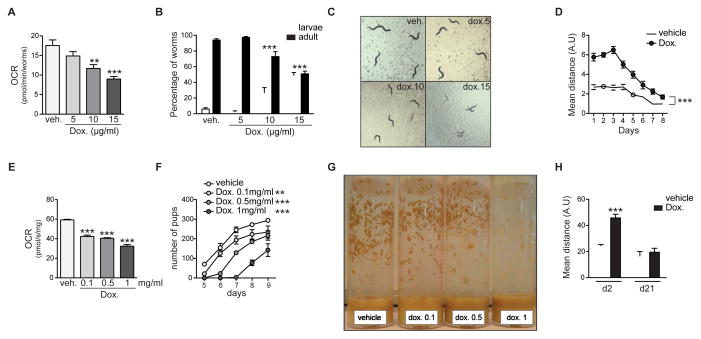
To assess the longer-term effect of doxycycline in intact organisms, we switched to the nematode C. elegans and the fruit fly D. melanogaster, commonly used models for functional genomic studies. In worms, we measured oxygen consumption using the Seahorse XF96 respirometer as a physiological proxy for metabolic activity (Houtkooper et al., 2013). Indeed, supplementing doxycycline to worms during their development from egg to adulthood (4 days) led to a dose-dependent reduction of oxygen consumption (Figure 4A). These doses also delayed worm development, leading to a higher percentage of worms staying in larval phases after 4 days of treatment (Figure 4B), and an overall smaller appearance (Figure 4C). Despite the lowered oxygen consumption, doxycycline-treated worms moved more than untreated worms (Figure 4D). Even though both treated and untreated worms displayed decreasing mobility after the third day of adulthood, doxycycline-treated worms maintained significantly higher mobility throughout life (Figure 4D), which is consistent with our previous findings that inhibition of mitochondrial translation—using mrps-5 RNAi—prevents age-related decline in physical fitness (Houtkooper et al., 2013).
Figure 4. Doxycyline inhibits mitochondrial function in worms and flies, delays their development, and increases mobility.
(A) Doxycycline treatment in C. elegans dose-dependently decreases oxygen consumption rate (OCR), as determined by Seahorse XF96 respirometry.
(B) Doxycycline delays larval development of worms, shown by the bigger proportion of worms in larval stages after 4 days of development.
(C) Representative images of worm populations after 4 days of treatment with the indicated concentration of doxycycline, showing an effect of doxycycline on worm development. Doxycycline-treated worms are significantly smaller than non-treated worms because they are still in the larval stages.
(D) Worms move less with age, but doxycycline-treated worms (30μg/ml) consistently moved more than untreated worms.
(E–G) Doxycycline decreases OCR (E) and delays larval development in D. melanogaster in a dose-dependent manner (F–G). Both curves and pictures show a delay in pupae apparition relative to the doxycycline concentration.
(H) Doxycycline-treated flies move more early in life (day 2), but reach similar activity profiles late in life (day 21).
Bar graphs are expressed as mean+SEM, ** p≤0.01; *** p≤0.001.
In flies, doxycycline supplementation in the food led to a similar decrease in oxygen consumption, as evidenced by Oroboros oxygraph analysis in fly homogenates (Figure 4E). Flies also showed delayed and reduced fecundity, producing less pupae (Figure 4F–G). Finally, doxycycline-treated flies moved more than untreated flies, although the difference disappeared at higher ages (Figure 4H). These data are in line with previous work showing how mild mitochondrial stress improves muscle fitness in flies (Owusu-Ansah et al., 2013), suggesting that similar mechanisms underlie the enhanced movement observed in doxycycline-treated flies.
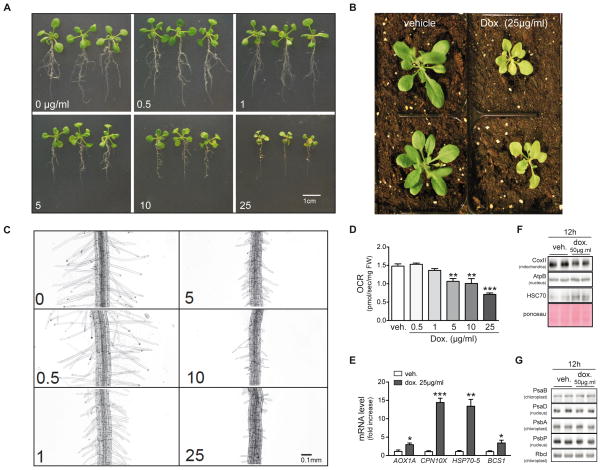
Doxycycline impairs plant growth through inhibition of mitochondrial function
Next, we tested the effects of doxycycline on growth and metabolism of the plant model A. thaliana. Even though plants rely on chloroplasts for glucose production, mitochondrial respiration provides the ATP for all cellular operations except photosynthetic carbon reduction (Jacoby et al., 2012). In addition, mitochondrial metabolism is intimately linked with photosynthesis through photorespiration and nitrogen assimilation (Schwarzlander and Finkemeier, 2013). So robust mitochondrial function is crucial to plant growth. Arabidopsis ecotype Col-0 plants were first grown in normal medium for 7 days, and then transferred to medium containing different concentrations of doxycycline for another 7 days. Doxycycline treatment caused mild to severe growth retardation of seedlings (Figure 5A) at doses that are found in soil of agricultural fields (Bowman et al., 2011; Xie et al., 2011). This effect became even more apparent after transferring the doxycycline-treated plants into soil for 7 days and watering with 25 μg/ml of doxycycline (Figure 5B). Furthermore, a closer observation of the root hairs in the maturation region shows that gradually increasing the doxycycline concentration inhibits root hair elongation (Figure 5C). To examine whether plants share the same mechanisms as worms and flies in response to doxycycline treatment, we measured oxygen consumption in Arabidopsis seedlings. Leaves from doxycycline-treated seedlings displayed a marked reduction in oxygen consumption (Figure 5D), which was accompanied by changes in the expression of the stress response genes AOX1A (AT3G22370), CPN10X (AT1G23100), HSC70-5 (AT5G09590) and BCS1 (AT3G50930) (Figure 5E). AOX1A (alternative oxidase 1a) is a plant-specific quinol oxidase that is activated when complex IV function is inhibited and may play an antioxidant role in plant mitochondria by the reduction of oxygen into water without proton translocation (Jacoby et al., 2012). CPN10X (putative 10kDa chaperonin), HSC70-5 (heat shock protein 70), and BCS1 (cytochrome BC1 synthesis) are all molecular chaperones involved in mitochondrial stress response in plants (Van Aken et al., 2009). Similarly, at the protein level, plants that are exposed to doxycycline display marked mitonuclear protein imbalance (mtDNA-encoded CoxII versus nDNA-encoded AtpB) and activation of the UPRmt marker HSC70 (Figure 5F). Importantly, exposing plants to doxycycline did not induce chloroplast-specific stress response, as the expression of chloroplast-specific chaperones PsaB and PsbA was unaltered upon doxycycline exposure (Figure 5G).
Figure 5. Doxycyline inhibits mitochondrial function in plants and impairs plant growth.
(A) Growing A. thaliana seedlings in the presence of doxycycline for 7 days disturbs both root and leaf growth in a dose-dependent fashion.
(B) Severe growth retardation in plants after 14 days of doxycycline (25μg/ml) treatment.
(C) A. thaliana in the presence of doxycycline for 7 days show a dose-dependent inhibition of root hair growth.
(D) Doxycycline treatment in A. thaliana dose-dependently reduces OCR.
(E) Doxycycline treatment induces the expression of mitochondrial stress genes AOX1A, CPN10X, HSP70-5, and BCS1.
(F) Doxycycline inhibited mitochondrial translation (COXII) but not nuclear translation (AtpB) in A. thaliana seedlings. The induction of the chaperone HSC70 reflects the accumulation of unfolded proteins in mitochondria.
(G) Doxycycline treatment did not induce significant chloroplast protein changes, e.g. photosystem proteins PsaB and PsbA encoded by chloroplast DNA, or PsaD and PsbP encoded by nDNA are shown. RbcI is shown as a loading control.
Bar graphs are expressed as mean+SEM, ** p≤0.01; *** p≤0.001.
Doxycycline impairs mitochondrial proteostasis and function in mice
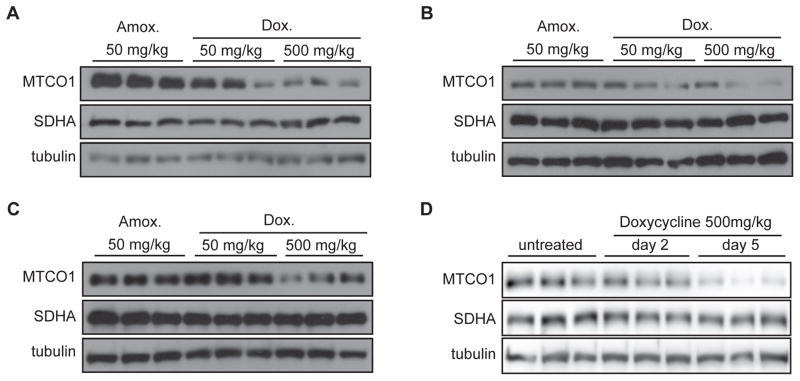
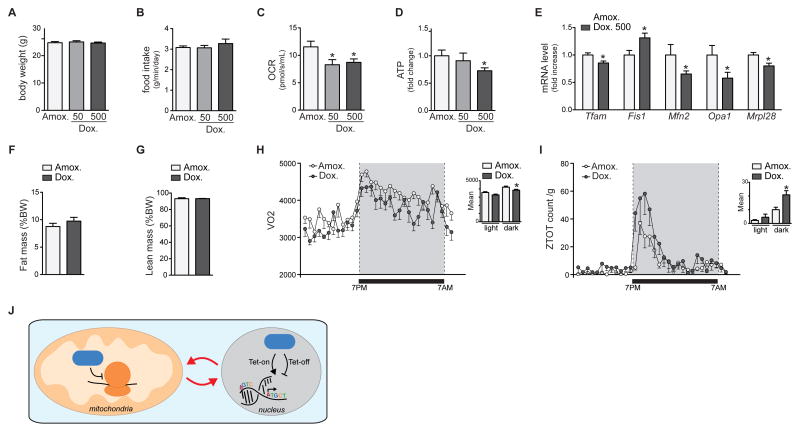
We then tested whether the doxycycline effects are conserved in mice in vivo. We gave male C57BL/6J mice 50 or 500 mg/kg/d of doxycycline dissolved in their drinking water for 14 days. We used 50 mg/kg/d amoxicillin as a control, since this antibiotic does not interfere with bacterial/mitochondrial translation but rather disrupts the bacterial cell wall. Similar to our results in cell lines, doxycycline dose-dependently induced mitonuclear protein imbalance in different mouse tissues, including the liver, the heart, and the brain (Figure 6A–C). We then performed a more detailed time course experiment to establish the dynamic induction of mitonuclear protein imbalance. While mitonuclear protein imbalance was apparent after two days of doxycycline treatment, the ratio between MTCO1 and SDHA became markedly increased at day 5 (Figure 6D). Altogether, these experiments highlight the marked time- and dose-dependent regulation of mitochondrial proteostasis by doxycycline. Finally, we analyzed the physiological consequences of doxycycline treatment in mice. The treatment did not affect body weight or food intake, suggesting no overt toxicity at these doses and within the treatment timeframe (Figure 7A–B). Nevertheless, when mitochondria were isolated from mouse livers, mitochondrial respiration was reduced (Figure 7C), which was associated with a depletion of mitochondrial ATP levels (Figure 7D) and changes in expression of different mitochondrial genes (Figure 7E), together demonstrating inhibition of mitochondrial function. Of note and in support of the significant mitochondrial fragmentation (Figure 2), we observed a decrease in the expression of genes involved in fusion, such as Opa1 and Mfn2, with a concomitant increase in the expression of Fis1 involved in fission (Figure 7E).
Figure 6. Doxycyline inhibits mitochondrial function in vivo in mice.
(A–C) Doxycycline (at 50 mg/kg/d and 500 mg/kg/d in the drinking water) induces mitonuclear protein imbalance in liver (A), heart (B) and brain (C), as shown by the reduced ratio between mtDNA-encoded MTCO1 and nDNA-encoded SDHA when compared to amoxicillin (at 50 mg/kg/d) after 14 days of treatment.
(D) Doxycycline at 500 mg/kg/d in the drinking water induces mitonuclear protein imbalance as evaluated by the ratio of MTCO1/SDHA expression after five days. After two days of treatment, the effect is still discrete.
Figure 7. Doxycyline affects physiological functions in vivo in mice.
(A–B) Doxycycline (at 50 mg/kg/d and 500 mg/kg/d in the drinking water) and amoxicillin (at 50 mg/kg/d) treatment in mice does not affect body weight (A) or food intake (B).
(C–D) Liver homogenates from doxycycline-treated mice (at 50 mg/kg/d and 500 mg/kg/d in the drinking water) display a marked decrease in OCR (C) and ATP content (D) compared to those isolated from livers of the amoxicillin-treated cohort (at 50 mg/kg/d).
(E) Doxycycline given for 14 days at a concentration of 500 mg/kg/d in the drinking water changes the expression of several genes involved in maintenance of mitochondrial function in the liver.
(F–G) Fat mass (F) and lean mass (G) were not affected by either doxycycline or amoxicillin treatment.
(H) Doxycycline treatment (500 mg/kg/day in drinking water) of mice reduced energy expenditure compared to that of mice treated with amoxicillin (at 50 mg/kg/d). The grey area shows the time when lights in the animal facility were switched off.
(I) Doxycycline-treated mice move more than amoxicillin-treated mice. Conditions are similar as in panel H.
(J) Doxycycline can increase (Tet-On) or decrease (Tet-Off) gene expression in the nucleus, but its use also impairs mitochondrial translation and function, leading to the generation of a retrograde signal that on its turn affects nuclear gene expression. The resulting phenotypic changes are hence not only the result of loss- or gain-of-function of the gene under study, but are the result of a double hit.
Bar graphs are expressed as mean+SEM, * p≤0.05.
At the whole body level, these short-term treatments did not affect body composition in either lean and fat mass between the amoxicillin and doxycycline groups (Figure 7F–G). The decreased oxygen consumption of doxycycline-treated mice, indicative for reduced energy expenditure (Figure 7H), and the marked increase in physical activity (Figure 7I) are, however, clear indicators for altered physiological fitness.
Discussion
The use of tetracyclines to control gene expression confounds research
Tetracyclines are a group of antibiotics that have found their way in biomedical research for their use in tetracycline-responsive control of gene expression. Such Tet-On/Tet-Off systems offer experimental flexibility that is often looked for, especially in cases where temporal control of gene expression is necessary for cell survival, and are therefore now widely used. However, tetracycline use is seldom questioned, even though classical literature describes tetracyclines as mitochondrial inhibitors (Clark-Walker and Linnane, 1966). Similarly, we identified widespread effects of the tetracycline, doxycycline, on mitochondrial morphology and function in worms and mammalian cells (Houtkooper et al., 2013). Worms furthermore lived longer upon low-dose doxycycline exposure, which was dependent on changes in mitochondrial proteostasis (Houtkooper et al., 2013). Even though the physiological outcome in this study was beneficial to worms, the potential confounding effects of doxycycline treatment are obvious.
Our cross-species data confirm a strong inhibitory effect of doxycycline on mitochondria in common cell lines, invertebrate animal models, mice, and in plants. The fact that these changes occur in cells derived from different tissues, and in organisms across kingdoms, confirms that the mechanisms underlying these alterations are highly conserved. Also, these effects occurred at doses that are typically used in the literature to control gene expression. Admittedly, some experimentalists use a doxycycline-treated control groups, but even in this context doxycycline may induce adverse effects on mitochondria. We here confirmed this effect in reanalyzing a global transcriptomics data set (Du et al., 2012). Doxycycline treatment in the RT112 cell line altered the expression of almost 10% of genes. These changes, even if just considered a mild stress, can sensitize cells and animals to the Tet-On/Tet-Off, inducing a “two-hit” model, where the effect of Tet-controlled gene expression in the nucleus are confounded by and occurring in a sensitized transcriptional milieu (Figure 7J). In fact, the stress inflicted by tetracyclines on the mitochondria induces a retrograde response—with signaling from the mitochondria to nucleus (Liu and Butow, 2006; Rhoads and Subbaiah, 2007). The phenotypic changes observed are hence the result of the synergistic actions of doxycycline and LOF or GOF of the gene of interest, rather than the singular effect of the gene of interest itself.
We observed a phenotypic imprint of doxycycline at several levels of organization. At the molecular and cellular level, doxycycline inhibits bacterial, but also mitochondrial translation. As a consequence, the 13 oxidative phosphorylation subunits encoded by mtDNA are not properly expressed. This likely leads to unstable oxidative phosphorylation complexes, and an adaptive state we previously termed mitonuclear protein imbalance (Houtkooper et al., 2013). Exposure to doxycycline indeed induced mitonuclear protein imbalance in cultured cells but also in tissues from mice treated with this antibiotic. Mitonuclear protein imbalance is the trigger for the UPRmt, an adaptive stress response in mitochondria that involves the upregulation of mitochondrial chaperones and proteases (Jovaisaite et al., 2014). In fact, proper UPRmt activation is essential for the lifespan extension observed in long-lived mitochondrial worm mutants (Durieux et al., 2011; Houtkooper et al., 2013). Concomitant with the induction of mitonuclear protein imbalance, we observed striking adaptive changes in global gene expression profiles of doxycycline-treated cells. Even though this state—mitonuclear protein imbalance and UPRmt—is adaptive and protective when induced early in life (Durieux et al., 2011; Houtkooper et al., 2013), we now show that prolonged or high-dose activation can in fact be maladaptive and harmful for homeostasis.
Tetracyclines impair organismal physiology and have a major environmental impact
The effects of tetracyclines are also evident at the physiological level of the entire organism. In worms, flies, and plants, treatment with doxycycline led to a marked reduction and delay of growth, and oxygen consumption was severely impaired. The latter was also observed in mice, both using isolated liver mitochondria and in vivo using indirect calorimetry in metabolic cages. Additionally, both flies and worms displayed reduced fertility. Interestingly, doxycycline treatment also caused increased mobility across organisms. Although the physiological reason for this change is not completely understood, we and others previously showed that low-level mitochondrial stress and UPRmt were associated with improved muscle function and fitness in worms and flies (Houtkooper et al., 2013; Owusu-Ansah et al., 2013).
The results we obtained in plants are a reflection of one of the potential environmental challenges that tetracyclines pose at the ecological level. According to FDA reports, tetracycline antibiotics account for about 41% of the ~13.74 million kg of antibiotics sold in 2011 in the US for use in livestock (versus ~3.3 million kg of antibiotics used per year in humans in the US) (FDA, 2011). More critically, a survey in 2007 estimated that almost half of the 210 million kg of antibiotics produced in China per year were used in animal feed and that these antibiotics are mostly tetracyclines (Hvistendahl, 2012). While plants are naturally away from tetracyclines, the increased use of these antibiotics in humans and especially livestock has led to widespread contamination of water and soil resources (Mathews and Reinhold, 2013), increasingly exposing plants to these toxins. As a consequence, tetracyclines accumulate in soils of agricultural fields at concentrations that are comparable to the concentrations we used in our study, and which cause a major growth delay in A. thaliana (Bowman et al., 2011; Xie et al., 2011). Even though the extent of ecological disturbance is poorly understood, our data warrant further exploration in this direction.
Conclusions and perspectives
Taken together, our results call for extreme caution when using tetracyclines for research purposes. This is particularly true if metabolism is the phenotype that is being studied. It is, however, not restricted to these applications since mitochondria have been implicated in a range of processes, including neurodegeneration, immunology, and cancer (Andreux et al., 2013). The widespread effects of antibiotics on mitochondrial function need also to be taken into account in clinical practice. Recently, much attention has been drawn to the relevance of microbiota in this respect, as the complete microbiome is estimated to be ten times larger than the number of cells in a human body (1014 bacteria versus 1013 cells), possibly explaining why early life exposure to antibiotics severely impacts metabolic traits through disruption of microbial homeostasis (Cho et al., 2012). One should keep in mind, however, that tetracyclines and some other antibiotic classes also inhibit the mitochondria—to be considered as bacteria within our cells—the population of which approximately exceeds the number of bacterial cells by a further order of magnitude (1015 mitochondria), thus providing a strong platform for adverse effects.
Experimental procedures
Cell experiments
The human cervical carcinoma HeLa, the human embryonic kidney HEK-293 and the mouse hepatoma Hepa 1-6 cell lines were obtained from ATCC (Manassas, VA, USA). Mouse hypothalamic GT1-7 cell lines were kindly provided by Dr. Pamela Mellon (UCSD, United States) (Wetsel et al., 1991). Cells were grown at 37°C in a humidified atmosphere of 5%CO2/95% air in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with non-essential amino acids and 10% fetal bovine serum without antibiotics. Doxycycline (Sigma) and amoxicillin (Mepha) were dissolved in water. Treatments were performed in DMEM complete medium without antibiotics, which was refreshed every 24 hours.
D. melanogaster experiments
The Drosophila melanogaster strain w− was provided by the Bloomington Drosophila Stock Center (University of Indiana). Flies were kept at 25°C, 60% humidity and on a 12/12h light dark cycle. The food was supplemented with doxycycline at the indicated concentration.
Developmental observation was performed with synchronized eggs. 2–3 day old flies were placed in a grape-agar medium housing tube supplemented with yeast paste. After 24 hours, tubes containing the non-synchronized eggs were replaced by new ones. Synchronized eggs were collected on the new tube after 22 hours. After washing in 1x PBS, 220 eggs were placed in each tube. Pictures were taken from day 4 (3rd instar larvae) and following. The number of pupae were recorded.
Mobility assays
C. elegans movement was recorded for 45 seconds at different days of adulthood using a Nikon DS-L2 / DS-Fi1 camera and controller setup, attached to both a computer and a standard bright field microscope. Five plates of worms, with 10 worms per plate were measured in each condition. Fly movement was recorded by placing flies in a sealed chamber, tapping the chamber, and recording their movement as they naturally climb towards the top. For the tapping test, flies at the corresponding ages were recorded using a standard SLR camera with a Leica macro objective. The experiment was performed 3 times for each cohort with one minute recordings each, with a “tap” sending the flies to the bottom of the chamber every 10 seconds.
Using these video recordings, the movement traces of both worms and flies during all recording periods were calculated by following the organism centroids using a modified version of the Parallel Worm Tracker for MATLAB (Ramot et al., 2008). The distance covered during the recording periods was then averaged per plate and per condition.
C. elegans experiments
C. elegans strains were cultured at 20°C on nematode growth media agar plates seeded with the E. coli strain HT115. The N2 wild type strain was provided by the Caenorhabditis Genetics Center (University of Minnesota). Doxycycline was added at the indicated concentration just before pouring the plates. Animals were exposed to compounds from eggs until the day of the experiment.
A. thaliana experiments
Arabidopsis (Col-0) seedlings were cultivated on ½ MS+agar medium (Caissonlabs) for 7 days. Doxycycline treatment was initiated by transferring the seedlings to ½ MS medium supplemented with different doxycycline concentration of 0.5, 1, 5, 10, 15, and 25 μg/mL. Plants were grown in a growth chamber at 22°C with a 16h/8h light/dark cycle, 100 μmol s−1 m−2 light intensity. Morphological phenotypes were recorded after 7 days of treatment. Root hair images were taken by using an Olympus AX70 microscope with a 4x objective, other parameters include sensitivity of ISO200, manual exposure time of 1/128 sec, and gray scale. Some of the seedlings were transferred into soil and watered with 25 μg/mL doxycycline 3 times per day to observe the longer effect of treatment.
Mouse experiments
Male C57BL/6J mice at 8 weeks old of age were treated for 2, 5 and 14 days with 50 or 500 mg/kg/day doxycycline (Sigma) or 50 mg/kg/day amoxicillin (Mepha) in drinking water. As doxycycline is bitter we supplemented the water for all the conditions (treatments and controls) with 50 g/L sucrose. Drinking water was changed every 48 hours. Indirect calorimetry to monitor O2 consumption, CO2 production, and measurement of activity was measured using Comprehensive Lab Animal Monitoring System (CLAMS) (Columbus Instruments, Columbus, OH) (Lagouge et al., 2006). Mouse experiments were performed in accordance with Swiss law and institutional guidelines.
Western blotting
Proteins were extracted from liver and cells in protein extraction buffer containing 25 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS with added protease inhibitor cocktail (Roche). We used 20 μg of total protein lysate to detect mitochondrial protein imbalance. Antibodies against MTCO1, SDHA (both from Abcam) and β tubulin (Santa-Cruz) were used for immunoblotting.
Respiration assays
For cultured cells, liver tissue homogenates, and flies, oxygen consumption was measured using the Oxygraph-2k (Oroboros Instruments). Cells are trypsinized, counted and resuspended in complete DMEM medium. Basal respiration was measured four times during 10 minutes at 37°C in the respiration chambers, values are normalized by the total number of cells in the chamber. Liver tissues and flies were homogenized with a pestle and mortar in the MiR05 respiration buffer (0,5 mM EGTA, 3mM MgCl2, 60mM K-lactobionate, 20mM taurine, 10mM KH2PO4, 20mM HEPES, 110mM sucrose, 1g/L BSA, 5mM pyruvate, 2mM malate, 10mM glutamate, 2,5mM ADP) at 4°C. Basal respiration was measured four times during 10 minutes at 37°C in the respiration chambers, and values are normalized by the total amount of protein determined using a Bradford assay.
For worms, oxygen consumption was measured using the Seahorse XF96 (Seahorse Bioscience) as described (Mouchiroud et al., 2013). Typically, 100 animals were recovered from NGM plates with M9 medium, washed three times to eliminate residual bacteria, and resuspended in 200 μl M9 medium. Worms were transferred in 96-well Seahorse plates (10 worms per well) and oxygen consumption was measured six times. Respiration rates were normalized to the number of worms in each individual well. Doxycycline treatments were performed at the indicated concentration.
A. thaliana leaf respiration was measured using the Oxygraph-2k (Oroboros Instruments, Austria) at 24°C in the dark. Leaf discs (10–15 mg) were first suspended in 2 mL of plant respiration buffer (Sew et al., 2013) and incubated in the dark for 30 min. Oxygen consumption rate for 15 min after input was recorded. The OCR was calculated accordingly to the fresh weight (FW) of the leaf discs.
Quantification of ATP Levels
Total ATP content was measured by the CellTiter-Glo luminescent cell viability assays (Promega). Typically, the luminescence was recorded with a Victor X4 plate reader (PerkinElmer) and values are normalized by the total protein concentration determined using a Bradford assay.
Quantitative real-time PCR for mRNA
For mouse tissues, total RNA was prepared using TRIzol (Invitrogen) according to the manufacturer’s instructions. RNA was treated with DNase, and 1 μg of RNA was used for reverse transcription (RT). 20X diluted cDNA was used for RT-quantitative PCR (RT-qPCR) reactions. The RT-qPCR reactions were performed using the Light-Cycler system (Roche) and a qPCR Supermix (Qiagen) with the indicated primers. Total RNA from plant samples was prepared using TRIzol as previously described (Connolly et al., 2006). 2 μg of RNA was used for RT using QuantiTect reverse transcription kit (Qiagen). 20X diluted cDNA was used for RT-qPCR reactions using the Light-Cycler system (Roche) and a LightCycler 480 SYBR Green I Master mix (Roche). Ubiquitin gene UBQ10 (AT4G05320) was used as a reference. The average of at least three technical repeats was used for each biological data point. Primer sequences are shown in Table S1 and S2.
Confocal microscopy and image processing
Cells were fixed with Formal-Fixx (Thermo Scientific) and permeabilized with 0.4% (v/v) Triton X-100 for 5 minutes. Images were acquired using a Zeiss LSM 700 Upright confocal microscope (Carl Zeiss AG) under non-saturating exposure conditions. For each condition multiple cells were observed and imaged. For the uniformity of the represented images, contrast and brightness were adjusted to eliminate undesirable background signal. Neither of these manipulations was affecting the mitochondrial shape. Image processing was performed with the Fiji software (http://imagej.nih.gov/ij; version 1.47b).
Bioinformatics analysis
The dataset generated in human bladder cancer cell line RT112 (Du et al., 2012) was downloaded from Gene Expression Omnibus under the accession number GSE41035 and GSE30746, respectively. Data were analyzed using the Gene Set Enrichment Analysis software (Subramanian et al., 2005).
Before the analysis, mitochondrial gene sets were defined by searching for all gene ontology terms containing the word “mitochondria” on the AmiGO website (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi). A total of 131 gene sets were defined using this method. During the analysis, 7 gene sets were filtered out according to the setting of geneset size (3–1000). The remaining 124 gene sets were then used for the analysis. Raw data from human RT112 were first normalized with RMAExpress software (http://rmaexpress.bmbolstad.com/), then loaded in the GSEA software (http://www.broadinstitute.org/gsea) and analyzed using the following parameters: Genesets: local “mitochondrial genesets”; Geneset size: 3–1000; Chip: Affymetrix HG_U133_Plus_2.chip for human RT112 dataset; Output plot no: 60.
Statistics
Differences between two groups were assessed using two-tailed t-tests. Analysis of variance was used when comparing more than two groups. To compare the interaction between age and treatment, two-way ANOVA tests were performed. GraphPad Prism 6 (GraphPad Software, Inc.) was used for all statistical analyses, and p<0.05 was considered significant.
Supplementary Material
Figure S1. Doxycycline effects in cell lines. Related to Figure 1. (A) Doxycycline induces Tet-controlled luciferase expression in the A549 cell line at the concentration of 5–10 μg/ml. (B) Doxycycline dose-dependently causes mitonuclear protein imbalance as evidenced by the altered ratio between mtDNA-encoded (MTCO1 and MTCO2) and nDNA-encoded (SDHA and SDHB) OXPHOS subunits. (C) Quantification of the mitonuclear protein imbalance observed in panel B. (D) Doxycycline treatment in A549 “Tet-on” cells at 1 and 5 μg/ml for 48 hours causes mitonuclear protein imbalance as evidenced by the altered ratio between mtDNA-encoded (MTCO1) and nDNA-encoded (SDHA) OXPHOS subunits. (E) Quantification of the mitonuclear protein imbalance observed in panel D. (F) EC50 determination of doxycycline on mitochondrial respiration in HeLa cells. Oxygen consumption rate (OCR) without treatment is used as 100%. (G) Ten days doxycycline treatment reduces OCR in Hela cells. Conversely, when cells were allowed to recover for the last 3 days in regular medium (+3D recov), this leads to restoration of oxygen consumption.
Figure S2. Geneset enrichment of doxycycline-treated RT112 cells. Related to Figure 3. Mitochondrial genesets modulated by doxycycline treatment in human bladder cancer cell line RT112. The normalized enrichment scores (NES) were used to compare analysis results across gene sets. Significant enriched gene sets after doxycycline treatment were indicated by a false discovery rate (FDR) of less than 25%, which is adjusted for gene set size and multiple hypotheses testing.
Table S1. Mouse primers list. Related to Figure 7.
Table S2. Plant primers list. Related to Figure 5.
Acknowledgments
We thank Prof. Exbrayat (EPHE, Lyon) for helpful discussions. JA is the Nestlé Chair in Energy Metabolism. Work in the JA laboratory is supported by the École Polytechnique Fédérale de Lausanne, the NIH (R01AG043930), the Swiss National Science Foundation (31003A-124713) and Systems X (51RTP0-151019). LM is supported by an FRM fellowship, and RHH by a VENI grant from ZonMw (#91613050).
Footnotes
Supplemental information Supplemental information includes supplemental results, experimental procedures, six figures and three tables, including detailed lifespan statistics.
Author contributions NM, LM, RHH and JA conceived and designed the project. NM performed the cellular and mouse experiments, LM the C. elegans experiments, XW the A. thaliana experiments and Gene Set Enrichment Analysis, and MVF the D. melanogaster experiments. DR, VJ, AM, EGW, PQM and BD performed and analyzed experiments. LM, RHH and JA wrote the manuscript, with contributions from all other authors.
The authors declare no competing financial interests related to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreux PA, Houtkooper RH, Auwerx J. Pharmacological approaches to restore mitochondrial function. Nat Rev Drug Discov. 2013;12:465–483. doi: 10.1038/nrd4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argmann CA, Chambon P, Auwerx J. Mouse phenogenomics: the fast track to “systems metabolism”. Cell Metab. 2005;2:349–360. doi: 10.1016/j.cmet.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Bowman SM, Drzewiecki KE, Mojica ER, Zielinski AM, Siegel A, Aga DS, Berry JO. Toxicity and reductions in intracellular calcium levels following uptake of a tetracycline antibiotic in Arabidopsis. Environmental science & technology. 2011;45:8958–8964. doi: 10.1021/es200863j. [DOI] [PubMed] [Google Scholar]
- Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- Brummett RE, Fox KE. Aminoglycoside-induced hearing loss in humans. Antimicrob Agents Chemother. 1989;33:797–800. doi: 10.1128/aac.33.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker GD, Linnane AW. In vivo differentiation of yeast cytoplasmic and mitochondrial protein synthesis with antibiotics. Biochem Biophys Res Commun. 1966;25:8–13. doi: 10.1016/0006-291x(66)90631-0. [DOI] [PubMed] [Google Scholar]
- Cole M, Elson S, Fullbrook PD. Inhibition of the beta-lactamases of Escherichia coli and Klebsiella aerogenes by semi-synthetic penicillins. Biochem J. 1972;127:295–308. doi: 10.1042/bj1270295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly MA, Clausen PA, Lazar JG. Preparation of RNA from Plant Tissue Using Trizol. Cold Spring Harbor Protocols. 2006;2006 doi: 10.1101/pdb.prot4105. pdb.prot4105. [DOI] [PubMed] [Google Scholar]
- Du X, Wang QR, Chan E, Merchant M, Liu J, French D, Ashkenazi A, Qing J. FGFR3 stimulates stearoyl CoA desaturase 1 activity to promote bladder tumor growth. Cancer Res. 2012;72:5843–5855. doi: 10.1158/0008-5472.CAN-12-1329. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA. Summary report on antimicrobials sold or distributed for use in food-producing animals 2011 [Google Scholar]
- Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao AW, Canto C, Houtkooper RH. Mitochondrial response to nutrient availability and its role in metabolic disease. EMBO Mol Med. 2014;6:580–589. doi: 10.1002/emmm.201303782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvistendahl M. China Takes Aim at Rampant Antibiotic Resistance. Science. 2012;336:795. doi: 10.1126/science.336.6083.795. [DOI] [PubMed] [Google Scholar]
- Jacoby RP, Li L, Huang S, Pong Lee C, Millar AH, Taylor NL. Mitochondrial composition, function and stress response in plants. J Integr Plant Biol. 2012;54:887–906. doi: 10.1111/j.1744-7909.2012.01177.x. [DOI] [PubMed] [Google Scholar]
- Jovaisaite V, Mouchiroud L, Auwerx J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J Exp Biol. 2014;217:137–143. doi: 10.1242/jeb.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Lelliott CJ, Lopez M, Curtis RK, Parker N, Laudes M, Yeo G, Jimenez-Linan M, Grosse J, Saha AK, Wiggins D, et al. Transcript and metabolite analysis of the effects of tamoxifen in rat liver reveals inhibition of fatty acid synthesis in the presence of hepatic steatosis. FASEB J. 2005;19:1108–1119. doi: 10.1096/fj.04-3196com. [DOI] [PubMed] [Google Scholar]
- Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA. Mitochondrial retrograde signaling. Annual review of genetics. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- Lopez M, Lelliott CJ, Tovar S, Kimber W, Gallego R, Virtue S, Blount M, Vazquez MJ, Finer N, Powles TJ, et al. Tamoxifen-induced anorexia is associated with fatty acid synthase inhibition in the ventromedial nucleus of the hypothalamus and accumulation of malonyl-CoA. Diabetes. 2006;55:1327–1336. doi: 10.2337/db05-1356. [DOI] [PubMed] [Google Scholar]
- Mathews S, Reinhold D. Biosolid-borne tetracyclines and sulfonamides in plants. Environ Sci Pollut Res Int. 2013;20:4327–4338. doi: 10.1007/s11356-013-1693-y. [DOI] [PubMed] [Google Scholar]
- Minamino T, Gaussin V, DeMayo FJ, Schneider MD. Inducible gene targeting in postnatal myocardium by cardiac-specific expression of a hormone-activated Cre fusion protein. Circ Res. 2001;88:587–592. doi: 10.1161/01.res.88.6.587. [DOI] [PubMed] [Google Scholar]
- Mingeot-Leclercq MP, Tulkens PM. Aminoglycosides: nephrotoxicity. Antimicrob Agents Chemother. 1999;43:1003–1012. doi: 10.1128/aac.43.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- No D, Yao TP, Evans RM. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramot D, Johnson BE, Berry TL, Jr, Carnell L, Goodman MB. The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PLoS One. 2008;3:e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DM, Subbaiah CC. Mitochondrial retrograde regulation in plants. Mitochondrion. 2007;7:177–194. doi: 10.1016/j.mito.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Ryding AD, Sharp MG, Mullins JJ. Conditional transgenic technologies. J Endocrinol. 2001;171:1–14. doi: 10.1677/joe.0.1710001. [DOI] [PubMed] [Google Scholar]
- Schwarzlander M, Finkemeier I. Mitochondrial energy and redox signaling in plants. Antioxid Redox Signal. 2013;18:2122–2144. doi: 10.1089/ars.2012.5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selimoglu E. Aminoglycoside-induced ototoxicity. Curr Pharm Des. 2007;13:119–126. doi: 10.2174/138161207779313731. [DOI] [PubMed] [Google Scholar]
- Sew YS, Stroher E, Holzmann C, Huang S, Taylor NL, Jordana X, Millar AH. Multiplex micro-respiratory measurements of Arabidopsis tissues. New Phytol. 2013;200:922–932. doi: 10.1111/nph.12394. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utomo AR, Nikitin AY, Lee WH. Temporal, spatial, and cell type-specific control of Cre-mediated DNA recombination in transgenic mice. Nat Biotechnol. 1999;17:1091–1096. doi: 10.1038/15073. [DOI] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Carrie C, Uggalla V, Paynter E, Giraud E, Whelan J. Defining the mitochondrial stress response in Arabidopsis thaliana. Molecular plant. 2009;2:1310–1324. doi: 10.1093/mp/ssp053. [DOI] [PubMed] [Google Scholar]
- Wetsel WC, Mellon PL, Weiner RI, Negro-Vilar A. Metabolism of pro-luteinizing hormone-releasing hormone in immortalized hypothalamic neurons. Endocrinology. 1991;129:1584–1595. doi: 10.1210/endo-129-3-1584. [DOI] [PubMed] [Google Scholar]
- Xie X, Zhou Q, Lin D, Guo J, Bao Y. Toxic effect of tetracycline exposure on growth, antioxidative and genetic indices of wheat (Triticum aestivum L.) Environmental science and pollution research international. 2011;18:566–575. doi: 10.1007/s11356-010-0398-8. [DOI] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Doxycycline effects in cell lines. Related to Figure 1. (A) Doxycycline induces Tet-controlled luciferase expression in the A549 cell line at the concentration of 5–10 μg/ml. (B) Doxycycline dose-dependently causes mitonuclear protein imbalance as evidenced by the altered ratio between mtDNA-encoded (MTCO1 and MTCO2) and nDNA-encoded (SDHA and SDHB) OXPHOS subunits. (C) Quantification of the mitonuclear protein imbalance observed in panel B. (D) Doxycycline treatment in A549 “Tet-on” cells at 1 and 5 μg/ml for 48 hours causes mitonuclear protein imbalance as evidenced by the altered ratio between mtDNA-encoded (MTCO1) and nDNA-encoded (SDHA) OXPHOS subunits. (E) Quantification of the mitonuclear protein imbalance observed in panel D. (F) EC50 determination of doxycycline on mitochondrial respiration in HeLa cells. Oxygen consumption rate (OCR) without treatment is used as 100%. (G) Ten days doxycycline treatment reduces OCR in Hela cells. Conversely, when cells were allowed to recover for the last 3 days in regular medium (+3D recov), this leads to restoration of oxygen consumption.
Figure S2. Geneset enrichment of doxycycline-treated RT112 cells. Related to Figure 3. Mitochondrial genesets modulated by doxycycline treatment in human bladder cancer cell line RT112. The normalized enrichment scores (NES) were used to compare analysis results across gene sets. Significant enriched gene sets after doxycycline treatment were indicated by a false discovery rate (FDR) of less than 25%, which is adjusted for gene set size and multiple hypotheses testing.
Table S1. Mouse primers list. Related to Figure 7.
Table S2. Plant primers list. Related to Figure 5.