SUMMARY
The NAD+-dependent protein deacetylase SIRT1 regulates energy metabolism, responses to stress, and aging by deacetylating many different proteins, including histones and transcription factors. The mechanisms controlling SIRT1 enzymatic activity are complex and incompletely characterized, yet essential for understanding how to develop therapeutics that target SIRT1. Here we demonstrate that the N-terminal domain of SIRT1 (NTERM) can trans-activate deacetylation activity by physically interacting with endogenous SIRT1 and promoting its association with the deacetylation substrate NF-κB p65. Two motifs within the NTERM domain contribute to activation of SIRT1-dependent activities, and expression of one of these motifs in mice is sufficient to lower fasting glucose levels and improve glucose tolerance in a manner similar to overexpression of SIRT1. Our results provide new insights into the regulation of SIRT1 activity and a rationale for pharmacological control of SIRT1-dependent activities.
INTRODUCTION
The NAD+-dependent deacetylase SIRT1 is a modifier of transcriptional outputs that regulate lipid and glucose metabolism, inflammatory signaling, and programmed cell death through its effects on chromatin structure and transcription factor activity (Satoh et al., 2011; Sinclair and Guarente, 2014). Increased SIRT1 activity enhances mitochondrial biogenesis, suppresses inflammation, prevents apoptosis following DNA damage, and generally promotes cell survival in degenerative conditions (Kang et al., 2009; Kim et al., 2007a; Satoh et al., 2011; Yuan et al., 2011). The deacetylase activity of SIRT1 is limited by cellular levels of NAD+, which fluctuate in response to cellular energy requirements and changing rates of NAD+ biosynthesis and consumption (Houtkooper et al., 2010; Revollo et al., 2004). Additionally, several protein regulators of SIRT1 have been identified, including the positive regulators AROS (active regulator of SIRT1) and Necdin (Hasegawa, 2008; Kim et al., 2008) and an inhibitory protein DBC1 (deleted in breast cancer 1) (Kim et al., 2008; Zhao et al., 2008). SIRT1 is phosphorylated at multiple sites, and some of these posttranslational modifications alter interactions with proteins that are deacetylated by SIRT1 (Back et al., 2011; Kang et al., 2009; Nasrin et al., 2009; Nin et al., 2012). Additionally, methylation, sumoylation and nitrosylation of SIRT1 have been reported (Revollo and Li, 2013), although the functional significance and mechanisms of these modifications are not fully understood. Expression of SIRT1 increases following DNA damage (Kang et al., 2009; Wang et al., 2006; 2008; Yuan et al., 2012) and a growing list of transcription factors have been shown to increase or decrease Sirt1 expression (Revollo and Li, 2013). It is well established that SIRT1 is broadly involved with regulating cellular metabolism and stress-dependent signaling. However, the molecular mechanisms that govern substrate selection by SIRT1 (Blander et al., 2005; Pan et al., 2012) and regulate its deacetylation activity (Sinclair and Guarente, 2014) are poorly understood.
Seven mammalian paralogs of the founder protein SIR2 in yeast, SIRT1 through SIRT7, share a conserved catalytic core but have distinctive biological activities that are ascribed to the nonconserved, flanking regions of each enzyme (Satoh et al., 2011). The ESA motif (Essential for SIRT1 Deacetylase Activity; Figure 1A) located C-terminal to the catalytic core of SIRT1 is suggested to physically interact with the core domain to enable deacetylase activity (Kang et al., 2011). SIRT1 also has a large, N-terminal flanking domain that interacts with AROS (Kim et al., 2007b), the active regulator of SIRT1, as well as small molecule activators of deacetylase activity (Hubbard et al., 2013). The numerous sites of posttranslational modification located in the N-terminal domain of SIRT1 and its interactions with protein and small molecule activators of deacetylase activity together suggested this domain functions as a regulator of SIRT1-dependent activities. Here, we report that overexpression of the N-terminal domain of SIRT1 (Figure 1A; NTERM, residues 1–221) activates SIRT1-dependent activities in mammalian cells. This trans-activation of SIRT1 function is mediated by two separate motifs within NTERM that interact with one another and stabilize complexes of SIRT1 with deacetylation substrates. Mice transduced with an adenovirus expressing one of these NTERM motifs show reduced fasting glucose levels and enhanced glucose tolerance, similar to mice overexpressing full-length SIRT1. Our findings suggest a mechanistic framework for examining substrate selection and the regulation of SIRT1 enzymatic activity.
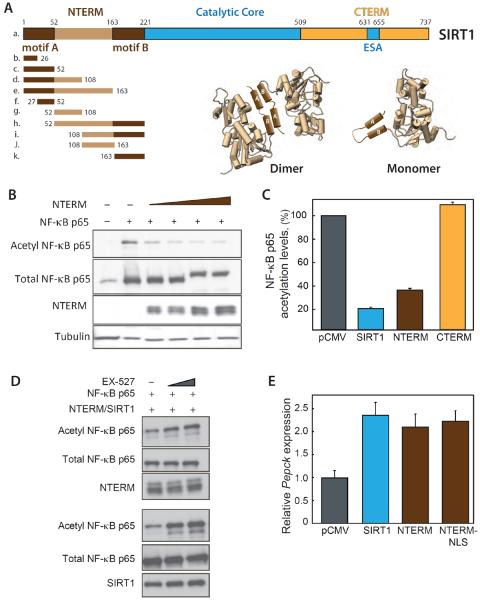
Figure 1. Expression of NTERM activates endogenous SIRT1.
(A) Schematic representation of SIRT1 N-terminal, catalytic core and C-terminal domains. (Bottom left) Peptides spanning the NTERM domain and including the activating motifs A and B (dark brown) were expressed in cells to monitor effects on p65 acetylation (cf. Figure 2). A region of predicted disorder (light brown) separates motifs A and B. (Bottom right) A model depicting the interaction of NTERM motifs A and B, functioning in trans as a dimerization interface (Dimer, left side), or in cis to stabilize the fold of the NTERM domain (Monomer, right side). (B) Acetylation of NF-κB p65 was monitored by Western blotting whole cell extracts from HepG2 cells co-expressing SIRT1, the NTERM domain, or CTERM. Expression of the catalytically inactive NTERM domain lowers acetyl-p65 to the same level as overexpression of enzymatically active SIRT1 (Figure S1). (C) The levels of acetyl-p65 were quantitated for experiments in cells transfected with 1 μg of each plasmid including SIRT1, CTERM, NTERM or empty vector. The means and standard errors are shown for 4 experiments normalized to total NF-κB and expressed as a percentage of the acetylated NF-κB in control cells. (D) NTERM overexpression decreases acetylation of p65 NF-κB levels and this effect is antagonized by the SIRT1 inhibitor, EX-527. HepG2 cells were transfected with NTERM (top 3 panels) or SIRT1 (bottom 3 panels) and grown for 48 hr in the presence of DMSO (2%) or EX-527 (10 μM or 25 μM) prior to Western blotting. NF-κB acetylation increases with increasing concentrations of the SIRT1 inhibitor EX-527 in HepG2 cells expressing NTERM (top) or SIRT1 (bottom), indicating the effect of NTERM overexpression is dependent upon SIRT1 enzymatic activity. (E) Phosphoenolpyruvate carboxykinase (Pepck) expression increases significantly in mouse primary hepatocytes expressing NTERM, similar to the effects of SIRT1 overexpression. The NTERM construct with the native nuclear localization signal (NLS) behaved similarly to the NTERM-NLS construct with an additional NLS added to its N-terminus. Pepck transcripts were quantitated by qRT-PCR as shown for 4 independent experiments.
RESULTS
NTERM expression increases SIRT1-dependent activities
We expressed the NTERM domain of SIRT1 (Figure 1A) in HepG2 cells to examine effects on protein acetylation and gene expression linked to SIRT1 enzymatic activity. Acetylation of Lys310 in the p65 subunit of NF-κB, a well characterized target of SIRT1 deacetylation (Yeung et al., 2004), was monitored in cells overexpressing the NTERM domain. Remarkably, the acetylation of p65(Lys310) decreased in response to increasing NTERM expression (Figures 1B and 1C). The maximal effect of NTERM expression, a 66% decrease in p65 acetylation, is comparable to that resulting from high level overexpression of enzymatically active SIRT1 (Figures 1C and S1A). In contrast to NTERM overexpression, the levels of acetyl-p65 were unaffected by overexpression of the SIRT1 C-terminal domain (Figure 1A; CTERM, residues 509 to 737) that includes the ESA motif required for enzymatic activity (Kang et al., 2011) (Figures 1C and S1B). We additionally examined the acetylation status of the transcription factor p53, another well characterized substrate of SIRT1 (Hasegawa, 2008; Kim et al., 2007b; Vaziri et al., 2001), and found that cellular levels of acetyl-p53 were coordinately decreased with increasing NTERM expression (Figures S1C and S1D).
The cellular concentration of SIRT1 protein was unaffected by overexpression of NTERM (Figure S1E), suggesting that NTERM functions by increasing the enzymatic activity of endogenous SIRT1. Consistent with this mechanism, the small molecule SIRT1 inhibitor EX-527 antagonized the effect of NTERM and returned p65 acetylation to control levels (Figure 1D). The expression of Pepck (phosphoenolpyruvate carboxykinase) mRNA was increased by NTERM overexpression in primary hepatocytes (Figure 1E), another hallmark of increased SIRT1 activity. Together, these molecular readouts indicate a widespread activation of SIRT1-dependent processes (Grimm et al., 2011; Rodgers et al., 2005) concordant with NTERM overexpression.
The recombinant NTERM domain interacts with SIRT1
To examine the mechanism of this change in SIRT1-dependent processes, we next measured the physical interactions of SIRT1 with NTERM and deacetylation substrates. Myc-tagged SIRT1 was efficiently co-immunoprecipitated by the HA-tagged NTERM domain expressed in human HepG2 cells (Figure 2A), and in the reciprocal pull-down HA-tagged SIRT1 was co-immunoprecipitated with Myc-tagged NTERM domain (not shown). Increasing SIRT1 expression resulted in more SIRT1 in complex with HA-NTERM. Deletion of the N-terminal domain of SIRT1 (SIRT1(Δ221) protein) eliminated the physical interaction with recombinant NTERM domain (Figure S2A), suggesting that the NTERM domains of both proteins either directly bind to one another or are incorporated into a larger complex. Indeed, two different NTERM domains tagged with Myc or HA could be co-immunoprecipitated from HEK293 cell lysates (Figure S2B), whereas the recombinant C-terminal domain of SIRT1 (SIRT1(509–737) protein, Figure S2A) did not interact with NTERM. Full-length SIRT1 proteins with Myc- and FLAG-tags were also efficiently co-immunoprecipitated (not shown).
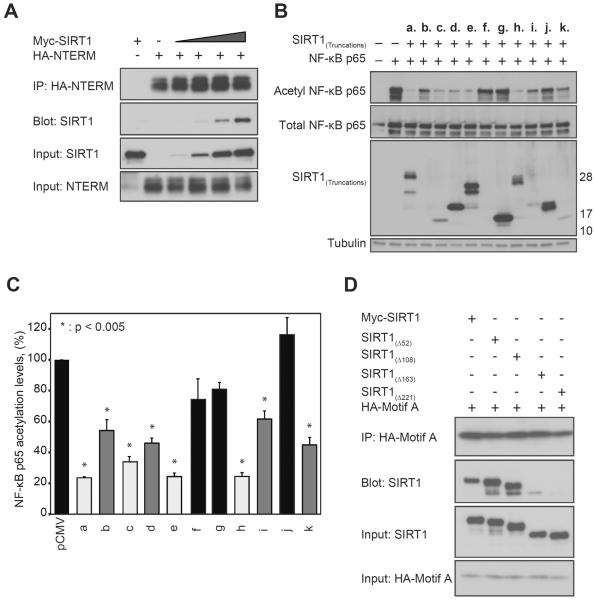
Figure 2. Two motifs within NTERM activate SIRT1 and physically interact.
(A) SIRT1 is efficiently co-immunoprecipitated with HA-tagged NTERM in HepG2 cells, and more SIRT1 is pulled down in the complex with HA-NTERM as SIRT1 expression is increased. (B) Acetylation levels of NF-κB were monitored in HepG2 cells expressing the p65 subunit of NF-κB together with the NTERM construct a. (residues 1–221) or one of the indicated constructs (b. – k.) shown in Figure 1A. The positions of molecular weight markers are shown at right. (C) Two independent motifs within NTERM promote deacetylation of NF-κB by endogenous SIRT1. Strongly activating peptides (light grey; peptides c., e. and h.) decreased p65 acetylation to 23–34% of the vector control. NTERM peptides b., d., i. and k. (medium grey) showed intermediate levels of activity (p65 acetylation levels 45–62% of the control values). Peptides f., g. and j. (black) had no significant effect on p65 acetylation levels relative to the pCMV control. The means and standard errors are shown for 4 independent experiments including the representative experiment shown in panel B. The results are normalized to total NF-κB and expressed as a percentage of the acetylated NF-κB in control cells. (D) NTERM motif A physically interacts with motif B. HEK293 cells were transfected with 1 μg of DNA encoding SIRT1(Δ52), SIRT1(Δ108), SIRT1(Δ163), or SIRT1(Δ221) together with 2 μg of DNA encoding HA-tagged motif A (residues 1–52). The motif A peptide co-immunoprecipitated proteins containing motif B (SIRT1(Δ52) SIRT1(Δ108), and SIRT1(Δ163), but not the SIRT1(Δ221) protein lacking the NTERM domain.
Two NTERM motifs independently stimulate SIRT1 activities
The amino acid sequence of NTERM consists of two regions of predicted α-helical character that are separated by low complexity sequence indicative of disorder (Figure 1A). Intrinsically, disordered regions of proteins are frequently involved in protein-protein interactions (Mészáros et al., 2011). Deletion mutagenesis identified two regions within NTERM that could trans-activate SIRT1 (Figure 2). A series of Myc-tagged peptides was expressed in HepG2 cells (Figure 1A) spanning NTERM residues 1–26 (b.), 1–52 (c.), 1–108 (d.), 1–163 (e.), 27–52 (f.), 52–108 (g.), 52–221 (h.), 108–221 (i.), 108–163 (j.), and 163–221 (k.). Most of these constructs were abundantly expressed, although we could not verify the expression of short peptides (b.) and (f.) by western blot analysis (Figure 2B). A pattern emerged in which peptides from two different regions of NTERM significantly decreased the acetylation of p65 NFκB (Figures 2B and 2C). Strongly activating peptides (c., e. and h.) decreased p65 acetylation to 23–34% of that seen in cells transfected with vector only. Peptides b., d., i. and k. showed intermediate levels of activity, decreasing p65 acetylation levels to 45–62% of control values, whereas constructs f., g., and j. had no significant effect on p65 acetylation. The results suggest that NTERM residues 1–52 (motif A; Figure 1A) and 163–221 (motif B) comprise independent motifs that when overexpressed can trans-activate SIRT1-dependent cellular activities (Figure 2C). Notably, these motifs correspond to the regions of NTERM that are predicted to have α-helical structure. Motif B overlaps with a region that is essential for activation of SIRT1 enzymatic activity by small molecule agonists (Hubbard et al., 2013).
We considered the possibility that helical motifs A and B might interact with one another either in trans or within the native fold of the NTERM domain (Figure 1A). The HA-tagged motif A peptide was co-expressed with a series of Myc-tagged, N-terminally truncated SIRT1 proteins and then immunoprecipitated with an anti-HA antibody. Motif A efficiently co-immunoprecipitated Myc-tagged SIRT1, SIRT1(Δ52), and SIRT1(Δ108) proteins, and exhibited a weak interaction with SIRT1(Δ163) (Figure 2D). However, the motif A peptide failed to interact with the SIRT1(Δ221) protein lacking motif B (Figure 2D). The motif B peptide (k.) bound nonspecifically to the beads and could not be tested for interactions by itself, although expression of this peptide consistently decreased acetylation levels of NF-κB (Figure 2B).
It was reported that recombinant SIRT1 dimerizes, and that dimerization is abolished by the phosphomimetic mutation Thr522Glu (Guo et al., 2012). However, we and others have examined the oligomeric state of recombinant SIRT1 protein purified from E. coli and find no evidence of dimer formation (Lakshminarasimhan et al., 2013b) (Figure S3). Sedimentation equilibrium experiments conducted over a range of SIRT1 concentrations (5–50 μM) and g-forces indicated that recombinant SIRT1 is monomeric with a buoyant mass of 90 kDa, consistent with the calculated mass of 86 kDa (Figure S3A). The amino acid sequence of the NTERM domain is predicted to be disordered (Dunker et al., 2008), and the recombinant SIRT1 protein has an anomalously small sedimentation coefficient (3.82 S) and large frictional coefficient that are indicative of an elongated conformation (Figure S3C) (Lakshminarasimhan et al. 2013). Consistent with the previous report (Guo et al., 2012), dynamic light scattering shows an anomalously large Mr value for SIRT1 (173 kDa, versus a calculated mass of 86 kDa) (Figure S3B). In contrast, a truncated SIRT1(Δ221) protein lacking the NTERM domain has an apparent mass (Mr = 73 kDa) in agreement with the expected value for a compact globular protein. These data indicate the slow diffusion and unusually large hydrodynamic radius of SIRT1 are properties of the NTERM domain, which may be intrinsically disordered. The NTERM region also shows high rates of hydrogen-deuterium exchange, a further indication of an unfolded or loosely structured domain (Hubbard et al., 2013). Disorder is frequently associated with protein interaction sites (Dunker et al. 2008), and correspondingly, the NTERM domain contributes to interactions with substrates for deacetylation and with protein and small molecule activators of SIRT1 activity (Kim et al., 2007b; Sinclair and Guarente, 2014).
Overexpression of the NTERM constructs described here allows us to detect interactions between motifs A and B in trans (Figures 2B and S2A) and the self-association of NTERM domains (Figure S2B). Co-immunoprecipitation of NTERM domains may require an elevated protein concentration, posttranslational modifications, and/or partner proteins that are absent from in vitro studies of recombinant SIRT1 protein. The results with purified, recombinant SIRT1 protein do not support a dimerization model. Instead, motifs A and B may pack together to stabilize the native fold of SIRT1 monomers (Figure 1A).
The NTERM domain promotes the interaction of SIRT1 with substrates
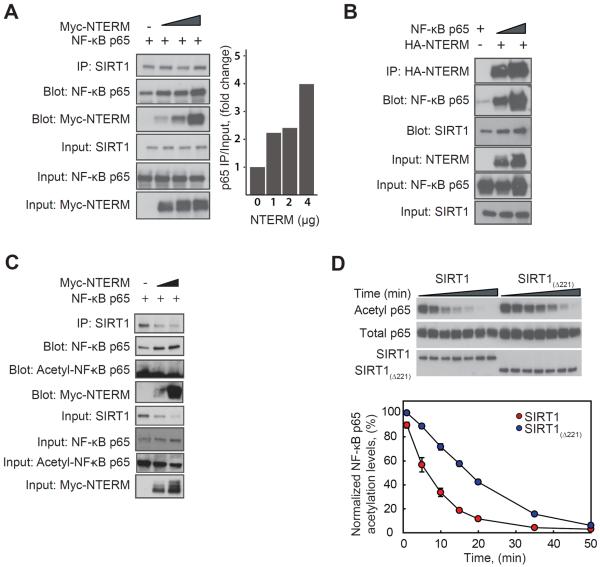
NTERM overexpression causes a decline in acetyl-p65 NF-κB levels (Figure 1B), suggestive of enhanced SIRT1 activity, perhaps from enhanced enzyme-substrate interactions. To examine this hypothesis, endogenous SIRT1 was pulled down from extracts of HEK293 cells overexpressing the deacetylation substrate NF-κB p65 that were co-transfected with increasing amounts of NTERM. SIRT1 was immunoprecipitated with an antibody directed at its C-terminus, and the resulting precipitates contained a ternary complex of SIRT1, p65, and NTERM. The amount of p65 in the complex increased concurrently with increased expression of NTERM (Figure 3A), suggesting that the NTERM domain does indeed promote the association of SIRT1 with the NF-κB p65 substrate. Reciprocal pull-downs confirmed the formation of a ternary p65/SIRT1/NTERM complex using antibodies directed at NTERM (Figure 3B) or p65 (not shown). To further probe the components of the SIRT1/NTERM/p65 complex, we attempted to measure the amounts of acetylated and deacetylated p65 in the complex. To our surprise, p65 in the complex was completely deacetylated, suggesting that once recruited into the complex, p65 is efficiently deacetylated and remains complexed to SIRT1 (Figure 3C). Consistent with these findings, the comparison of the deacetylation activities of full-length SIRT1 and SIRT1(Δ221) on an in vitro acetylated p65 (1–325) substrate shows that the N-terminal domain of SIRT1 significantly enhances the rate of NF-κB p65 deacetylation in vitro (Figure 3D). Because both SIRT1 and SIRT1(Δ221) exhibit similar kinetics towards a peptide substrate (not shown), these results suggest that the NTERM domain likely enhances deacetylation activity towards protein substrates by promoting the enzyme-substrate interaction.
Figure 3. NTERM enhances the interaction of SIRT1 with its substrate NF-κB p65.
(A) Overexpression of NTERM in HEK293 cells increases the interaction between endogenous SIRT1 and its substrate, the p65 subunit of NF-κB. HEK293 cells were co-transfected with p65 together with increasing concentrations of NTERM (1, 2, or 4 μg of plasmid DNA), and endogenous SIRT1 protein was immunoprecipitated with an antibody directed at a C-terminal epitope of human SIRT1. Increasing expression of HA-NTERM resulted in greater association of SIRT1 with p65 and with NTERM in a ternary complex (right panel). (B) In a reciprocal pull-down experiment, HA-NTERM was coimmunoprecipitated in a complex with SIRT1 and p65. (C) p65 in the complex with SIRT1 and NTERM is fully deacetylated at Lys310. Immunoprecipitation was performed as in panel A and blots were probed with antibodies for total p65 and acetyl-(Lys310)-p65. (D) In vitro deacetylation of the purified p65 protein is enhanced by the presence of the NTERM domain. Deacetylation of lysine 310 was monitored by Western blotting for p65 acetylated in vitro by pretreatment with the histone acetyltransferase domain of p300. The p65 protein substrate is deacetylated at a significantly faster rate by SIRT1 than by SIRT1(Δ221) lacking the NTERM domain. The means and standard errors of 3 experiments are shown, after normalization to the activity of SIRT1.
The co-immunoprecipitation of the NTERM domain with the enzyme-substrate complex is consistent with a direct effect of NTERM on the substrate binding activity of SIRT1. Alternatively, the overexpressed NTERM domain could sequester a negative regulator of SIRT1 such as the DBC1 protein (Deleted in Breast Cancer 1), a competitive inhibitor of SIRT1 substrate binding activity (Kim et al., 2008; Zhao et al., 2008). In this scenario, a physical interaction between NTERM and DBC1 would antagonize DBC1-dependent inhibition of SIRT1. We examined this hypothesis by co-expressing FLAG-tagged DBC1 and HA-tagged NTERM proteins in HEK293 cells and pulling down the NTERM domain with an anti-HA antibody. DBC1 was efficiently co-immunoprecipitated by NTERM (Figure S4A), and the reciprocal immunoprecipitation confirmed this interaction (Figure S4B). The DBC1 binding site was previously localized to the catalytic core of SIRT1 (Kim et al., 2008; Zhao et al., 2008). However, protein constructs described as the catalytic core in these studies included all (Kim et al., 2008) or part of (Zhao et al., 2008) the region we have defined as motif B of the NTERM domain. The NTERM domain of SIRT1 is sufficient for interaction with DBC1 (Figure S4B), and a SIRT1 deletion mutant lacking the NTERM domain does not interact with DBC1 (Figure S4C). Constructs that include motif B (residues 163–221; Figure 1A) also bind to DBC1 (Figure S4D), whereas the motif A peptide (residues 1–52) does not bind (Figure S4D).
Although overexpression of DBC1 caused a modest increase in acetyl-p53 levels, this effect was overcome by low-level expression of the NTERM domain (Figure S4E). Increasing the level of DBC1 expression did not antagonize the effect of NTERM on acetyl-p53 levels (Figure S4E). The interaction of DBC1 with SIRT1 is regulated by phosphorylation of both proteins (Nin et al., 2012; Yuan et al., 2012), and DBC1 may not be the primary determinant of SIRT1 activity in HepG2 cells under our experimental conditions. Other investigators have similarly reported that DBC1 does not consistently function as an inhibitor of SIRT1 activity in every cell line tested (Nam, 2012). The co-immunoprecipitation of NTERM and SIRT1 (Figure 2A) is consistent with a direct effect of the NTERM domain on the substrate binding and enzymatic activities of SIRT1.
The motif A peptide improves glucose metabolism in mice
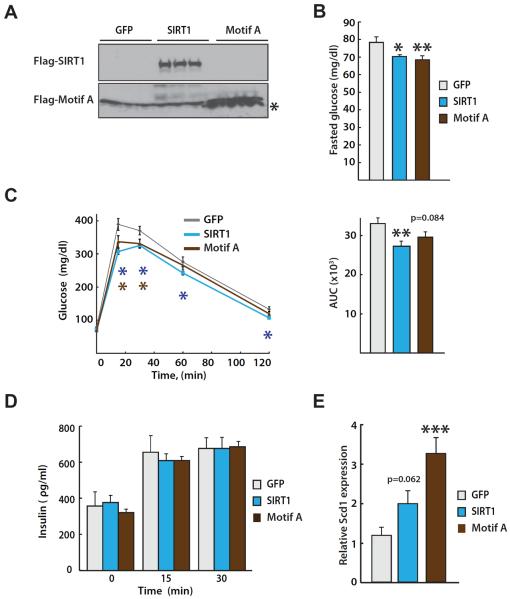
To examine the metabolic effects of NTERM expression in vivo, we constructed recombinant adenoviruses expressing SIRT1 and the motif A peptide (residues 1–52). Despite repeated attempts, we were unable to recover virus from an expression construct encoding the NTERM domain. Male C57BL/6 mice at approximately two months of age were infected with the other two recombinant viruses by tail vein injection, and overexpression of SIRT1 and the motif A peptide were confirmed four days post-infection by western blot analysis (Figure 4A). In agreement with previous reports (Rodgers and Puigserver, 2007; Rodgers et al., 2005), animals overexpressing SIRT1 had lower fasting glucose levels (Figure 4B) and significantly improved glucose tolerance measured by an intraperitoneal glucose tolerance test (IPGTT; Figure 4C). Remarkably, overexpression of the motif A peptide had a similar effect of reduced fasting glucose levels and improved glucose tolerance in comparison to control animals overexpressing the green fluorescent protein (Figures 4B and 4C). Insulin levels measured during the IPGTT were similar for the control animals and those expressing SIRT1 or the motif A peptide (Figure 4D). These findings suggest that expression of the motif A peptide likely improves insulin sensitivity, an effect similar to overexpressing enzymatically active SIRT1 (Rodgers and Puigserver, 2007). Enhanced SIRT1 activity in the liver of the transduced animals was also evidenced by increased expression of stearoyl-CoA desaturase 1 (Scd1) in mice overexpressing the motif A peptide (Figure 4E), an effect comparable to overexpression of SIRT1 (Ramsey et al., 2008).
Figure 4. Expression of NTERM improves glucose metabolism in vivo.
(A) Representative immunoblot of Flag-tagged SIRT1 and Flag-tagged motif A (residues 1-52) expression from 3 mouse livers in each group. (B) Fasted glucose levels were measured in mice 4 days post-infection with adenoviruses expressing GFP, SIRT1, or NTERM motif A and after fasting animals for 16 hr. Expression of SIRT1 or the motif A peptide caused a significant reduction in fasting glucose. (C) An intraperitoneal glucose tolerance test (IPGTT) was performed as previously described (Ramsey et al., 2008) in mice four days post-infection with the adenoviruses (left) and the areas under the curves (AUC) were calculated for each group (right). The AUC values are presented as the mean and standard error of each group. (D) Plasma insulin levels measured during the IPGTT were not significantly different for the 3 groups of animals, suggesting that the motif A peptide increases insulin sensitivity in mice but does not alter insulin secretion. (E) Mouse liver mRNA was isolated and expression levels of stearoyl-CoA desaturase 1 (Scd1) were evaluated by qRT-PCR. The means and standard errors are shown (n= 9–11). The three groups were analyzed by a one-way ANOVA and Fisher's PLSD post hoc test; *p<0.05, **p<0.01, ***p<0.001.
DISCUSSION
SIRT1 broadly regulates cellular metabolism, growth, and responses to stress by deacetylating transcription factors and histones, yet we have limited knowledge of mechanisms governing substrate selection by SIRT1 and the control of deacetylation activity (Satoh et al., 2011; Sinclair and Guarente, 2014). Acetyl-lysine containing peptides are promiscuously deacetylated in vitro by SIRT1 with little sequence selectivity (Blander et al., 2005; Lakshminarasimhan et al., 2013a), suggesting that additional binding interactions with protein substrates contribute to the biological specificity of deacetylation in a cellular context. Here we identify protein interactions mediated by the N-terminal domain of SIRT1 that increase SIRT1-dependent activities, decreasing acetylation of the NF-κB p65 and p53 proteins, and increasing expression of Pepck (Figure 1E) and Scd1 (Figure 4E). The physical interaction of NTERM with SIRT1 promotes the recruitment of a protein substrate NF-κB p65 into the enzyme-substrate complex, suggesting a mechanism for enhancing deacetylation of this protein. Additionally, the NTERM domain interacts with DBC1, a negative regulator of SIRT1 activity (Kang et al., 2011; Kim et al., 2008; Zhao et al., 2008). The binding of DBC1 to the NTERM domain may additionally contribute to enhanced SIRT1 deacetylase activity.
Several models can be envisioned for the regulation of SIRT1 enzymatic activity by the NTERM domain. The self-association of overexpressed NTERM domains through an interaction of motifs A and B is consistent with SIRT1 dimerization (Guo et al., 2012), although recombinant SIRT1 proteins purified from E. coli or Sf9 insect cells are monomeric (our results, Lakshminarasimhan et al., 2013b). SIRT1 oligomerization may require additional posttranslational modifications, or partner proteins that assemble into a multiprotein complex. Alternatively, the NTERM domain may function in cis to enhance the substrate binding and deacetylase activities of the SIRT1 catalytic core. The interacting motifs A and B could pack together in the folded conformation of the NTERM domain, an interaction that could be stabilized in complex with a SIRT1 substrate. In addition, sites of phosphorylation within activating motifs A and B (Back et al., 2011; Kang et al., 2009; Nasrin et al., 2009) may affect the conformation of NTERM or its interactions with protein partners. It is also notable that motif B corresponds to a region of SIRT1 that is essential for activation of deacetylase activity by small molecules (Hubbard et al., 2013; Sinclair and Guarente, 2014).
Viral transduction of a 52-residue peptide corresponding to motif A of the NTERM domain improved glucose metabolism in mice, coincident with overexpression of the peptide in the liver. The precise mechanism for these in vivo effects is still being investigated but may be related to the down regulation of NF-κB signaling by enhancing SIRT1 activity. So far, we have not been able to reconstitute the activation event of SIRT1 enzymatic activity in vitro upon addition of the purified recombinant NTERM domain, or the interaction of NTERM with the purified p65 Rel homology domain (residues 1–325). These results are consistent with the proposal that SIRT1 may be a part of a larger complex in which additional factors physically interact with NTERM, SIRT1 and substrate proteins in cells. The active regulator of SIRT1 (AROS) enhances the SIRT1-dependent deacetylation of p53 by binding to the N-terminal domain of SIRT1 (Kim et al., 2007b) in a region (residues 114–217) that includes the activating motif B. It has been suggested that AROS may induce a conformational change of SIRT1 to stimulate deacetylase activity. One could envision positive regulators of SIRT1 activity stabilizing the interaction of motifs A and B to promote interactions with substrates, whereas DBC1 and negative regulators might stabilize an inactive conformation of the NTERM domain. To address such possibilities, we are currently attempting to identify additional regulators of SIRT1 activity that could potentially affect the conformation and/or the function of the NTERM domain. Structural studies of SIRT1 in complex with those regulatory proteins will also be important for understanding the mechanisms of regulating its enzymatic activity by proteins and by small molecules.
In conclusion, we have identified functional interactions of the SIRT1 NTERM domain, and specifically the activating motifs A and B that enhance SIRT1-dependent activities in cells and in mice. Our results provide new insights into possible interventions that modulate SIRT1 activity through the binding interactions of the NTERM domain.
EXPERIMENTAL PROCEDURES
Acetylation levels of SIRT1 substrates
HepG2 cells were transfected with expression plasmids constructed from pCMV-HA and pCMV-Myc vectors (Clontech) and encoding NF-κB p65 (Chen Lf et al., 2001) or p53, and SIRT1, NTERM, SIRT1(Δ221), or SIRT1(509–737) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. 24 hr later, the media was replaced with DMEM containing deacetylase inhibitors (10 μM trichostatin A, 10 mM nicotinamide) and cells were incubated overnight. Cells were treated for 48 hr with the SIRT1 inhibitor EX-527 at a concentration of 10 μM or 25 μM. For immunoprecipitation, cells were harvested in cold PBS including deacetylase inhibitors, pelleted at 1000g, and lysed in IP buffer as previously described (Grimm et al., 2011). After normalizing the protein concentrations, the samples were analyzed by SDS-PAGE and Western blotting as described below. The band intensities were quantitated using Image J (NIH) and plotted as the normalized mean and standard error.
Co-immunoprecipitation and Western blotting
Cells were transfected with the following constructs as indicated: Myc-SIRT1, HA- SIRT1(1–221), HA-SIRT1(1–52), Myc- SIRT1(Δ221), Myc- SIRT1(509–737), Myc-SIRT1(Δ52), Myc- SIRT1(Δ108), Myc-SIRT1(Δ163), Myc- SIRT1(1–221), or DBC1 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Immunoprecipitation was performed as previously described (Grimm et al., 2011). In brief, cell extracts were pre-cleared with a 50% slurry of protein A agarose beads (Millipore) in IP buffer for 2 hr at 4 °C. For Flag, HA, or Myc IP, 30 μl anti-Flag-conjugated agarose beads (Sigma), anti-HA-conjugated agarose beads (Sigma), or anti-Myc-conjugated agarose beads (Clontech) were added, and the samples were rocked overnight at 4 °C. Endogenous SIRT1 was immunoprecipitated by incubating cell extracts overnight with anti-SIRT1 (Santa Cruz B-7, C-terminal antibody) followed by protein G beads (Sigma) for 2–3 hr at 4 °C. Western blotting was performed as previously described (Revollo et al., 2004). To visualize SIRT1 peptides, the membranes were incubated with 10% glutaraldehyde (Sigma) in PBS for 30 min immediately before blocking. Primary antibodies included α-SIRT1 (1:10,000, Upstate Biotechnology), α-SIRT1 (1:5,000, Sigma AS-16), α-hSIRT1 (1:1,000, Abcam 19A7AB4), α-hSIRT1 (1:1,000, Santa Cruz, B-7), α-Myc (1:500, Clontech), α-NF-κB (1:1000, Santa Cruz), α-tubulin (1:1000, Santa Cruz), α-acetyl-NF-κB (1:1000, Cell Signaling), α-HA (1:1000, Covance), α-Flag (1:1000, Sigma) and α-acetyl-p53 Lys-379 (1:1000, Cell Signaling). Secondary antibodies included the HRP-conjugated α-rabbit IgG or α-mouse IgG (1:10,000, GE Healthcare) and α-rabbit IgG (1:1,000, Cell Signaling).
Animal experimentation
All animal studies were approved by the Washington University Animal Studies Committee and performed in accordance with NIH guidelines. Recombinant adenoviruses expressing GFP, SIRT1 or motif A (residues 1–52) proteins were prepared following the manufacturer's protocol (Invitrogen AddEasy) and used to infect 8–10 week old male C57BL/6J mice (Taconic Laboratories) by tail vein injection of 5×109 virus particles. After 4 days, intraperitoneal glucose tolerance test (IPGTT) was performed as previously described (Ramsey et al., 2008). Significant differences between groups of animals were determined using a one-way ANOVA with the Fisher's PLSD test. For the detection of SIRT1 and the motif A peptide, mice were sacrificed by carbon dioxide asphyxiation after IPGTT experiments. Livers were immediately collected, minced, and flash frozen in liquid nitrogen. Liver samples were homogenized in 2X Laemmli's sample buffer and boiled for 5 minutes. Samples were then centrifuged to remove debris and protein concentration was measured by the Bradford assay.
Analytical ultracentrifugation
Experiments were performed at 25 °C using a Beckman Instruments Optima XL-A analytical ultracentrifuge. For sedimentation velocity (SV) experiments, sample cells were loaded with 380 μL of SIRT1 or 400 μL of appropriate reference buffer then centrifuged at 40,000 rpm. Data were collected in continuous mode at a wavelength of 280 nm and fitted to a continuous sedimentation coefficient [c(s)] distribution model using the program SEDFIT (NIH). The sedimentation coefficient for SIRT1 was determined at a protein concentration of 40 μM in a buffer containing 50 mM NaCl, 50 mM HEPES pH 8.0, 2 mM MgCl2, 1 mM TCEP and 0.1 mM EDTA. Sedimentation coefficients were determined in the presence of one or both substrates, NAD+ (1 mM) and/or a thio-acetyl lysine peptide (400 μM) corresponding to lysine 382 of the p53 protein, by monitoring absorbance at 295 nm. For sedimentation equilibrium (SE) experiments an Epon charcoal-filled six-sector centerpiece was used allowing three concentrations to be run simultaneously (5 μM, 10 μM, 50 μM). Cells were loaded with 120 μL of protein sample and 130 μL of appropriate reference buffer. Optical absorbance values were monitored at 280 nm and runs were performed at speeds of 12,000, 15,000, 18,000 and 22,000 rpm until equilibrium was achieved (~24 hr determined from overlap of scans at 4 hr separation). Sedimentation equilibrium data were processed and analyzed using SEDFIT/SEDPHAT (Lebowitz et al., 2002), and values for partial specific volume and solvent density were calculated using SEDNTERP.
Supplementary Material
ACKNOWLEDGMENTS
We thank Roberto Galletto, James Doom, and Valentina Ghisays for advice and technical assistance, Darin Jones for the thioacetyl-lysine peptide, and members of the Ellenberger, Imai, and Janetka laboratories for valuable discussions. This work was supported by grants from the National Institutes of Health (R01 GM052504 to T.E.; R01 AG024150 to S.I.; R01 CA154697 to D.T.C.) and the Ellison Medical Foundation to S. I.
Footnotes
AUTHOR CONTRIBUTIONS FG, HK, SI and TE designed experiments, and FG, CB, SY, HK, KM, EK, ID, and TE conducted experiments. SI and DC also designed and supervised experiments. FG, SI, and TE wrote the manuscript.
SUPPLEMENTAL INFORMATION Supplemental Information includes four figures, Supplemental Experimental Procedures, and Supplemental References.
REFERENCES
- Back JH, Rezvani HR, Zhu Y, Guyonnet-Duperat V, Athar M, Ratner D, Kim AL. Cancer cell survival following DNA damage-mediated premature senescence is regulated by mammalian target of rapamycin (mTOR)-dependent inhibition of sirtuin. J Biol Chem. 2011;286:19100–19108. doi: 10.1074/jbc.M111.240598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Olejnik J, Krzymanska-Olejnik E, McDonagh T, Haigis M, Yaffe MB, Guarente L. SIRT1 shows no substrate specificity in vitro. J Biol Chem. 2005;280:9780–9785. doi: 10.1074/jbc.M414080200. [DOI] [PubMed] [Google Scholar]
- Chen Lf, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Grimm AA, Brace CS, Wang T, Stormo GD, Imai SI. A nutrient-sensitive interaction between Sirt1 and HNF-1α regulates Crp expression. Aging Cell. 2011;10:305–317. doi: 10.1111/j.1474-9726.2010.00667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Kesimer M, Tolun G, Zheng X, Xu Q, Lu J, Sheehan JK, Griffith JD, Li X. The NAD(+)-dependent protein deacetylase activity of SIRT1 is regulated by its oligomeric status. Sci Rep. 2012;2:640. doi: 10.1038/srep00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K. Necdin Regulates p53 Acetylation via Sirtuin1 to Modulate DNA Damage Response in Cortical Neurons. The Journal of Neuroscience. 2008 doi: 10.1523/JNEUROSCI.3052-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. The secret life of NAD+: An old metabolite controlling new metabolic signaling pathways. Endocr. Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard BP, Gomes AP, Dai H, Li J, Case AW, Considine T, Riera TV, Lee JE, E SY, Lamming DW, et al. Evidence for a Common Mechanism of SIRT1 Regulation by Allosteric Activators. Science. 2013;339:1216–1219. doi: 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Jung J-W, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. 2009;4:e6611. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Suh J-Y, Jung Y-S, Jung J-W, Kim MK, Chung JH. Peptide switch is essential for sirt1 deacetylase activity. Mol Cell. 2011;44:203–213. doi: 10.1016/j.molcel.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. Embo J. 2007a;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E-J, Kho J-H, Kang M-R, Um S-J. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007b;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Kim J-E, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- Lakshminarasimhan M, Rauh D, Schutkowski M, Steegborn C. Sirt1 activation by resveratrol is substrate sequence-selective. Aging (Albany NY) 2013a;5:151–154. doi: 10.18632/aging.100542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshminarasimhan M, Curth U, Moniot S, Mosalaganti S, Raunser S, Steegborn C. Molecular architecture of the human protein deacetylase Sirt1 and its regulation by AROS and resveratrol. Biosci. Rep. 2013b;33:395–404. doi: 10.1042/BSR20120121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mészáros B, Simon I, Dosztányi Z. The expanding view of protein–protein interactions: complexes involving intrinsically disordered proteins. Phys. Biol. 2011;8:035003. doi: 10.1088/1478-3975/8/3/035003. [DOI] [PubMed] [Google Scholar]
- Nam S. DBC1 does not function as a negative regulator of SIRT1 in liver cancer. Oncol Lett. 2012;4:873–877. doi: 10.3892/ol.2012.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, Bordone L. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS ONE. 2009;4:e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nin V, Escande C, Chini CC, Giri S, Camacho-Pereira J, Matalonga J, Lou Z, Chini EN. Role of deleted in breast cancer 1 (DBC1) protein in SIRT1 deacetylase activation induced by protein kinase A and AMP-activated protein kinase. Journal of Biological Chemistry. 2012;287:23489–23501. doi: 10.1074/jbc.M112.365874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M, Yuan H, Brent M, Ding EC, Marmorstein R. SIRT1 contains N- and C-terminal regions that potentiate deacetylase activity. Journal of Biological Chemistry. 2012;287:2468–2476. doi: 10.1074/jbc.M111.285031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, K M, Mills KF, Satoh A, Imai S-I. Age-associated loss of Sirt1-mediated enhancement of glucose-stimulated insulin secretion in beta cell-specific Sirt1-overexpressing (BESTO) mice. Aging Cell. 2008;7:78–88. doi: 10.1111/j.1474-9726.2007.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo JR, Li X. The ways and means that fine tune Sirt1 activity. Trends Biochem Sci. 2013;38:160–167. doi: 10.1016/j.tibs.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA, Imai S-I. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci USA. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nat Cell Biol. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Satoh A, Stein L, Imai S. The role of mammalian sirtuins in the regulation of metabolism, aging, and longevity. Handb Exp Pharmacol. 2011;206:125–162. doi: 10.1007/978-3-642-21631-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Small-molecule allosteric activators of sirtuins. Annu Rev Pharmacol Toxicol. 2014;54:363–380. doi: 10.1146/annurev-pharmtox-010611-134657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- Wang R-H, Sengupta K, Li C, Kim H-S, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. Embo J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F, Xie Q, Wu J, Bai Y, Mao B, Dong Y, Bi W, Ji G, Tao W, Wang Y, et al. MST1 promotes apoptosis through regulating Sirt1-dependent p53 deacetylation. J Biol Chem. 2011;286:6940–6945. doi: 10.1074/jbc.M110.182543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Luo K, Liu T, Lou Z. Regulation of SIRT1 activity by genotoxic stress. Genes Dev. 2012;26:791–796. doi: 10.1101/gad.188482.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Kruse J-P, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.