Abstract
Light-responsive proteins have been used in the field of optogenetics to control cellular functions. However, surprisingly, analogous approaches to regulate and alter the functions of RNA molecules by light remain underdeveloped. RNA aptamers and RNA devices can perform diverse intracellular functions and are important tools in synthetic biology. This report explores the challenges of and potential strategies for engineering light regulation into functional RNAs in cells. We discuss approaches for using existing light-regulated proteins and small molecules to control RNA function in living cells. Additionally, applications of light-regulated RNAs for synthetic biology and for studying functions of endogenously expressed RNAs are discussed.
Keywords: optogenetics, RNA aptamer, photoswitching, genetic encoding, synthetic biology
Introduction: applications of optogenetically controlled RNA for exploring RNA biology and controlling cellular processes
Light is an important tool that can be used to manipulate the function of proteins and thereby control cellular function and intracellular signaling pathways. The use of light to control heterologously expressed proteins that are designed to be light-responsive is generally termed optogenetics and implies the control of a protein with both spatial and temporal precision. For example, in the case of the channelrhodopsins and related light-activated proteins, application of light results in selective opening or closing of these channels, thereby altering neuronal excitability and function.1-3 This technology has provided considerable insight into the functions of specific classes of neurons that are engineered to specifically express these channels.
On the other hand, approaches to regulate and alter the functions of RNA molecules by light remain underdeveloped. The lack of optogenetic techniques for controlling RNA is surprising considering the highly useful nature of synthetic RNA molecules for influencing cellular function. For example, RNA aptamers can be synthesized that inhibit diverse classes of intracellular proteins.4, 5 Indeed, many RNA aptamers have been described that inhibit proteins that are otherwise not able to inhibited by small molecules. Thus, RNA aptamers provide the only tool for inhibiting the function of these proteins. Additionally, although developing a small-molecule inhibitor that targets a specific protein can be a difficult process, creating a highly selective protein-inhibiting RNA aptamer is relatively straightforward and rapid. RNAs can have other useful functions as well. For example, certain RNAs show enzymatic activity and are termed ribozymes.6 Most notably, these ribozymes perform diverse reactions, such as oligonucleotide hydrolysis, as well as diverse chemical reactions, such as Diels–Alder cycloaddition, Michael addition, phosphoryl transfer, transesterification, and peptide bond formation.7 Importantly, RNA sequences that exhibit these functionalities can be created using selection approaches, such as systematic evolution of ligands by exponential enrichment (SELEX), from random pools of RNAs.8,9 Thus, highly useful RNAs can be readily generated, and the ability to control these RNAs by light would extend their utility by enabling their activity to be regulated with considerable precision within cells or tissues.
Meanwhile, in addition to synthetic RNAs that influence cellular function, there are numerous endogenous RNAs with unknown functions. Indeed, the genome is thought to encode at least 40,000 different noncoding RNAs. Some of these RNAs appear to bind to genomic promoters and regulate epigenetic state or transcription rates.10,11 However, in many cases, these noncoding RNAs bind to cytosolic or other proteins and do not appear to directly influence transcription.12,13 Indeed, the vast majority of noncoding RNAs are mysterious, and studies are needed to uncover their biological functions.
One potential mechanism for understanding their function is to regulate their activity by light. Although their precise activity may not be known, light may be useful for rapidly controlling the expression levels or subcellular localization of these RNA species, thereby enabling researchers to link changes in cellular functions with changes in the intracellular availability of these noncoding RNAs. These approaches may facilitate the discovery of the physiological function of diverse noncoding RNAs.
Thus, there are diverse applications in which control of RNA functionality by light can be valuable for researchers. General approaches that can be applied to diverse types of RNAs are likely to have broad applicability and could further enhance the use and understanding of RNA in cells.
Genetic encodability and reversible photoswitching are desirable properties
A particularly important aspect of these tools is that they should be genetically encoded. Some light-responsive tools have been described that contain chemical modifications that confer light responsiveness. For example, some proteins are chemically modified to contain photolabile caging groups that inactivate amino acid side chains, but which are then removed by light to restore protein function.14 These modifications require chemical synthesis, which is not easily performed by the average laboratory. Additionally, the process of microinjecting proteins into cells is a specialized technique that is also technically challenging. However, genetic encodability bypasses the complexity of chemical synthesis and the toxicities associated with cellular injection. Genetically encoded tools can be expressed directly within cells after a suitable DNA element is introduced in the cell and also enables these light-regulated proteins to be expressed in specific cell types or tissues in animals. Thus, genetic encoding is a highly valuable property that maximizes the utility of these novel technologies.
Another desirable property of light-regulated tools is that they can undergo light-dependent reversible switching. In other words, one wavelength of light can be used to alter the function of the optogenetic tool, while another wavelength can restore that function. For example, specific light-regulated channels can be opened with one wavelength and closed with another.15 Additionally, reversibility provides a mechanism by which the effects of the light-dependent change in cellular function can be definitively attributed to the light-activated RNA or protein and not to an unanticipated effect of light exposure. Lastly, reversibility allows a single cell or tissue to be repeatedly activated and then inactivated to obtain multiple measurements. The strategies that enable reversibility confer additional versatility to light-regulated technologies.
Extrapolating from light-responsive proteins to light-responsive RNAs
A major goal is to have RNAs that switch between two conformations in response to light. For example, an RNA can be unfolded and inactive in one state, and then upon irradiation with light, the RNA can switch to a folded, active form. Thus, a ribozyme can switch from off to on, and an aptamer that inhibits a protein can switch from an inactive form to an inhibitory form in response to light. In order to achieve this, the RNA needs to undergo a conformational change (i.e., structure switching) in response to light.
Strategies for engineering light-mediated regulation of RNA function in cells are likely to draw upon existing light-regulated molecules. In the case of light-regulated proteins, many contain chromophores that interact with light and undergo cis-trans isomerizations. For instance, light-sensitive rhodopsins contain covalently bound retinal, which undergoes an isomerization in response to light, altering the structure and signaling activity of these proteins.16 The phytochromes mediate signal activity by light-regulated cis-trans isomerization of phytochromobilin.17,18 Similarly, the xanthopsin photoreceptors contain the trans-p-coumaric acid chromophore to regulate biological signal transfer.19,20 Conceivably, RNAs can be designed to bind similar fluorophores and undergo conformational changes that are induced by cis-trans isomerizations of bound chromophores.
However, it should be noted that biological chromophores are not the exclusive set of compounds that could be used by RNAs. Indeed, numerous small molecules can undergo structure switching within cells. For example, stilbenes, diazobenzenes, spiropyrans, diarylethene, fulgide, and others have been shown to undergo photo-induced isomerizations or ring closure/openings.21 Importantly, in many of these cases, reversal of these interactions can be achieved by a different wavelength, which can enable selective control of the isomerization state of the small molecule. Conceivably, RNAs that bind these molecules and are sensitive to their isomerization state could allow the RNAs to be regulated by light.
Another way that light can induce structure switching is by light-dependent formation of thymidine cross-links, as well as light-induced cross-linking with adjacent biomolecules.22 Conceivably, RNA sequences can be generated that are highly susceptible to forming these covalent adducts. However, some drawbacks of these approaches are that these reactions are unlikely to be reversible and that reactions that exploit photochemistry of nucleotide bases require wavelengths in the ultraviolet range, which can be damaging to cells. Conceivably, this can be overcome by using photo-sensitizing molecules that bind RNA and enhance the rate of base reactivity, thereby shifting the wavelength needed to enable chemical reactivity of nucleotide bases. Additionally, two-photon excitation strategies are now generally available with nonlinear sample excitation that use lower intensity radiation and potentially bypass toxicities associated with ultraviolet light.23
It is important to keep in mind that light could potentially control RNAs in ways besides structure switching. These mechanisms are less intuitive but could potentially be useful in synthetic biology applications. For instance, light affects the dimerization status of the green fluorescent protein (GFP)-like Dronpa. A Dronpa mutant can be used to reversibly multimerize different proteins.24 Additionally, light can be used to induce GFP to act as electron donors within photochemical reactions.25 The recent description of RNAs that mimic of GFP and comprise RNA aptamers bound to GFP-like fluorophores26 raises the possibility that similar GFP-like functionalities can be built into these RNAs.
Realizing light-regulated RNA using photoswitchable proteins
One strategy to photo-regulate cellular RNA is to take advantage of photoswitchable natural proteins. This is particularly important since small molecules have problems that can limit their usefulness. For example, small molecules readily diffuse throughout cells and can diffuse out of cells and into other cells. Thus, photoactivation in one part of the cell (e.g., in the nucleus) will result in the photoactivated molecule localized throughout the cell within seconds. Additionally, since the molecule can potentially diffuse out of the cell and into neighboring cells, spatial precision at the level of the specific photoactivated cell can be lost. This contrasts greatly with photoactivated proteins. The spatial localization and diffusion of proteins inside cells can be highly controlled. Thus, RNAs that take advantage of photoactivated proteins as their mechanism of light regulation will be more readily controlled in a spatially and temporally specific manner. As mentioned above, RNA aptamers can be synthesized that bind to diverse classes of intracellular proteins; therefore, it should be straightforward to create RNA aptamers that selectively bind to the light-activated or inactivated version of the photoswitchable proteins.
Recent studies show that RNAs can be switched from one conformation to another by protein binding. We showed this using protein-binding aptamers that regulate Spinach, an RNA mimic of GFP27 that binds DFHBI, a GFP-like fluorophore, and switches it from a nonfluorescent state to a green fluorescent form.26,28 We synthesized an RNA that contains the Spinach sequence as well as the sequence of the RNA aptamer that binds to the MS2 coat protein (MCP).27 The MCP aptamer is largely unfolded in the absence of MCP. The MCP aptamer was fused to Spinach in such a way that the Spinach structure was unfolded due to the unfolded nature of the MCP aptamer. However, upon binding MCP, the MCP aptamer folded and allowed Spinach to fold as well (Fig. 1A), resulting in MCP-induced fluorescence, which could be induced in living cells.27 These experiments showed that a protein can be used to switch Spinach from an unfolded to a folded state.
Figure 1.
Light-dependent regulation of RNA using photoswitchable proteins. (A) Protein-induced folding of a Spinach-based sensor to activate the DFHBI fluorescence. The sensor comprises a Spinach module (black), a transducer module (orange) and a protein-recognition module (blue). (B) An RNA aptamer (blue) that binds the light-activated conformation of a light-responsive protein is fused to a protein-inhibiting aptamer (black). Upon binding the light-activated protein, the aptamer (blue) then folds, enabling the protein-inhibiting aptamer (black) to fold and inhibit its target protein. The active aptamer is indicated with a green check mark.
A similar strategy can be employed to confer light-sensitivity to other aptamers, such as a protein-inhibiting aptamer. If an aptamer that only binds the light-activated conformation of light-responsive protein is generated, it can be fused to an aptamer of interest in such a way that it impairs its folding. However, upon binding the light-activated protein, the aptamer then folds, enabling the aptamer of interest to inhibit its target protein (Fig. 1B).
Riboswitches can be made light sensitive in this manner as well. Importantly, the general concept of using aptamers that switch from an unfolded to folded conformation and affect the function of another RNA was first demonstrated by Breaker and colleagues with the development of a ribozyme that was regulated by adenosine triphosphate (ATP).29 In this case, an ATP-binding aptamer was fused to the hammerhead ribozyme in such a way that the unfolded ATP aptamer disrupted the ribozyme structure. Upon ATP binding, the ribozyme structure was restored. To make this light responsive, an aptamer that binds a light-induced conformation of a light-responsive protein could similarly be used to switch on the ribozyme.
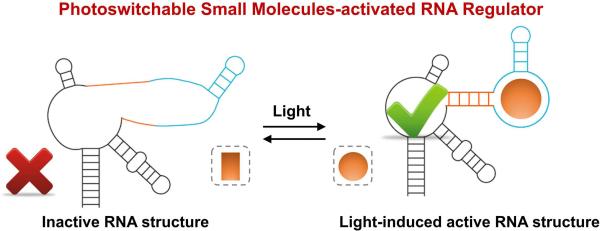
Realizing light-regulated RNA using photoswitchable small molecules
In the example above, the light-induced switch in aptamer or ribozyme function was mediated by a light-responsive protein. However, a similar approach can be performed with a photoswitchable small molecule. Instead of using an aptamer that binds a light-induced conformational state of a protein, an aptamer that binds a photoswitched small molecule can be used (Fig. 2). These aptamers can be fused to protein-inhibiting aptamers or to ribozymes, thus inducing folding only when the photoswitched form of the molecule is present.
Figure 2.
Realizing light-regulated RNA using photoswitchable small molecules. An aptamer (blue) that binds a photoswitched small molecule (golden circle) can be used to regulate the folding and activation of a protein-inhibiting aptamer (black). The active aptamer is indicated with a green check mark.
Indeed, recent studies have begun to utilize this type of approach to create light-regulated RNA-based switches.30 For instance, RNA aptamers that only bind the trans form of an arginine-substituted azobenzene were generated.31, 32 The secondary structure of these aptamers is regulated by photo-irradiation with different wavelengths of light. Similarly, an RNA aptamer that specifically binds to one isomerization state of spiropyran has been identified.33 In another interesting example, a light-controlled dihydropyrene isomer–aptamer interaction has been used to reversibly regulate the catalytic efficiency of a hammerhead ribozyme.34
However, so far, these reported systems have never been applied to regulate processes in living cells. In order to achieve efficient cellular regulation, the target chromophores should be able to achieve high conversion efficiency with fast kinetics by irradiating with a low dosage of visible-to-infrared light, which would confer low cellular toxicity. Additionally, the compounds should show no effects on cellular function in the absence of light.
As mentioned above, a challenge of using isomerizable small molecules is that their spatial localization and diffusion inside and outside of cells is difficult to control. Typically, it is desirable to control the activity of a biomolecule at specific sites within the cell. However, if isomerization is induced at any site in the cell, the isomerized molecule can normally diffuse throughout the cell within seconds. As a result, RNAs throughout the cell that are regulated by these isomerized molecules will be activated. One strategy to overcome this challenge can be to use chromophores with low membrane permeability or cellular mobility after photo-irradiation (Fig. 3), as evidenced by photoactivatable deoxycycline derivatives for the Tet-On system.35 An additional problem is that the isomerized molecules can potentially diffuse inside and outside of the cell. Thus, a signaling pathway activated by the isomerized molecule will be self-limited by diffusion. Conceivably, this could be an advantage because it would limit the activity of the light-responsive RNA to the duration of time that the light is applied. Indeed, this can reduce the requirement for identifying molecules that are able to be reversibly photoswitched by a separate wavelength. Nevertheless, these issues need to be considered when designing RNA devices regulated by photoswitchable small molecules.
Figure 3.
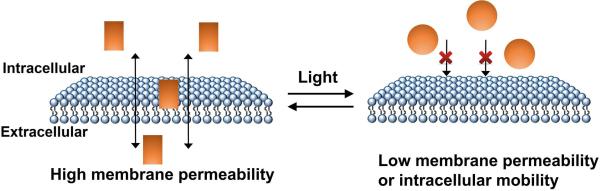
Light-controlled spatial localization and diffusion of photoswitchable small molecules. Chromophores with low membrane permeability or cellular mobility after photo-irradiation (golden circle) can be used to improve the spatial resolution of RNA regulation.
Challenges
An important issue that needs to be addressed for all synthetic biology applications is the development of expression systems that enable accumulation of high levels of RNA inside cells. Typically, RNA transcripts are present at nanomolar concentrations in the cell, while proteins are often present at micromolar concentrations. Indeed, each mRNA can encode hundreds of protein molecules. Thus, the copy number of an encoded protein is significantly higher than the corresponding transcript. In the case of aptamers that are designed to selectively inhibit a target protein, the number of copies of the aptamer has to at least match the number of copies of protein. This requires the availability of expression systems that enable RNAs to accumulate to levels normally seen with proteins.
The low expression level is related to the problem that RNA is typically rapidly degraded in cells. Numerous degradation pathways exist to detect and degrade RNAs from both the 5’ and 3’ ends, as well as at internal sites. In order to use RNAs for synthetic biology applications, the propensity of RNA to be degraded needs to be resolved. Several strategies are likely to help resolve this issue. For example, recent work has established the basis for RNA stability in flaviviruses; these studies have identified short sequence elements that are present at the 5’ end of the mature viral RNA that prevent its degradation by 5’→3’ exonucleases. These elements form a knot structure that essentially results in tightening as the 5’ end of the transcript is pulled into the Xrn1 exonuclease.36 Both viral elements as well as endogenous noncoding RNA utilize a strategy to protect RNAs from 3’→5’ exonucleases. In this case, the 3’ poly(A) tail of the RNA is selectively recognized by sequence elements that form a triple helix sequestering the 3’ hydroxyl. By sequestering this element, the RNA becomes inaccessible to 3’→5’ exonucleases.37, 38 Another possibility to achieve high-level expression is to utilize circular RNAs, which have been shown by recent studies to be present in diverse organisms. Because these RNAs lack both a 5’ and 3’ end, they are expected to be resistant to all exonucleases. Thus, systems that enable the high-level expression of circular RNAs will be a major advance for achieving high-level RNA in cells.39, 40 Lastly, selection techniques, such as SELEX, can be performed in living cells.41 Selection for RNAs that remain intact and can allow for an unbiased discovery of sequence elements enable the stability of synthetic RNAs inside cells.
Acknowledgements
This work was supported by NIH Grant EB010249 to S.R.J.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Deisseroth K. Optogenetics. Nat. Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagel G, Szellas T, Huhn W, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zemelman BV, Lee GA, Miesenbock G, et al. Selective photostimulation of genetically chARGed neurons. Neuron. 2002;33:15–22. doi: 10.1016/s0896-6273(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 4.Lee JH, Canny MD, Erkenez AD, et al. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc. Natl. Acad. Sci. USA. 2005;102:18902–18907. doi: 10.1073/pnas.0509069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem. Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kruger K, Grabowski PJ, Zaug AJ, et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 7.Breaker RR, Joyce GF. The Expanding View of RNA and DNA Function. Chem. Biol. 2014;21:1059–1065. doi: 10.1016/j.chembiol.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 9.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 10.Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brieke C, Rohrbach F, Gottschalk A, et al. Light-controlled tools. Angew. Chem. Int. Ed. Engl. 2012;51:8446–8476. doi: 10.1002/anie.201202134. [DOI] [PubMed] [Google Scholar]
- 15.Zhang F, Wang L, Brauner M, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 16.Stehfest K, Ritter E, Berndt A, et al. The branched photocycle of the slow-cycling channelrhodopsin-2 mutant C128T. J. Mol. Biol. 2010;398:690–702. doi: 10.1016/j.jmb.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 17.van der Horst MA, Hellingwerf KJ. Photoreceptor proteins, “star actors of modern times”: a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc. Chem. Res. 2004;37:13–20. doi: 10.1021/ar020219d. [DOI] [PubMed] [Google Scholar]
- 18.Quail PH, Boylan MT, Parks BM, et al. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 19.Kort R, Hoff WD, van West M, et al. The xanthopsins: a new family of eubacterial blue-light photoreceptors. EMBO J. 1996;15:3209–3218. [PMC free article] [PubMed] [Google Scholar]
- 20.Perman B, Srajer V, Ren Z, et al. Energy transduction on the nanosecond time scale: early structural events in a xanthopsin photocycle. Science. 1998;279:1946–1950. doi: 10.1126/science.279.5358.1946. [DOI] [PubMed] [Google Scholar]
- 21.Russew MM, Hecht S. Photoswitches: from molecules to materials. Adv. Mater. 2010;22:3348–3360. doi: 10.1002/adma.200904102. [DOI] [PubMed] [Google Scholar]
- 22.Friedberg EC. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 23.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 24.Zhou XX, Chung HK, Lam AJ, et al. Optical control of protein activity by fluorescent protein domains. Science. 2012;338:810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogdanov AM, Mishin AS, Yampolsky IV, et al. Green fluorescent proteins are light-induced electron donors. Nat. Chem. Biol. 2009;5:459–461. doi: 10.1038/nchembio.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song W, Strack RL, Jaffrey SR. Imaging bacterial protein expression using genetically encoded RNA sensors. Nat. Methods. 2013;10:873–875. doi: 10.1038/nmeth.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paige JS, Nguyen-Duc T, Song W, et al. Fluorescence imaging of cellular metabolites with RNA. Science. 2012;335:1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang J, Breaker RR. Rational design of allosteric ribozymes. Chem. Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 30.Jäschke A. Genetically encoded RNA photoswitches as tools for the control of gene expression. FEBS Lett. 2012;586:2106–2111. doi: 10.1016/j.febslet.2012.05.040. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi G, Hagihara M, Nakatani K. RNA aptamers that reversibly bind photoresponsive azobenzene-containing peptides. Chem. Eur. J. 2009;15:424–432. doi: 10.1002/chem.200800936. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi G, Hagihara M, Dohno C, et al. Photoregulation of a peptide-RNA interaction on a gold surface. J. Am. Chem. Soc. 2007;129:8678–8679. doi: 10.1021/ja071298x. [DOI] [PubMed] [Google Scholar]
- 33.Young DD, Deiters A. Light-regulated RNA-small molecule interactions. ChemBioChem. 2008;9:1225–1228. doi: 10.1002/cbic.200800051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HW, Robinson SG, Bandyopadhyay S, et al. Reversible photo-regulation of a hammerhead ribozyme using a diffusible effector. J. Mol. Biol. 2007;371:1163–1173. doi: 10.1016/j.jmb.2007.06.042. [DOI] [PubMed] [Google Scholar]
- 35.Cambridge SB, Geissler D, Calegari F, et al. Doxycycline-dependent photoactivated gene expression in eukaryotic systems. Nat. Methods. 2009;6:527–531. doi: 10.1038/nmeth.1340. [DOI] [PubMed] [Google Scholar]
- 36.Chapman EG, Costantino DA, Rabe JL, et al. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science. 2014;344:307–310. doi: 10.1126/science.1250897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitton-Fry RM, DeGregorio SJ, Wang J, et al. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244–1247. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown JA, Bulkley D, Wang J, et al. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 2014;21:633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 40.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 41.Filonov GS, Moon J, Svensen N, et al. Broccoli: Rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 2014 doi: 10.1021/ja508478x. DOI: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]