Abstract
Most RNAs transcribed in mammalian cells lack protein-coding sequences. Among them is a vast family of long (>200 nt) noncoding (lnc)RNAs. LncRNAs can modulate cellular protein expression patterns by influencing the transcription of many genes, the post-transcriptional fate of mRNAs and ncRNAs, and the turnover and localization of proteins. Given the broad impact of lncRNAs on gene regulation, there is escalating interest in elucidating the mechanisms that govern the steady-state levels of lncRNAs. In this review, we summarize our current knowledge of the factors and mechanisms that modulate mammalian lncRNA stability.
1. Introduction
Eukaryotic cells transcribe large numbers of noncoding (nc)RNAs. The Encyclopedia of DNA Elements (ENCODE) project recently revealed that while 2–3% of the mammalian genome is transcribed into protein-coding RNAs (mRNAs), the vast majority of our genome (up to 80%) is transcribed into noncoding (nc)RNAs [1,2]. Some ncRNAs are processed to generate small RNAs [3,4], but most ncRNAs are larger than 200 nucleotides in their mature forms and thus are designated long noncoding (lnc)RNAs [5–8]. LncRNAs are generally a few hundred to a few thousand nucleotides in length, but some very long noncoding RNAs (vlncRNAs) can reach one million nucleotides in length. Mammalian lncRNAs can be expressed from intergenic regions (lincRNAs), from introns of annotated genes (long intronic ncRNAs), from the promoter regions of coding mRNAs (promoter-associated lncRNAs), from the opposite strand of mRNAs (antisense lncRNAs), or from pseudogenes [6]. They can also be generated by the splicing machinery (circular RNAs) [9–11].
The molecular functions of lncRNAs are quite varied. Some nuclear lncRNAs can regulate gene expression epigenetically by recruiting chromatin-modification factors to activate or inactivate different loci [12,13]. LncRNAs can also regulate transcription by assembling transcriptional activators and repressors to modulate the initiation of transcription [14]. They can influence the nuclear architecture and the structure of nuclear speckles, paraspeckles, and interchromatin granules [15]. Additionally, lncRNAs can regulate gene expression post-transcriptionally by modulating the translation and/or stability of partially complementary mRNAs and by interfering with RNA-binding proteins to influence splicing and translation [16–20]. Competing endogenous RNAs (ceRNAs) and circular RNAs are stable lncRNAs that accumulate in the cell and modulate gene expression by acting as decoys or sponges for microRNAs [21]. Finally, some lncRNAs function post-translationally to control protein turnover by facilitating ubiquitination [22,23]. Given the recognition that lncRNAs robustly regulate gene expression [14,24], there is mounting interest in understanding how lncRNA expression is regulated.
Like the transcription of protein-coding genes, the transcription of lncRNAs is driven by promoter elements, in some cases by distinct histone modification signatures [25,26]. Akin to mRNAs, lncRNA precursor transcripts are subsequently regulated by splicing, 5′ capping, and 3′ polyadenylation to generate mature lncRNAs which can eventually reside in the nucleus, the cytoplasm, or both of these compartments. Unlike mRNAs, however, cytoplasmic lncRNAs are generally not translated [27,28], even though recent reports have uncovered an association of lncRNAs with ribosomes and have identified small peptides generated from lncRNAs [29–32]. The mechanisms that regulate lncRNA stability are poorly understood. However, since modulating the abundance of a lncRNA is a rapid and effective way to regulate its function, the mechanisms that control lncRNA turnover are attracting much attention. In this review, we discuss our emerging understanding of the mechanisms that influence mammalian lncRNA stability.
2. Mechanisms of RNA decay
2.1. Global lncRNA stability
Recent studies using custom RNA arrays to measure the half-lives of ~800 lncRNAs in mouse Neuro-2a cells revealed that ~29% of lncRNAs were unstable when considering a half-life (t1/2) of 2 h or less, compared with 17% of unstable mRNAs [33]. Intergenic, cis-antisense, and spliced lncRNAs were generally more stable than intronic lncRNAs and unspliced lncRNAs comprising a single exon, while cytoplasmic lncRNAs were found to be more stable than nuclear lncRNAs [33]. The regulators of lncRNA turnover have not been identified systematically, but we are beginning to discover that some of the machineries same machineries that control mRNA decay also govern lncRNA decay.
2.2. Decapping, deadenylation, exonucleolytic and endonucleolytic degradation
The control of mRNA stability is a major mechanism of post-transcriptional gene regulation in eukaryotic cells, affecting the abundance of a given mRNA by increasing or decreasing its half-life [34,35]. The canonical mRNA decay follows sequential ribonucleolytic activities although the order of the enzymatic events vary depending on the target mRNA and the specific cellular conditions. Three main mechanisms can trigger mRNA decay: 1) removal of the 3′-polyadenosine [poly(A)] tail by a deadenylase complex initiating 3′-to-5′decay, 2) removal of the 7mGpppG 5′ cap structure by decapping enzymes, inducing 5′-to 3-exoribonucleolytic decay, and 3) endoribonuclease cleavage of a target mRNA in the middle of the transcript. The 3′ deadenylation can be triggered by a multi-subunit complex consisting of poly(A) tail-specific ribonucleases and deadenylases, including PAN2, PAN3, CNOT6/CCR4A, CNOT6L/CCR4B, PARN and other enzymatic activities (reviewed in [34–36]). Deadenylated mRNA is targeted to the exosome complex, which contains the 3′-to-5′ exonucleases RRP44 and EXOSC10. The deadenylated RNA is also a substrate of 5′ decapping enzymes such as DCP2, followed by 5′-to-3′ exonucleolytic degradation via XRN1 [34,37]. Endonucleases such as RMP1, ZC3H12A and IRE1 cleave in the middle of a transcript, followed by degradation of the resulting fragments via 3′-to-5′exosomal and 5′-to-3′ exoribonucleolytic enzyme activities, respectively [38]. Since many lncRNAs are structurally similar to mRNAs, with 5′ caps and 3′ poly(A) tails, the enzymes that control mRNA decapping, deadenylation, and exo- and endonucleolytic degradation are believed to modulate the turnover of many lncRNAs.
2.3. Translation-associated RNA decay: NMD, SMD, NGD, NSD
mRNA decay is also tightly associated with the translation machinery, serving as a quality-control mechanism to ensure the production of adequate protein products. External stimuli and intrinsic properties in mRNA sequences can block mRNA translation during initiation and elongation. Inhibition of translation initiation leads to the accumulation of 43S preinitiation complex, translation repressors, and mRNA decay enzymes in cytoplasmic granules that contain bulk mRNAs targeted for translation repression and stabilization [39–41]. Nonsense-mediated mRNA decay (NMD) is a mechanism that prevents the synthesis of aberrant protein products that can induce cellular toxicity [42]. NMD is triggered by the termination of translation upstream of a splicing-derived exon junction complex (EJC) during a pioneer round of translation. Binding of UPF1 to the EJC triggers UPF1 phosphorylation by SMG1 [34]. The interaction of p-UPF1 and SMG5-7 or SMG-6 ultimately cleaves the RNA with the PTC (premature termination codon) and promotes 5′-to-3′ decay through deadenylation and decapping [43]. STAU1-mediated mRNA decay (SMD) degrades mRNAs bearing intra- or inter-molecular double-stranded structures in the 3′UTR [16,44]. After the termination of translation, UPF1 binds to the mRNA-associated STAU1 and triggers mRNA degradation, likely following events similar to those in conventional NMD [44]. Ribosome stalling on the mRNA during elongation triggers no-go mRNA decay (NGD) and bypassing a stop codon induces non-stop mRNA decay (NSD) which triggers dissociation of ribosomes from mRNA and removes unfavorable mRNA and peptide products [45]. As much of protein-coding mRNA degradation is closely linked to mRNA translation, these decay mechanisms may in principle be distinct from those that govern lncRNA decay, as they do not codify proteins. On the other hand, given the accumulating evidence that ribosomes can bind to some lncRNAs and may generate small peptides [46,47], the full spectrum of translation-independent and -dependent mechanisms control lncRNA decay remains to be elucidated.
2.4. Recruitment of RNA decay machineries by microRNAs and RBPs
The degradation machinery can be recruited to selective groups of mRNAs. This recruitment can be promoted by small RNAs like microRNAs (miRNAs), small interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs), which accelerate target mRNA decay via complementarity sequences within the mRNA 3′UTR [48]. The best-studied small RNAs, microRNAs, associate with several proteins including argonaute 2 (AGO2) to form the RNA-induced silencing complex (RISC), which mediates the decay of the target mRNA. MicroRNA biogenesis typically begins with the synthesis of a primary miRNA (pri-miRNAs) containing a 5′ cap and a 3′ poly(A) tail. Subsequent ‘cropping’ of the pri-miRNA by the type III RNase Drosha releases a small stem-loop structure, the precursor miRNA (pre-miRNA). Following export of the pre-miRNA to the cytoplasm by the nuclear exporting factor 5 (EXP5), cytoplasmic pre-miRNA is further processed by another type-III RNase, Dicer. Dicer cleaves the pre-miRNA loop structure and degrades one strand, releasing a ~22 nt-long mature miRNA which is loaded onto AGO1-AGO4 proteins [3]. The mature miRNA directs RISC to a target mRNA, with which it interacts via imperfect base-pairing. This interaction triggers mRNA decay (by promoting deadenylation, decapping, and exonucleolytic degradation), translational repression, or both of these processes [49–52]. The recruitment of mRNAs to the decay machinery can also be modulated through select RNA-binding proteins (RBPs) that associate with specific sequences on the mRNA. Some RBPs enhance the recruitment of target mRNAs to the degradation machinery, as seen for decay-promoting RBPs like tristetraprolin, UPF1, and AUF1; other RBPs prevent this recruitment, as seen for stabilizing RBPs such as members of the Hu/elav family, changing the relative half-life of the mRNA accordingly [53].
Similar to the decay of mRNAs, lncRNA degradation is driven by RBPs and small RNAs that promote or inhibit the interaction of lncRNAs with the decay machinery, via recruitment of decapping enzymes or by mobilization of the mRNA to the exosome. Also similar to mRNA turnover, cis-regulatory elements recognized by microRNAs and by RBPs are present in lncRNAs and influence their rate of decay. Here, we summarize our current understanding of lncRNA degradation pathways.
3. microRNA-triggered lncRNA decay
Several recent reports highlight a role for microRNAs in modulating lncRNA stability, and consequently lncRNA abundance and function.
3.1. Let-7
A recent study revealed that the stability of LINCRNAP21 is controlled by the microRNA let-7b [18]. Knockdown of AGO2 stabilized human LINCRNAP21, while targeted deletion of AGO2 in mouse embryonic fibroblasts (MEFs) increased the abundance of mouse Lincrnap21, suggesting that microRNAs were involved in the decay of this lncRNA. Among several miRNAs predicted to target human LINCRNAP21 (using the software RNA22), microRNA let-7b was shown to regulate its stability. Ectopic expression of let-7b promoted LINCRNAP21 decay in human cervical carcinoma HeLa cells, while antagonization of let-7b prevented it, indicating that let-7b accelerated LINCRNAP21 degradation [18]. Interestingly, the RBP HuR promoted LINCRNAP21 turnover in a let-7b-dependent manner, as the decay of LINCRNAP21 by overexpression of HuR was rescued by antagonizing let-7b. When LINCRNAP21 was stabilized, it formed partial hybrids with CTNNB1 and JUNB mRNAs and suppressed their translation.
Let-7 was also found to interact with HOTAIR, an antisense lncRNA transcribed from the HOX locus, and reduced HOTAIR levels [22]. Similar to LINCRNA21, HOTAIR was destabilized by let-7 (in this case let-7i), and was stabilized when let-7i or AGO2 were suppressed. Also like LINCRNAP21 decay, HOTAIR degradation occurred when HuR promoted the interaction of let-7i-AGO2 with HOTAIR. Accordingly, in cells that expressed low levels of HuR (for example by reaching replicative senescence), HOTAIR was stable, accumulated, and served as a scaffold for E3 ubiquitin ligases (as shown for Mex3b and Dzip3) and their respective ubiquitination substrates (Snurportin-1 and Ataxin-1) [22].
Additional lncRNAs are also targeted by the let-7 family. One of the long-known lncRNAs, H19, contains RNA sequences targeted by let-7 family microRNAs (let-7a, -b, -g, -i [54]). H19 levels declined after let-7a overexpression in an AGO2-dependent manner. Although it remains to be determined if microRNAs affect H19 stability, a reporter construct containing H19 sequences was selectively inhibited after let-7b overexpression. As a target of let-7, H19 functions as competing endogenous RNA (ceRNA) of mRNAs encoding proteins that are critical for myotube formation [54].
3.2. miR-9
miR-9 was shown to target MALAT1 in the human primary glioblastoma cell line U87MG and the human Hodgkin lymphoma cell line L428 [55]. MALAT1 levels increased when miR-9 was inhibited using an antagomir, while they decreased when miR-9 was overexpressed. Since MALAT1 is exclusively nuclear, miR-9 may promote MALAT1 degradation in the nucleus via nuclear decay-promoting factors.
3.3. miR-34a
Very recently, miR-34a was found to lower the stability of UFC1, a lncRNA that is more highly expressed in human hepatocellular carcinoma (HCC) than in healthy liver [56]. Interestingly, the lincRNA UFC1 appeared to export HuR to the cytoplasm, thereby enhancing the stability of the HuR target CTNNB1 mRNA (encoding β-catenin), in this manner possibly contributing to enhancing tumorigenesis [56].
3.4. miR-211
The lncRNA LOC285194, a p53-inducible tumor suppressor, was found to be a target of miR-211 in the human colon cancer cell line HCT-116 [57]. Transfection of a miR-211 precursor reduced the expression of LOC285194, although the contribution of LC285194 destabilization to this reduction was not studied. The lower expression of LOC285194 in human colon cancer was linked to the accumulation of miR-211 in this cancer type. In this paradigm, an interesting reciprocal regulation of miR-211 was postulated, as LC285194 reduces steady-state level of mature miR-211 but not pri-miR-211 or pre-miR-211 levels, further implicating LOC285194 in the regulation of miR-211 stability [57].
3.5. miR-574-5p
miR-574-5p interacts with and lowers the levels of the tumor suppressor lncRNA PTCSC3 in the human thyroid cancer cell lines BCPAP, FTC133, and 8505C. Ectopic expression of miR-574-5p in these cell lines decreased PTCSC3 abundance [58]. Since PTCSC3 arrests cells in G0/G1, miR-574-5p was proposed to enhance cell proliferation by maintaining low levels of PTCSC3.
3.6. miR-124
miR-124 binds to and lowers the levels of the retropseudogene LNCSCA7 [59]. Overexpression of miR-124 in mouse N2A cells decreased Lncsca7 abundance, whereas introduction of miR-124 antagomir increased it. Analysis of reporter constructs further supported the notion that miR-124 specifically promoted Lncsca7 decay. Since Lncsca7 was proposed to function as ceRNA for Atxn7 mRNA, which encodes a transcriptional repressor of miR-124 itself, this auto-regulatory loop may help to titrate the level of Lncsca7, Atxn7, and miR-124 during neurodegeneration [59].
4. RBP-mediated lncRNA decay
Several RBPs are involved in lncRNA decay. Through their ability to bind specific RNA sequences, they can promote or inhibit the degradation of lncRNAs. These RBPs include classic ARE-binding proteins, RNA helicases, and RBPs that function in RNA transport from the nucleus.
4.1. HuR
This ubiquitous RBP recognizes AU- or U-rich RNA sequences in many mRNAs and promotes their stability [60]. Interestingly, however, HuR instead lowered the stability of some lncRNAs [18,22]. As mentioned above, HuR promoted the decay of LINCRNAP21, a lncRNA present in both the nucleus and the cytoplasm, by promoting its interaction with AGO2-let-7b complexes in HeLa cells. In keeping with this mode of action, HuR overexpression decreased the steady-state levels of LINCRNAP21, while a let-7b antagomir rescued the effect of HuR and increased LINCRNAP21 abundance. Conversely, HuR depletion increased LINCRNAP21 abundance, and co-expression of let-7b precursor lowered LINCRNAP21 levels. These results suggest that HuR promotes LINCRNAP21 decay in let-7b-dependent manner. Similarly, HuR promoted decay of lncRNA HOTAIR (also present in both the nucleus and the cytoplasm) by enhancing the recruitment of let-7i-AGO2 to HOTAIR [22].
4.2. AUF1
This RBP, which exists in four isoforms (p37, p40, p40, p45), was found to have strong affinity for U- and GU-rich RNAs, and promoted decay of the nuclear lncRNA NEAT1 [61]. In contrast to HuR, which generally stabilizes mRNA but destabilized lncRNAs, AUF1 displayed the same degradation-promoting influence on lncRNAs as that reported for mRNAs. This influence was evidenced by the fact that silencing AUF1 (all four isoforms) increased the steady-state levels of NEAT1. Although NEAT1 is mainly localized in nuclear speckles, it is not known at present whether AUF1 promoted NEAT1 degradation in the nucleus or the cytoplasm. In turn, the accumulation of nuclear NEAT1 led to increased non-clustered (random) nuclear speckles and to the retention of NEAT1 target mRNAs in the nucleus [61].
4.3. HuD
The neuronal RBP HuD associated with BACE1AS, an antisense lncRNA which shuttles between the nucleus and the cytoplasm depending on growth conditions, and partly complements BACE1 (β-site APP-cleaving enzyme 1) mRNA [17]. HuD was shown to bind 5′ and 3′ segments of BACE1AS but not the central double-stranded RNA segment complementary to BACE1 mRNA [62]. Depletion of HuD in neuroblastoma SK-N-F1 destabilized BACE1AS and reduced its steady-state level. As the microRNA miR-485-5p targets BACE1 mRNA [62], one of the proposed mechanisms through which BACE1AS increases BACE1 expression is by blocking the interaction of miR-485-5p with BACE1 mRNA [63]. A recent study suggests that HuD prevents the interaction of eIF4A with microRNA-RISC [64].
4.4. PABPN1
Poly(A)-binding protein nuclear 1 (PABPN1), an RBP involved in the addition of poly(A) tails to the 3′ end of mRNAs, reportedly protects lncRNAs from degradation [65]. Depletion of PABPN1 in HeLa cells did not affect the steady-state levels of many mRNAs. Interestingly, however, PABPN1 silencing increased the stability and abundance of nuclear lncRNAs NEAT1, SHG60, and SHG104. Moreover, mutation in the AAUAAA sequence, required for polyadenylation, reduced the steady-state level of lncRNA SHG60, implicating this sequence in polyadenylation-dependent lncRNA decay. Further analysis revealed that SHG60 accumulated in human embryonic kidney (HEK)293T cells after silencing RRP40, a protein component of exosome, but not UPF1, a core protein in nonsense-mediated mRNA decay (NMD) [65].
4.5. IGF2BP
Microarray analysis of 60 human hepatocellular carcinoma (HCC) samples found that the lncRNA HULC (highly up-regulated in liver cancer) was overexpressed in many HCCs compared to normal liver [66]. HULC interacts with several RBPs, including the IGF2 mRNA-binding protein (IGF2BP) 1, -2, and -3 in the hepatocellular carcinoma cell line HepG2. Notably, depletion of IGF2BP1 (but not IGF2BP2 or -3), stabilized HULC, increasing its abundance. Since IGF2BP1 interacts with CNOT1, a core protein of the deadenylase complex, it may recruit CNOT1 to promote HULC deadenylation [66].
4.6. UPF1
A recent study using BrU RNA labeling measured lncRNA stability globally in mammalian cells [67]. UPF1 depletion stabilized GAS5 (growth arrest-specific 5), a lncRNA found in both the nucleus and the cytoplasm, as well as cIAP2 and SGK1 mRNAs, which are involved in the glucocorticoid response. Serum depletion increased the stability and abundance of GAS5, whereas it decreased the abundance of cIAP2 and SGK1 mRNAs. Strikingly, suppression of GAS5 accumulated cIAP2 and SGK1 mRNAs in HEK293T cells, suggesting that GAS5 may, in turn, promote the degradation of cIAP2 and SGK1 mRNAs by interacting with UPF1.
5. Decapping and deadenylation of lncRNA
5.1. XRN1
Mammalian XRN1 can degrade long noncoding RNA generated by arthropod-borne flaviviruses [68]. The Dengue virus 3′UTR is degraded by XRN1, HeLa cell extract, or mosquito (Aedes C6/36) extracts to generate a short noncoding RNA (sfRNA). In turn, accumulation of the sfRNA in mammalian cells inhibits XRN1 function, allowing the accumulation of many cellular mRNAs, including FOS and TUT1 mRNA. These results indicate that inhibition of XRN1 can result in the accumulation of host mRNA as well as long noncoding RNAs.
5.2. CNOT1
As mentioned above, the lncRNA HULC is degraded by CNOT1, a core protein component of the mammalian deadenylase complex, as silencing CNOT1 increased HULC stability and steady-state levels [66]. The RBP IGF2BP1 interacts with CNOT1 and may recruit it to HULC for deadenylation and subsequent degradation. In agreement with this idea, silencing IGF2BP1 stabilizes HULC, although deadenylation and IGF2BP1-mediated decay may also occur independently.
6. Cis-regulatory elements in lncRNAs
6.1. 3′ end processing
Although mature MALAT1 is exclusively nuclear, the 3′end processing product from nascent MALAT1 resides in the cytoplasm after cleavage by RNases P and Z. The resulting 61 nt-long 3′end product is processed further through the tRNA biogenesis pathway to generate a tRNA-like cytoplasmic RNA called mascRNA [69]. Further processing adds CCA to the 3′end of mascRNA resulting in its destabilization and export to the cytoplasm [70].
6.2. RNA triple helix
Although mature MALAT1 lacks a poly(A) tail, it is much more stable than mRNAs. The 3′end of MALAT1 forms a conserved triple-helix that prevents access of 3′-to-5′ exonucleases [71]. The crystal structure of the human MALAT1 ENE (expression and nuclear retention element) A-rich tract reveals a bipartite triple helix [72]. Indeed, mutations that disrupt this blunt-ended triple helix abolish the stability of MALAT1 in HeLa cells. Similar RNA triple helix characteristics were identified in lncRNA PAN (polyadenylated nuclear) from Kaposi’s sarcoma-associated herpes virus (KSHV) [73]. Triple helix formation with a poly(A) tail inhibits its rapid decay in the nucleus. Taken together, these studies indicate that evolutionally conserved RNA triple-helix complexes protect lncRNAs from rapid degradation.
7. Concluding remarks and perspectives
LncRNAs regulate mammalian protein expression programs by influencing the transcription of protein-coding transcripts, as well as pre-mRNA splicing, mRNA decay, protein translation, and protein degradation. In order to ensure proper lncRNA function, the levels and localization of lncRNAs must be carefully orchestrated. Here, we have reviewed the mechanisms of lncRNA degradation by focusing on how lncRNA stability is affected by trans-interacting factors (proteins and RNAs) and by cis-elements in the lncRNA.
Although this review focused on mammalian lncRNA turnover, the decay of yeast lncRNAs [74] likely shares many similarities and may guide future studies. In one report, yeast Dcp2 and Xrn1 were found to be necessary for the decay of lncRNAs implicated in the response to galactose availability [75], notably the lncRNA GAL10; accordingly, stabilization and accumulation of GAL10 repressed GAL1 mRNA transcription via changes in chromatin structure. Whether a similar mechanism operates in mammalian cells is not yet known.
As discussed in this review, lncRNA and mRNA decay pathways share many similarities. The main difference between lncRNA decay and mRNA decay pathways may reside in the fact that mRNA metabolism is closely tied to its interaction with ribosomes for translation. However, recent ribosome density mapping (also called ribosome profiling) revealed that a large proportion (43.1% [30]) of lncRNAs do interact with ribosomes [46,47]. Nonetheless, lncRNAs are ~13-fold less likely to produce peptides than mRNAs, and in the two cell lines analyzed by GENCODE v7, the majority of lncRNAs (~92%) were not translated [27]. Further analysis of a collection of 79,333 peptides revealed only 85 peptides matching to open reading frames in a handful of lncRNAs (<1% of total lncRNAs) [27]. Whether the translation machinery-associated decay pathways differ for coding and noncoding RNAs needs more detailed examination.
With the exception of HuR, which generally promotes mRNA decay but enhances lncRNA decay [18,22], the rest of lncRNA decay mechanisms follow similar rules as mRNAs for decay mediated by trans-factor binding. As discussed here, many miRNAs promote lncRNA degradation by seed-dependent complementarity and several RBPs (AUF1, IGF2BP1, and UPF1) promote lncRNA decay [61,66,67]. Likewise, mRNA-stabilizing RBPs such as HuD and PABPN1 also protect lncRNAs from degradation [62,65]. LncRNAs also appear to utilize general mRNA decay factors (Dcp2, Dcp1, Xrn1, and CNOT1) for decapping, deadenylation, and degradation 5′-to-3′ and 3′-to-5′ [66,68,75]. In distinction with mRNAs, however, cis-regulatory sequences embedded in lncRNAs can determine their stability by affecting 3′ end processing or by forming RNA triple helices exclusively in lncRNAs, but not mRNAs [70-73].
A comprehensive understanding of the mechanisms that control lncRNAs await much additional work. In the same way that regulation of mRNA decay contributes to the temporal availability of mRNA for translation, modulation of lncRNA decay affects the abundance and distribution of lncRNAs. The local and temporal control of lncRNA abundance can affect chromatin remodeling, target mRNA fate, and protein stability. By changing lncRNA levels, cells may rapidly and effectively titrate the amount of mRNA decay factors, RBPs, and miRNAs. For instance, just as ceRNAs compete with mRNAs for binding miRNAs [76], circular lncRNAs may bind and sequester RBPs [77]. Linear lncRNAs can also interact with decapping enzymes, deadenylases, exoribonucleases, and RBPs, thus modulating their availability in cells [77]. Given the unexpected functions of many lncRNAs, elucidating in full the mechanisms of lncRNA decay will increase our understanding of these regulatory RNAs and unveil new ways in which we can target lncRNAs in order to modulate gene expression patterns.
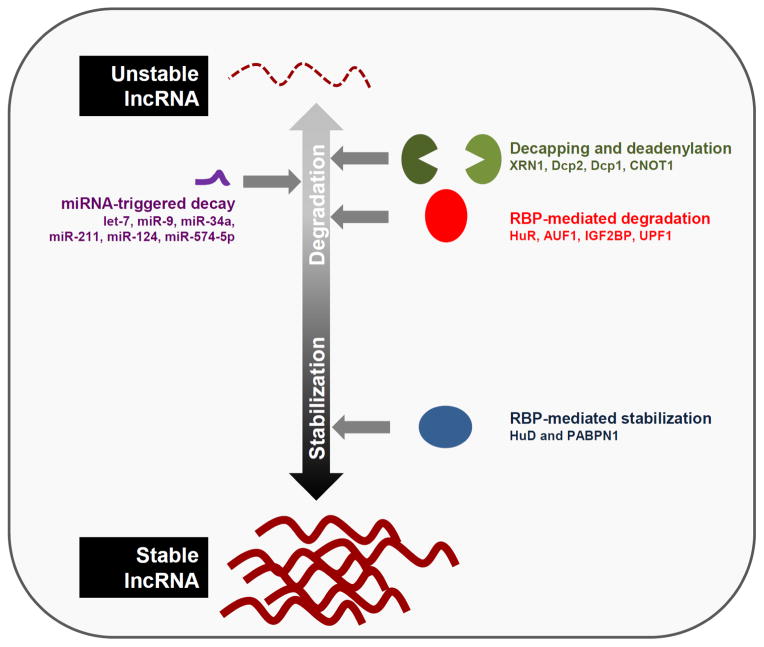
FIGURE 1. Modes of lncRNA degradation.
Schematic of the major mechanisms of lncRNA decay. Top, LncRNAs are degraded by many miRNAs, decay-promoting RBPs (HuR, AUF1, IGF2BP, and UPF1), and general decapping enzymes and dadenylases (Dcp2, Dcp1, and CNOT1). Bottom, lncRNAs are protected from degradation by stabilizing RBPs (HuD and PABPN1).
Table 1. Examples of mammalian lncRNA degradation.
Decay factors (column 1) affecting the stability and/or level of lncRNAs (column 2) in various species (column 3). The consequences on lncRNA stability are indicated (column 4).
| Turnover Factor | lncRNA | Species | Consequences | References |
|---|---|---|---|---|
| microRNAs | ||||
|
| ||||
| let-7b | LINCRNAP21 | Human, Mouse | Degradation | [18] |
| let-7i | HOTAIR | Human, Mouse | Degradation | [22] |
| let7a,b,g,i | H19 | Human, Mouse | Degradation | [54] |
| miR-9 | MALAT1 | Human | Degradation | [55] |
| miR-34a | UFC1 | Human | Degradation | [56] |
| miR-211 | LOC285194 | Human | Degradation | [57] |
| miR-574-5p | PTCSC3 | Human | Degradation | [58] |
| miR-124 | Lncsca7 | Mouse | Degradation | [59] |
|
| ||||
| RNA-binding proteins | ||||
|
| ||||
| HuR | LINCRNAP21 | Human, Mouse | Degradation | [18] |
| HuR | HOTAIR | Human, Mouse | Degradation | [22] |
| AUF1 | NEAT1 | Human | Degradation | [61] |
| HuD | BACE1AS | Human | Stabilization | [62] |
| PABPN1 | NEAT1, SHG60, SHG104 | Human | Stabilization | [64] |
| IGF1BP | HULC | Human | Degradation | [66] |
| UPF1 | GAS5 | Human | Degradation | [67] |
|
| ||||
| Decapping & deadenylation | ||||
|
| ||||
| XRN1 | Dengue virus 3′UTR | Flavivirus hosts | Degradation | [68] |
| CNOT1 | HULC | Human | Degradation | [66] |
|
| ||||
| Cis-regulatory elements | ||||
|
| ||||
| 3′end CCA | mascRNA | Mouse | Cytoplasmic export | [69] |
| RNA triple helix | Malat1 | Mouse | Stabilization | [71,72] |
| RNA triple helix | PAN | KSHV hosts | Stabilization | [73] |
LncRNAs control gene expression transcriptionally and posttranscriptionally
Changes in lncRNA turnover effectively alter mammalian lncRNA abundance
LncRNA turnover is largely regulated by the general mRNA decay machinery
RNA-binding proteins interacting with lncRNAs can enhance or lower lncRNA half-life
MicroRNAs associate with lncRNAs and promote lncRNA degradation
Acknowledgments
JHY, JK, and MG were supported by the National Institute on Aging Intramural Research Program, National Institutes of Health.
Abbreviations
- lncRNA
long noncoding RNA
- lincRNA
long intergenic noncoding RNA
- miRNA
micro RNA
- RBP
RNA-binding protein
- RISC
RNA-induced silencing complex
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.Tuck AC, Tollervey D. RNA in pieces. Trends Genet. 2011;27:422–432. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulitsky I, Bartel DP. LincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nam JW, Bartel DP. Long noncoding RNAs in C. elegans. Genome Res. 2012;22:2529–2540. doi: 10.1101/gr.140475.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DC, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 10.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 11.Lasda EI, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;12:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quinodoz S, Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24:651–663. doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonasio R, Shiekhattar R. Regulation of Transcription by Long Noncoding RNAs. Annu Rev Genet. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;3:456–461. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St-Laurent G, III, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 20.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, Wilson GM, Gorospe M. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Kundaje A, Jr, Gunawardena HP, Yu Y, Xie L, Krajewski K, Strahl BD, Chen X, Bickel P, Giddings MC, Brown JB, Lipovich L. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guttman M, Russell P, Ingolia NT, Weissman JX, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, Saghatelian A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM. Long non-coding RNAs as a source of new peptides. Elife. 2014;3:e03523. doi: 10.7554/eLife.03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, Giraldez AJ. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta-Huspenina J, Boese JH, Bantscheff M, Gerstmair A, Faerber F, Kuster B. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 33.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191:671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley Interdisc Rev RNA. 2010;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip. Rev RNA. 2010;1:253–265. doi: 10.1002/wrna.15. [DOI] [PubMed] [Google Scholar]
- 38.Schoenberg DR. Mechanisms of endonuclease-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2011;4:582–600. doi: 10.1002/wrna.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchan JR. mRNP granules: Assembly, function, and connections with disease. RNA Biol. 2014;11 doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagrm.2014.11.009. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 42.Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Okada-katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. N-and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMGg-5:Smg7 during NMD. Nucleic Acids Res. 2012;40:1251–1266. doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim YK, Furic L, Desgroseillers L. Mamailian staufen 1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 45.Lykke-Andersen J, Bennett EJ. Protecting the proteome: Eukaryotic cotranslational quality control pathways. J Cell Biol. 2014;204:467–476. doi: 10.1083/jcb.201311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, Hao W, Macinnes AW, Cuppen E, Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biology. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen CY, Dinghai Z, Zhenfang X, Shyu AB. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, Chen CY, Shyu AB, Yates JR, III, Hannon GJ, Filipowicz W, Duchaine TF, Sonenberg N. Mammalian miRNA RISC Recruits CAF1 and PABP to Affect PABP-Dependent Deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 52.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 53.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress. Biol Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, Min W, Bennett AM, Gregory RI, Ding Y, Huang Y. The imprinted H19 LncRNA antagonizes Let-7 MicroRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leucci E, Patella F, Waage J, Holmstrøm K, Lindow M, Porse B, Kauppinen S, Lund AH. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep. 2013;3:2535. doi: 10.1038/srep02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao C, Sun J, Zhang D, Guo X, Xie L, Li X, Wu D, Liu L. The long intergenic noncoding RNA UFC1, a target of microRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of β-catenin in HCC cells. Gastroenterology. 2015;148:415–426. doi: 10.1053/j.gastro.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 57.Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, Wu F, Mo YY. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan M, Li X, Jiang W, Huang Y, Li J, Wang Z. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp Ther Med. 2013;5:1143–1146. doi: 10.3892/etm.2013.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan JY, Vance KW, Varela MA, Sirey T, Watson LM, Curtis HJ, Marinello M, Alves S, Steinkraus BR, Cooper S, Nesterova T, Brockdorff N, Fulga TA, Brice A, Sittler A, Oliver PL, Wood MJ, Ponting CP, Marques AC. Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7. Nat Struct Mol Biol. 2014;21:955–961. doi: 10.1038/nsmb.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoon JH, De S, Srikantan S, Abdelmohsen K, Grammatikakis I, Kim J, Kim KM, Noh JH, White EJ, Martindale JL, Yang X, Kang MJ, Wood WH, 3rd, Noren Hooten N, vans MKE, Becker KG, Tripathi V, Prasanth KV, Wilson GM, Tuschl T, Ingolia NT, Hafner M, Gorospe M. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun. 2014;5:5248. doi: 10.1038/ncomms6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang MJ, Abdelmohsen K, Hutchison ER, Mitchell SJ, Grammatikakis I, Guo R, Noh JH, Martindale JL, Yang X, Lee EK, Faghihi MA, Wahlestedt C, Troncoso JC, Pletnikova O, Perrone-Bizzozero N, Resnick SM, de Cabo R, Mattson MP, Gorospe M. HuD regulates coding and noncoding RNA to induce APP → Aβ processing. Cell Rep. 2014;7:1401–1409. doi: 10.1016/j.celrep.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G, III, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fukao A, Mishima Y, Takizawa N, Oka S, Imataka H, Pelletier J, Sonenberg N, Thoma C, Fujiwara T. MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Mol Cell. 2014;56:79–89. doi: 10.1016/j.molcel.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 65.Beaulieu YB, Kleinman CL, Landry-Voyer AM, Majewski J, Bachand F. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet. 2012;8:e1003078. doi: 10.1371/journal.pgen.1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hämmerle M, Gutschner T, Uckelmann H, Ozgur S, Fiskin E, Gross M, Skawran B, Geffers R, Longerich T, Breuhahn K, Schirmacher P, Stoecklin G, Diederichs S. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1) Hepatology. 2013;58:1703–1712. doi: 10.1002/hep.26537. [DOI] [PubMed] [Google Scholar]
- 67.Tani H, Torimura M, Akimitsu N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS One. 2013;8:e55684. doi: 10.1371/journal.pone.0055684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, Khromykh AA, Wilusz J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 2012;18:2029–2040. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilusz JE, Whipple JM, Phizicky EM, Sharp PA. tRNAs marked with CCACCA are targeted for degradation. Science. 2011;334:817–821. doi: 10.1126/science.1213671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, Steitz JA. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol. 2014;21:633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tycowski KT, Shu MD, Borah S, Shi M, Steitz JA. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep. 2012;2:26–32. doi: 10.1016/j.celrep.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Geisler S, Lojek L, Khalil AM, Baker KE, Coller J. Decapping of long noncoding RNAs regulates inducible genes. Mol Cell. 2012;45:279–291. doi: 10.1016/j.molcel.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013;32:923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]