Abstract
The molecular factors involved in the development of Post-Traumatic Stress Disorder (PTSD) remain poorly understood. Previous transcriptomic studies investigating the mechanisms of PTSD apply targeted approaches to identify individual genes under a cross-sectional framework lack a holistic view of the behaviours and properties of these genes at the system-level. Here we sought to apply an unsupervised gene-network based approach to a prospective experimental design using whole-transcriptome RNA-Seq gene expression from peripheral blood leukocytes of U.S. Marines (N=188), obtained both pre- and post-deployment to conflict zones. We identified discrete groups of co-regulated genes (i.e., co-expression modules) and tested them for association to PTSD. We identified one module at both pre- and post-deployment containing putative causal signatures for PTSD development displaying an over-expression of genes enriched for functions of innate-immune response and interferon signalling (Type-I and Type-II). Importantly, these results were replicated in a second non-overlapping independent dataset of U.S. Marines (N=96), further outlining the role of innate immune and interferon signalling genes within co-expression modules to explain at least part of the causal pathophysiology for PTSD development. A second module, consequential of trauma exposure, contained PTSD resiliency signatures and an over-expression of genes involved in hemostasis and wound responsiveness suggesting that chronic levels of stress impair proper wound healing during/after exposure to the battlefield while highlighting the role of the hemostatic system as a clinical indicator of chronic-based stress. These findings provide novel insights for early preventative measures and advanced PTSD detection, which may lead to interventions that delay or perhaps abrogate the development of PTSD.
Keywords: Post-traumatic stress disorder, gene co-expression network, innate immunity
INTRODUCTION
The study of the molecular factors that determine risk and subsequent development of Post-traumatic stress disorder (PTSD) are at the forefront of molecular psychiatric research. A significant number of men and women exposed to severe emotional trauma and loss emerge from these events with persistent PTSD symptoms, such as intrusive imagery, avoidance and hyperarousal, as well as other long-term physical health problems. PTSD affects 7–8% of the general United States (US) population, and is higher among troops recently returned from the wars in Iraq and Afghanistan, with estimates of prevalence as high as 20%1. Annual health care costs associated with PTSD in the US have been estimated to be 180 million dollars2. Heterogeneity in susceptibility to PTSD suggests that differences at the molecular level (i.e. gene-expression level) may influence an individual’s physiological and psychological response to trauma and thus the development of PTSD. A clear understanding of the molecular mechanisms underlying this aberrant response to trauma is required to reduce the substantial morbidity and mortality associated with this disorder.
A number of studies have analyzed blood gene expression and glucocorticoid activity to build more effective models for identifying molecular factors associated to PTSD(3–12). These studies were recently reviewed by Heinzlemann and Gill2, who summarized that the increased expression of inflammatory genes and decreased expression of the genes that regulate inflammation contribute to the onset of PTSD. Specifically, when considering the overlap in results from transcriptomic studies, the decreased expression of FKBP5 and STAT5B, which regulate inflammation, is evident(4,6,7,9). The majority of these reviewed studies(3–8,11,12) centered transcriptomic analyses on subjects already diagnosed with PTSD, and thus lacked a prospective study design, as well as independent datasets for validation purposes. These studies employ gene expression analysis on pre-determined targets, focusing analyses on the individual gene-level and the putative clinical utilities of the resulting gene-list, without studying the connectivity of these genes at the system-level.
Recent gene-expression network analyses, such as weighted gene co-expression network analysis (WGCNA), aim to integrate expression data across thousands of genes into a higher-order system-level context to identify groups of genes within a network whose expressions are highly correlated (i.e. co-expression modules)13. In doing so, WGCNA provides a powerful unsupervised approach to tackle the molecular complexity that occurs in neurodevelopmental and psychophysiological disorders(14–19), although has never before been applied to PTSD.
We applied WGCNA to RNA-Seq and microarray peripheral blood leukocyte (PBL) gene expression taken from two independent groups of U.S. Marines, both pre- and post-deployment to conflict zones. The primary goal of this analysis was to best characterise the prognostic and diagnostic molecular signatures defining both ‘PTSD risk’ and ‘PTSD’ states, while demonstrating the robustness and reproducibility of WGCNA findings across datasets. Instead of identifying differentially expressed genes on a gene-by-gene basis, we constructed unsupervised gene co-expression networks from a combination of case and control data and identified gene co-expression modules within these networks. Modules were first assessed for containing differentially expressed genes, tested for their association with PTSD, and finally subjected to functional enrichment analysis. In this manner, we then assessed whether the PTSD-associated modules were detected in our second non-overlapping dataset of U.S. Marines to demonstrate a significant and consistent association of our findings. We conclude that prospectively profiling the transcriptome of U.S. Marines pre- and post-deployment to conflict zones, using a co-expression analysis approach is a promising strategy for identifying and studying the functions of causal and consequential molecular factors in PTSD development, with particular value in reproducing results across independent datasets of U.S. Marines.
SUBJECTS AND METHODS
Sample Collection and Datasets
All subjects were male and participants in either the Marine Resiliency Study (MRS) or the Marine Resiliency Study II (MRS II), prospective studies of well-characterized U.S. Marines scheduled for combat deployment to Iraq or Afghanistan, with longitudinal follow-up to track the effect of combat stress.
Dataset 1 - Whole blood was obtained from 124 MRS II U.S. Marine participants who served a seven month deployment. Blood was drawn 1-month prior to deployment and again at 3-months post-deployment for each participant. Each blood sample (10ml) was collected into an EDTA-coated collection tube, RNA was isolated from peripheral blood leukocytes using LeukoLOCK Total RNA isolation and sequenced using the Illumina Hi-Seq 2000.
Dataset 2 - For validation, data were compared to an independently generated gene expression data-set from a separate, non-overlapping, group of 50 MRS Marine participants (Glatt et al. 2013, previously published pre-deployment data12). Blood samples were treated in an identical fashion as described above, however final RNA was hybridized to the Affymetrix Hu-Gene 1.0 ST Array.
PTSD Diagnosis
At the time of each blood draw, PTSD symptoms were assessed using a structured diagnostic interview, the Clinician Administered PTSD Scale (CAPS)(20–23). Using the criteria from the Diagnostic and Statistical Manual of Mental Disorders, fourth edition (2000)24, diagnosis for partial or full PTSD was defined as a threat to life, injury, or physical integrity (Criterion A1) and the presence of at least one re-experiencing symptom and either three avoidance symptoms or two hyperarousal symptoms, or two avoidance symptoms plus two hyperarousal symptom(25–27). Symptoms must have occurred at least once within the past month (frequency ≥ 1) and caused a moderate amount of distress (intensity ≥ 2).
Subject selection
A subset of MRS study participants were pre-selected for RNA-Seq analysis. First, at pre-deployment, all participants had to be symptom free, with no PTSD diagnosis and a CAPS ≤ 25. Second, at post-deployment, participants who fulfilled criteria for partial or full PTSD diagnosis were designated the PTSD group. Third, participants with post-deployment CAPS ≤ 25 that matched the post-deployment PTSD group on variables of combat exposure, age and ethnicity were designated the “control” group. Under these criteria, all paired subjects were stratified into two groups based upon CAPS scores at 3-months post-deployment (Table 1, Supplementary Table 1). If a Marine participant developed PTSD following trauma-exposure at 3-months post-deployment, their pre-deployment sample would be included in the ‘PTSD-risk’ group. Likewise, if a subject avoided PTSD symptoms at 3 months post-deployment their sample at pre-deployment was included in the ‘control’ group.
Table 1.
Recorded clinical parameters from U.S. Marines assessed at pre- and post-deployment for Dataset 1.
| Time point | Pre-Deployment | Post-Deployment | ||||
|---|---|---|---|---|---|---|
| PTSD Cases (N=47) |
Controls (N=47) |
P-Value | PTSD Cases (N=47) |
Controls (N=47) |
P-Value | |
| Age | 22.15 ± 2.53 | 22.42 ± 3.92 | 0.682 | 23.14 ± 2.52 | 23.42 ± 3.92 | 0.685 |
| Alcohol | 2.08 ± 1.55 | 1.62 ± 1.33 | 0.124 | 1.79 ± 1.32 | 1.54 ± 1.11 | 0.318 |
| Tobacco | 1.75 ± 1.62 | 0.97 ± 1.51 | 0.02 | 1.69 ± 1.69 | 1.02 ± 1.47 | 0.042 |
| WC adj. | 1.65 ± 0.13 | 1.72 ± 0.13 | 0.015 | 1.68 ± 0.14 | 1.75 ± 0.12 | 0.012 |
| PCL | 21.29 ± 4.72 | 18.33 ± 2.27 | 0.0001 | 42.38 ± 11.09 | 20.94 ± 3.87 | 5.37E-22 |
| CAPS total | 11.39 ± 7.23 | 6.75 ± 6.90 | 0.002 | 53.17 ± 15.08 | 10.04 ± 7.26 | 5.99E-32 |
| CAPSBs | 1.00 ± 1.91 | 0.54 ± 1.92 | 0.245 | 14.9 ± 7.25 | 1.54 ± 2.37 | 6.29E-21 |
| CAPSCAs | 0.54 ± 1.11 | 0.10 ± 0.51 | 0.015 | 5.31 ± 4.57 | 0.85 ± 2.08 | 1.88E-08 |
| CAPSCN1s | 1.10 ± 2.23 | 0.97 ± 2.88 | 0.813 | 9.17 ± 5.32 | 1.19 ± 2.87 | 1.21E-14 |
| CAPSDs | 8.39 ± 5.66 | 4.58 ± 4.98 | 0.001 | 22.6 ± 6.7 | 6.42 ± 4.79 | 5.97E-24 |
| CAPSCs | 2.00 ± 2.73 | 1.62 ± 3.66 | 0.571 | 15.67 ± 7.23 | 2.08 ± 3.66 | 7.15E-20 |
| Prior Deployment | 19 | 16 | 0.6699 | - | - | - |
| TBI | - | - | - | 30 | 21 | 0.097 |
| CES PBE mean | - | - | - | 0.63 ± 0.25 | 0.53 ± 0.12 | 0.02 |
| Caucasian | 26 | 26 | 1 | - | - | - |
| African American | 4 | 4 | 1 | - | - | - |
| Native American Mexican | 13 | 15 | 0.822 | - | - | - |
| Asian & Other | 5 | 3 | 0.714 | - | - | - |
Abbreviations: Alcohol = alcohol consumption; Tobacco = tobacco use; WC adj. = waist circumference was adjusted for height; PCL = PTSD symptom check list, CAPS total = CAPS total score, CAPSBs = re-experiencing subscale, CAPSCAs = symptoms of avoidance, CAPSCN1s = symptoms of numbing, CAPSCs = subtotal C subscale, CAPSDs = hyper-arousal subscale, TBI = traumatic brain injury, CES = combat exposure scale, PBE = post battle experience; -, not applicable. Significance was assessed with a Student’s two-tailed t test for continuous variables and fishers exact
Data Pre-Processing
All data were pre-processed by normalization, filtering genes with low expression values, and removing any outliers which may bias down-stream analysis. Final subject numbers resulted in 94-paired subjects (47 paired cases and 47 paired controls) in Dataset 1 and 48 paired subjects (24 paired cases and 24 paired controls) in Dataset 2. To compare findings from RNA-Seq data in Dataset 1 to microarray data in Dataset 2, genes found only on both platforms (N=10,184) passed into our subsequent analysis (see Supplementary File for more detailed information).
Differential Gene Expression Analyses
Differentially expressed genes were assessed using the moderated t-test in edgeR28 and LIMMA29 packages for RNA-Seq and microarray data, respectively, and unless otherwise specified, the significance threshold was a nominal p-value < 0.05. A nominally significant p-value was used to yield a reasonable number of genes to include within network analyses. Differential expression analyses were performed on 10,184 genes between pre-deployment PTSD case and control groups, and again between post-deployment PTSD case and control groups (see Supplementary File for more detailed information).
Gene network construction and module detection
Signed co-expression networks were built using weighted gene co-expression network analysis (WGCNA)13 in R. A total of 10,184 genes were used to construct each network. To construct the networks, the absolute values of Pearson correlation coefficients were calculated for all possible gene pairs and resulting values were transformed so that the final matrix followed an approximate scale-free topology (see Supplementary File for detailed information). The WGCNA dynamic tree-cut algorithm was used to detect network modules. In order to determine which modules, and corresponding processes were most associated to PTSD related states, we ran singular value decomposition on each module’s expression matrix and used the resulting module eigengene (ME), which is equivalent to the first principal component13, to represent the overall expression profiles for each module.
For each gene in a module, module membership (kME) was defined as the correlation between gene expression values and ME expression. Genes with high kME inside co-expression modules are labeled as hub genes13. GS was calculated as the −log10 of the p-value generated for each gene within a particular module using a moderated t test and is a measure of the strength of differential gene expression between PTSD cases and controls. MS was calculated as the average GS within each module (see Supplementary File for more information).
Statistical Analyses
All gene-set overlap analyses were performed by assessing the cumulative hypergeometric probability using the phyper function in R.
Enrichment Analyses
Module enrichment was assessed three ways. First, general module enrichment categories were obtained using gene ontology biological processes from the DAVID database30 (http://david.abcc.ncifcrf.gov/). Second, specific module enrichment categories were obtained using the WGCNA function userlistEnrichment31 using modules as input-lists and curated Reactome NCBI Biosystems pathways and terms32 as user-defined lists. Finally, we downloaded the highly expressed, cell specific (HECS) gene expression database compiled by Shoemaker et al.33 to assess cell-type specific enrichment results, here cell-type marker lists were used as a user-defined lists. All module genes were used for enrichment analyses using a FDR corrected p-value < 0.05 as significant.
Data Availability
RNA-Seq and microarray gene expression data are freely available at the Gene Expression Omnibus under the SuperSeries accession number GSE64814 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64814).
Full Methods and any associated references are available in Supplementary Methods.
RESULTS
We analyzed two different gene expression datasets generated from RNA-Seq (Dataset 1, Table 1) and microarray (Dataset 2, Supplementary Table 1) using peripheral blood leukocyte (PBL) samples taken from U.S. Marines pre- and post-deployment. Following a set of differential gene expression analyses (Supplementary Figure 1), we aimed to characterise the prognostic and diagnostic molecular signatures of PTSD by studying transcriptional differences at the systems-level at pre-deployment and post-deployment separately. Initially, WGCNA was used in Dataset 1 to assess module preservation between PTSD cases (N = 47) and controls (N = 47) for the pre- and then the post-deployment time point (see Supplementary File for complete description). This analysis identifies large differences in gene co-regulatory patterns, as being disrupted or created in PTSD cases relative to controls, or vis-versa. However, we observed strong preservation statistics between the two groups indicating similar fundamental gene co-regulation within PTSD cases and controls, suggesting that major changes in the underlying gene-gene connectivity are not a basis for the pathology of this disorder (Supplementary Table 2). As a result we used the higher confidence and completeness of a combined network of case and control data.
Differential module expression post-deployment in Dataset 1
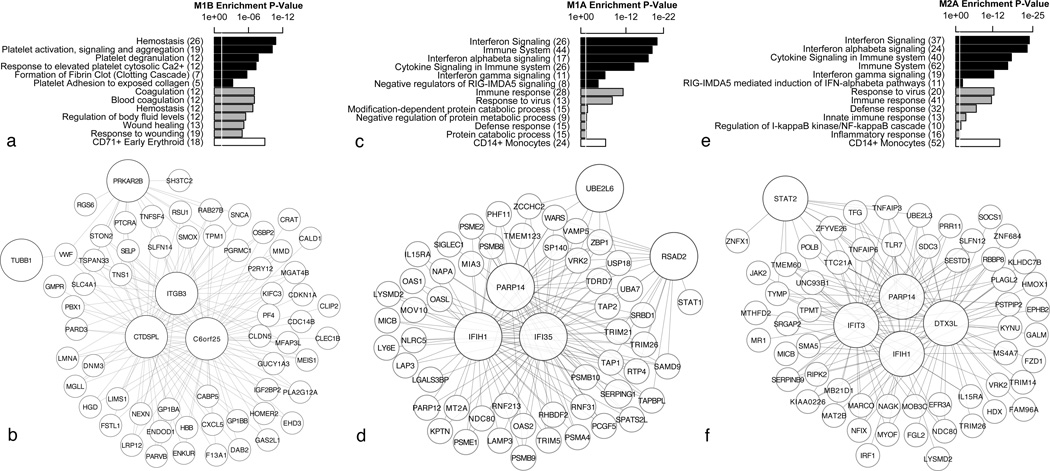
We constructed a gene co-expression network from a combination of PTSD cases (N = 47) and controls (N = 47) post-deployment using RNA-Seq expression data from Dataset 1 (Figure 1). This analysis identified nine modules (fully characterised in Supplementary Table 3) that were first examined for enrichment of differentially expressed genes. Two modules (M1A and M1B) were enriched for genes identified as differentially expressed between PTSD cases and controls, reflected by an elevated module significance (MS) value (Figure 2A). To determine if the overall expression of modules M1A and M1B were significantly associated with PTSD group status, we calculated differences in module expression using module eigengene (ME) values (See Materials and Methods for complete description of ME). Consistent with results using MS, expression of module M1B was significantly higher in the PTSD resilient control group (p=0.004 and Figure 2B) suggesting a positive correlation to PTSD resiliency, meanwhile expression of module M1A was significantly higher in the PTSD group (p=0.02, Figure 2B). Subsequently, ME values for each module were correlated to all clinical parameters, found in Table 1, to determine module-trait relationships. The ME for module M1B was significantly correlated to post-deployment PTSD resilient controls (r=0.29, p=0.005), negatively correlated to post-deployment CAPs and PCL (CAPs, r =−0.27, p =0.009; PCL r=−0.28, p=0.007) and negatively correlated other measures of CAPS (Supplementary Table 3) but not correlated to any other measured clinical variable, suggesting that differential gene expression in M1B was not confounded by recorded measurements such as body-mass-index, smoking, or alcohol consumption. Genes in M1B were expressed to a greater extent in PTSD resilient controls (Figure 2B) while enrichment analysis revealed a significant association with hemostasis, platelet activation and wound healing (Figure 3A). Further, enrichment for cell-type specificity revealed on over-representation of erythroid expression markers (blood platelets). Hub genes are those most strongly correlated to the ME value for a particular module and represent possible disease associated markers13, in this case putative PTSD-resiliency markers. The top 5 hub genes in M1B (C6orf25, CTDSPL, ITGB3, PRKAR2B and TUBB1) were are all associated with hemostasis and in particular, with platelet regulation and function34–37 (Figure 3B).
Figure 1.
Hierarchical cluster tree and post-deployment module structure in Dataset 1. Hierarchical cluster tree (dendrogram) of the combine post-deployment network of PTSD cases (N=47) and controls (N=47) comprising 10,184 genes. Each line represents a gene (leaf) and each low-hanging cluster represents a group of co-expressed genes with similar network connections (branch) on the tree. The first band underneath the tree indicates the nine detected, and subsequently analyzed, network modules. Genes shaded in grey were not assigned to a particular module and represent background noise. For a comprehensive functional annotation of each module and calculation of all significant module-trait relationships see Supplementary Table 3.
Figure 2.
Module significance (MS) and module eigengene (ME) expression boxplots. MS was measured across all pre- and post-deployment modules in Dataset 1. WGCNA detected ten modules post-deployment from a combination of PTSD cases and control (a) and twenty-two modules at pre-deployment from a combination of PTSD risk cases and controls (c). The y-axis indicates MS by calculating the average −log10 p-values, generated by a moderated t test, for each gene within a particular module, when assessing differential expression between PTSD cases and controls. Here, a kruskal-wallis p-value was used only for descriptive purposes and not inferential. Modules denoted with an asterisk (*) have ME values significantly correlated to conditional states (i.e. PTSD cases or controls). Representative modules with high MS at post-deployment and pre-deployment were investigated for module expression differences. Differences in ME expression were measured using a two-tailed student’s t test on and a p-value < 0.05 is considered significant. Boxplots are displayed for each main group. Significant differences in ME expression were observed in post-deployment modules M1B and M1A (b) and in pre-deployment module M2A (d).
Figure 3.
Module characterization for Dataset 1. Enrichment analysis and correlation networks for modules M1B (a & b) and M1A (c & d) identified post-deployment, and module M2A (e & f) identified pre-deployment in Dataset 1. Enrichment analysis was used to identify the top 6 REACTOME ontology terms (black bars), the top 6 DAVID ontology terms (grey bars) and the most significant cell-type signature (white bar) over-represented in the list of genes within each module. All terms were deemed significant as assessed by a hypergeometric test FDR corrected p-value <0.05 displayed as a white line. The total number of genes within each significant term is denoted within the brackets associated with that term. Gene-networks were constructed selecting the top 150 most significant connections ranked by kME. Nodes represent genes and edges represent correlations. The top 5 hub genes, those most correlated to ME values, are shown in larger sizes.
The ME for module M1A was significantly correlated to PTSD cases (r = 0.23, p = 0.03), post-deployment CAPs criteria of avoidance (CAPSCA, r = 0.32, p = 0.002) and post-deployment CAPs criteria of re-experiencing (CAPSBs, r=0.2, p=0.05) but to no other variables (Supplementary Table 3). Genes in M1A were over-expressed in PTSD cases (Figure 2B) while enrichment analysis revealed a significant association with immune response as exemplified by innate responses mediated by interferon (IFN) signalling (Figure 3C), as well as with monocyte specific markers. The top 5 hub genes in M1A included IFI35, IFIH1, PARP14, RSAD2 and UBE2L6; all well described interferon stimulated genes38 and here considered putative PTSD-associated markers (Figure 3D).
Differential module expression pre-deployment in Dataset 1
It is unclear whether the modules identified post-deployment are causal of PTSD development or are simply a consequence of the disorder. To determine if any post-deployment modules could be re-identified and thus associated as causal modules, we constructed a gene co-expression network combining RNA-Seq gene expression data from PTSD-risk cases (N = 47) and controls (N = 47) pre-deployment in Dataset 1. Twenty-two pre-deployment modules were identified (fully characterised in Supplementary Table 4) whereby a single module (M2A) was enriched for differentially expressed genes between PTSD-risk participants and controls as reflected by an elevated MS value (Figure 2C). Along the same lines, M2A module expression was significantly higher in the PTSD risk group (p=0.001 and Figure 2D). Module M2A ME was significantly correlated to one variable, PTSD-risk (r=0.32, p=0.002, Supplementary Table 4). Similar to module M1A that was identified post-deployment, enrichment analysis of genes in M2A revealed a significant association with innate immune responses, IFN signalling and monocyte specificity (Figure 3E). The top 5 hub genes were again associated with IFN signalling (DTX3L, IFIH1, IFIT3, PARP14 and STAT2) (Figure 3F). Gene-set overlap analysis compared all of the genes in M2A pre-deployment (n = 245) to those in M1A post-deployment (n =115) to reveal a significant overlap (∩ = 108, p = 6.7e-181, Figure 4).
Figure 4.
Venn Diagram of Innate Immune Modules across Dataset 1 and Dataset 2. Venn Diagram (a) depicting significant overlap in genes belonging to modules M1A post-deployment and M2A pre-deployment in Dataset 1 as well as modules M3A post-deployment and M4A pre-deployment in Dataset 2. Gene overlap (∩) with associated hypergeometric p-value, in italics, are depicted for all pairwise comparisons of module genes (b). The overlap identified 51 genes found across all four analyses (c) which are displayed in the table along with the corresponding kME rank (i.e. rank of connectivity) for each gene within a particular module. A high rank indicates hub gene status (i.e. PTSD risk and PTSD associated markers). Numbers in bold outline the top 10 hub genes across each module, respectively. Genes are ordered accordingly to M2A kME. All 51 genes are displayed via heatmap in Supplementary Figure 4.
Validation of differential module expression post-deployment in Dataset 2
To validate post-deployment findings in Dataset 1 we assessed Dataset 2 for similar network properties in a combined network analysis of PTSD cases (N = 24) and controls (N = 24) post-deployment. Out of 8 modules (full characterisation Supplementary Table 5), a single module (M3A) contained an enrichment of differentially expressed genes (Supplementary Figure 2A) demonstrating a modest, yet insignificant, increase in module expression within the PTSD group (p = 0.1, Supplementary Figure 2B). The ME was significantly correlated to post battle experience (r = 0.4, p = 0.004), post-deployment CAPS (r=0.32, p=0.03) and weakly correlated to a PTSD cases (r = 0.21, p = 0.1, Supplementary Table 5). The genes in this module were over-expressed in PTSD cases relative to controls (Supplementary Figure 2B) and enrichment analysis revealed a significant association with innate immune responses, IFN signalling and monocytes (Supplementary Figure 3A). The top 5 hub genes (DDX58, IFI35, IFIT5, PARP9 and ZBP1) were again all associated with IFN signalling (Supplementary Figure 3B). A highly significant overlap in post-deployment module genes across M1A (n=115) in Dataset 1 and M3A (n=83) in Dataset 2 (∩ = 63, p = 2.0E-105, Figure 4B) confirmed the identification of a dysregulated innate immune module related to PTSD cases across two independent datasets.
Validation of differential module expression pre-deployment in Dataset 2
To re-confirm pre-deployment findings from Dataset 1, PTSD-risk cases (N=24) and controls (N=24) pre-deployment were combined from Dataset 2 and subjected to network analysis which identified 11 modules (full characterisation in Supplementary Table 6). A single module (M4A) was enriched for differentially expressed genes between PTSD-risk cases and controls (Supplementary Figure 2C). The PTSD-risk group displayed a significant over-expression of module expression (p = 0.01, Supplementary Figure 2D). The ME for M4A was significantly correlated to PTSD-risk (r = 0.36, p = 0.01) and CAPs (r=0.44, p=0.002, Supplementary Table 6). Moreover, enrichment analysis of M4A revealed a significant association with innate immune responses, IFN signalling and monocytes (Supplementary Figure 3C), and the top 5 hub genes (PARP9, UBE2L6, STAT2, TRIM22 and GBP1) were again all associated with IFN signalling (Supplementary Figure 3D). All pairwise gene-set overlap analyses across modules M1A, M2A, M3A and M4A revealed a highly significant overlap (Figure 4B) and hub gene expression for these modules showed elevated expression in PTSD groups when compared to controls both pre- and post-deployment across both datasets (Supplementary Figure 4). These results demonstrate the association of a dysregulated innate immune module, related to IFN signalling, which appears to define at least part of the pathophysiology of PTSD through causal association to PTSD development.
DISCUSSION
We investigated the high-order system-level properties of PTSD using an unsupervised network-based approach (WGCNA) to identify differences at the gene co-expression level, rather than investigating at the individual gene level. Gene expression data were generated by RNA-Seq (Dataset 1) and microarray (Dataset 2) using PBL samples isolated from U.S. Marines pre- and post-deployment to conflict zones (i.e. Iraq and Afghanistan). Our comprehensive and prospective experimental design allowed the investigation of both biological processes that define PTSD and those driving the development of this disorder, and further, allowed the re-confirmation of findings in an independent dataset. This is the first time dysregulated gene networks specific for innate immunity have been used to characterise causal and consequential molecular signatures of PTSD and then to further replicated these findings across independent datasets.
A novel finding from our network analyses was the identification of modules related to hemostasis and wound responsiveness expressed to a greater extent post-deployment in US Marines who did not develop PTSD (Figure 2B), as in module M1B (Figure 3A). Interestingly, the three other network analyses also detected modules related to hemostasis and wound response with significant overlap (M16 pre-deployment Dataset 1; M7 and M6 indented post- and pre-deployment in Dataset 2; Supplementary Figure 5, Supplementary Tables 4–6). These other modules revealed patterns of heterogeneous gene expression irrespective of group status and time-point suggesting that these modules and corresponding processes may infer wound resilience in only a small subset of individuals. Along these lines, it has been well documented that different degrees of stress will elicit different stress responses (review39), and in particular, a response involving blood platelets, has been shown to be a critical biomarker of hemostatic, thrombotic, and inflammatory challenges to an organism and a key player in cardiovascular disease and chronic based stress, as in PTSD(40,41). Moreover, in a review of a large number of studies examining various tissue types, it was found that different types of psychological stress were associated with impaired wound healing42. A meta-analysis found an inverse correlation (r = −0.42) between psychological stress and wound healing43 supporting the positive association between wound healing and PTSD resilience (r =0.29, p =0.005) found in this study. This suggests that high levels of stress may hinder proper wound healing during/after battlefield trauma, although the degree of such stress appears to be a key factor for establishing associations with the hemostatic system.
Our central finding was the identification of a dysregulated innate immune module associated with the development of PTSD (Figure 2,3, Supplementary Figure 3,4), illuminated by the replication of modules post-deployment (M1A and M3A) and those pre-deployment (M2A and M4A) that could be associated with PTSD. These findings suggest that differences in innate immunity modules were not simply a consequence of the PTSD state post-deployment but also have causal relevance for PTSD development and explain at least part of the pathophysiology of the disorder, exemplified by their identification pre-deployment. These results highlight our differential expression analyses (Supplementary Figure 1) and our previous reports of C-reactive protein (CRP), a general marker of immune activation and inflammation, and 5’-oligoadenylate synthetase genes (i.e. OAS1, OAS2, OAS3) as markers of the antiviral interferon response, that were associated with an increased risk of developing PTSD(44,12). However, our current findings dramatically extend these results by showing that the IFN response is being modulated to a much greater extent than previously thought pre- and post-deployment. Notably, a number of single case studies have reported that treatment of hepatitis C virus (HCV) infected PTSD subjects with recombinant interferon (IFN- α2b) precipitated PTSD symptoms(45,46). In our study, where subjects were not receiving IFN therapy, it is unclear what is stimulating the IFN response.
Our observations lead to several fundamental questions and some putative solutions. First, how does one interpret the over-expression of innate immunity genes found prior-to trauma? One possible explanation is that both acute and severe stress, predictors in their own right for PTSD, are also associated with the hyper-activation of the immune system and subsequent inflammation(47,48). An alternative hypothesis is that stress, pathogens and/or high viral loads may ‘prime’ the immune system, driving the IFN response, altering a subsequent response to trauma. Along these lines, studies focusing on the gut-brain barrier have shown that intestinal mucosal dysfunction, defined as increased translocation of gram-negative bacteria (leaky gut), plays a role in the inflammatory pathophysiology of depression suggesting that differences in gut flora may stimulate an IFN response49. Second, does a dysregulated innate immune module pre-deployment hold predictive value? Our previous work constructing a prognostic classifier from Dataset 2 pre-deployment participants12 suggests that immune-related genes do hold predictive value although these results have not yet been replicated across larger datasets using machine-learning methods. Inferring the prognostic relevance of network-based applications remains challenging. However, cross-referencing our findings with this previous work suggests that network statistics, and our innate immune modules, do have potential to contain predictive value. Third, out of the entire network of pairwise correlations between genes across the transcriptome, are the most informative genes interconnected within similar modules or spread out across numerous modules? A possible limitation of this study was that by analyzing co-regulated modules of genes we may have missed individual genes, which do not correlate within our modules of interest although are of functional relevance to PTSD. For example, previous reports specifically target FKBP5 and STAT5B as differentially expressed biomarkers(3–8,11,12) although they were not assigned to co-expressed modules nor found to be significantly differentially expressed between PTSD cases and controls. Finally, of what relevance is PBL gene expression for a disorder primarily associated with the brain? In this study we identify innate immunity and IFN signalling genes whose expression was elevated in PBLs both before and after the development of PTSD (Figure 2 and Supplementary Figure 4). Although the recruitment of such signalling could be triggered by various factors, they ultimately release toxic compounds including degradative enzymes and reactive oxygen species that can impair cellular processes(50–53). It could be hypothesized that the accumulation of these compounds in the blood prior-to-deployment may be detrimental to the brain if the integrity of the blood-brain-barrier (BBB) was then compromised by injury (e.g. TBI). An increasing body of evidence indicates that changes in the blood may seed pathology in the brain across various disorders. In a recent Multiple Sclerosis study, Minagar and Alexander54 investigate the association of INF with the BBB suggesting that IFN-γ and other proinflammatory cytokines (TNF-α and IL-1β) disrupt the BBB through a variety of mechanisms. Further, Alzheimer’s disease models suggest that breaches in the BBB lead to leakage into the brain of blood-borne molecules that are toxic to neurons and cause neurodegenerative changes55. Future studies investigating the role of the BBB in PTSD may provide a detailed explanation for a specific course of PTSD development.
In summary, our data provide a global framework for previously unknown molecular aspects of PTSD and describe a new context concerning the complex pathophysiological nature of PTSD development. Specifically, modules of co-expressed genes associated with the innate immune response and IFN signalling appear to be implicated in the development of PTSD and continue to persist once the disorder is established. Modules associated with hemostasis and wound healing may contribute to resilience against developing PTSD. It is hoped that this study will lead to future work confirming the importance of differences in innate immune factors to the development of PTSD and the role of platelets in the stress response. Ideally, these findings will allow for advanced PTSD detection, which could delay or abrogate PTSD development by identifying susceptible service members prior to deployment to conflict zones by either removing the causal path (i.e. trauma exposure) or through early intervention of new therapies to modulate the interferon signature.
Supplementary Material
Differential gene expression analyses were performed using a moderated t statistic within Dataset 1 and Dataset 2. A cross-sectional analysis compared PTSD cases to controls post-deployment in Dataset 1 (47 PTSD cases vs. 47 controls) and subsequently in Dataset 2 (24 PTSD cases vs. 24 controls) to reveal 294 and 61 differentially expressed genes, respectively, with a significant overlap (a). The top 3 most significant biological processes (annotated with DAVID) based on over-expressed and under-expressed genes identified from Dataset 1 are reported (b). Subsequently, the same paired data were analyzed pre-deployment for Dataset 1 and Dataset 2 revealing 662 and 178 differentially expressed genes, respectively, with a significant overlap (c). The top 3 most significant biological processes from Dataset 1 are reported (d). Utilizing, the paired structure of the data, a longitudinal contrast analysis was applied to identify genes behaving differently across time within the PTSD and control groups. This analysis revealed a total of 177 genes in Dataset 1 and 110 genes in Dataset 2, with minimal overlap (e) and no significant functional annotation (f). Up and down symbols are relative to the PTSD group. Significance threshold for genes was set to a nominal p < 0.05 where as we used a more strict threshold for functional annotations with a Bonferroni corrected p-value < 0.05.
Module Significance (MS) across (a) pre- and (c) post-deployment modules in Dataset 2. WGCNA detected nine modules post-deployment from a combination of PTSD cases and control (a) and eleven modules at pre-deployment from a combination of PTSD risk cases and controls (b). The y axis indicates MS by calculating the average −log10 p-values, generated by a moderated t-test, for each gene within a particular module, when assessing differential expression between PTSD cases and controls. Here, a kruskal-wallis p-value was used only for descriptive purposes and not inferential. Modules denoted with an asterisk (*) have module eigengenes values (ME) significantly correlated to group status (i.e. PTSD cases or controls). Differences in ME expression were measured using a two-tailed student’s t-test on and a p-value < 0.05 is considered significant. Boxplots are displayed for each main group. Post-deployment module M3B displayed a moderate, yet insignificant, over-expression (b) and pre-deployment M4A displayed a significant over-expression (d).
Enrichment analysis and correlation networks for module M3A (a & b) identified post-deployment and module M4A (c & d) identified pre-deployment in Dataset 2. Enrichment analysis was used to identify the top 6 Reactome ontology terms (black bars), all DAVID ontology terms (grey bars) and the top cell-type signature (white bar) over-represented in the list of genes in each module. All terms were deemed significant as assessed by a hypergeometric test (FDR corrected p-value <0.05). Gene-networks were constructed selecting the top 150 most significant connections. Nodes represent genes and edges represent correlations. The top 5 hub genes with the highest correlation with ME are shown in larger sizes.
Heatmaps representation of the 51 overlapping genes from module M1A, M2A, M3A, and M4A. Heatmaps are divided into four main parts. Dataset 1 modules (a), M1A on the top row and M2A on the bottom row, are split into PTSD cases (left column) and controls (right column). Dataset 2 modules (b), M3A on the top row and M4A on the bottom row, are split into PTSD cases (left column) and controls (right column). All subjects have been sorted in an identical fashion in order to properly visualize differences in expression as occurring within each individual sample across the two time-points (i.e. Subject 1 post-deployment is directly above Subject 1 pre-deployment). Red represents over-expression, green represents under-expression and black represents the mean.
Venn Diagram (a) depicting significant overlap in hemostasis and blood coagulation genes belonging to modules M1B post-deployment and Module 16 pre-deployment in Dataset 1 as well as modules Module 7 post-deployment and Module 6 pre-deployment in Dataset 2. The overlap identified 46 genes found across all four analyses which are displayed in table format (b) along with the corresponding kME rank (i.e. rank of connectivity) for each gene within a particular module. Higher rank indicates hub gene status, the top 10 genes for each module are in bold. Heatmaps representing these 46 genes are shown across both datasets and are divided into four main parts. Dataset 1 modules (c), M1B on the top row and Module 16 on the bottom row, are split into PTSD cases (left column) and controls (right column). Dataset 2 modules (d), Module 7 on the top row and Module 6 on the bottom row, are split into PTSD cases (left column) and controls (right column). All subjects have been sorted in an identical fashion in order to properly visualize differences in expression as occurring within each individual sample across the two time-points (i.e. Subject 1 post-deployment is directly above Subject 1 pre-deployment). Red represents over-expression, green represents under-expression and black represents the mean according to the scale bar according to scale bar.
Acknowledgements
We thank and extend our deepest gratitude to Dr. Daniel T. O’Connor whose passion and intellect of clinical, translational, and basic research brought considerable knowledge and insight to various realms of this research project and numerous others. This work was supported in part by the Naval Medical Research Center's Advanced Medical Development program (Naval Medical Logistics Command Contract #N62645-11-C-4037, for MRS II (D.G.B), and this Demonstration Project (C.N. and D.O.C.). Replication of findings on a non-overlapping cohort was supported in part by both the National Institute of Mental Health R21 (MH085240) and R01 (MHO85560). Further support was provided by R01 (MH085521), the Gerber Foundation, the Sidney R. Baer, Jr. Foundation and NARSAD: The Brain and Behavior Research Foundation. We acknowledge assistance of the MRS-II administrative core, A. Patel, A. De La Rosa and other members of the MRS-II Team. Likewise, we acknowledge administrative support from the Veterans Medical Research Foundation (VMRF) and valuable input from M.E. Polak, D. Baldwin and A. Collins who assisted in critical reading of the manuscript. We also thank the Marine and Navy volunteers for their military service and for their participation in this study.
Footnotes
Author Contributions: D.G.B., C.N., C.H.W and D.O.C. obtained the funding for this study. A.X.M curated clinical information regarding all participants. S.J.G., D.S.T. and S.D.C. generated microarray data. M.S.B conducted the study which entailed generating RNA-Seq data, writing code for quality testing and computational interrogation of both RNA-Seq and microarray data. M.S.B. drafted and wrote the manuscript with the participation of remaining authors.
The authors declare no conflict of interest.
References
- 1.Ramchand R, Schell TL, Karney BR, Osilla KM, Burns RM, Caldarone LB. Disparate prevalence estimates of PTSD among service members who served in Iraq and Afghanistan: possible explanations. J. Trauma. Stress. 2010;23:59–68. doi: 10.1002/jts.20486. [DOI] [PubMed] [Google Scholar]
- 2.Heinzelmann M, Gill J. Epigenetic Mechanisms Shape the Biological Response to Trauma and Risk for PTSD: A Critical Review. Nursing Research and Practice. 2013;2013:1–10. doi: 10.1155/2013/417010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zieker J, Zieker D, Jatzko A, Dietzsch J, Niesel K, Schmitt A, et al. Differential gene expression in peripheral blood of patients suffering from post-traumatic stress disorder. Molecular Psychiatry. 2007;12.2:116–118. doi: 10.1038/sj.mp.4001905. [DOI] [PubMed] [Google Scholar]
- 4.Yehuda R, Holsboer F, Buxbaum JD, Miller-Myhsok B, Schmeidler J, Rein T, et al. Gene Expression Patterns Associated with Posttraumatic Stress Disorder Following Exposure to the World Trade Center Attacks. Biological Psychiatry. 2009;66.7:708–711. doi: 10.1016/j.biopsych.2009.02.034. [DOI] [PubMed] [Google Scholar]
- 5.Neylan TC, Sun B, Rempel H, Ross J, Lenoci M, O’Donovan A, et al. Suppressed monocyte gene expression profile in men versus women with PTSD. Brain, Behavior, and Immunity. 2011;25.3:524–531. doi: 10.1016/j.bbi.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarapas C, Cai G, Bierer LM, Golier JA, Galea S, Ising M, et al. Genetic Markers for PTSD Risk and Resilience Among Survivors of the World Trade Center Attacks. Disease Markers. 2011;30.2–3:101–110. doi: 10.3233/DMA-2011-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehta D, Gonik M, Klengel T, Rex-Haffner M, Menke A, Rubel J, et al. Using Polymorphisms in FKBP5 to Define Biologically Distinct Subtypes of Posttraumatic Stress Disorder: Evidence From Endocrine and Gene Expression Studies. Archives of General Psychiatry. 2011;68.9:901–910. doi: 10.1001/archgenpsychiatry.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pace TW, Wingenfeld K, Schmidt I, Meinlschmidt G, Hellhammer DH, Heim CM. Increased peripheral NF-KB pathway activity in women with childhood abuse-related posttraumatic stress disorder. Brain, Behavior, and Immunity. 2012;26.1:13–17. doi: 10.1016/j.bbi.2011.07.232. [DOI] [PubMed] [Google Scholar]
- 9.van Zuiden M, Heijnen CJ, Maas M, Amarouchi K, Vermetten E, Geuze E, et al. Glucocorticoid sensitivity of leukocytes predicts PTSD, depressive and fatigue symptoms after military deployment: A prospective study. Psychoneuroendocrinology. 2012;37.11:1822–1836. doi: 10.1016/j.psyneuen.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 10.van Zuiden M, Geuze E, Willemen HL, Vermetten E, Maas M, Amarouchi K, et al. Glucocorticoid Receptor Pathway Components Predict Posttraumatic Stress Disorder Symptom Development: A Prospective Study. Biological Psychiatry. 2012;71.4:309–316. doi: 10.1016/j.biopsych.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 11.Matić G, Milutinović DV, Nestorov J, Elaković I, Jovanović SM, Perišić T, et al. Lymphocyte glucocorticoid receptor expression level and hormone-binding properties differ between war trauma-exposed men with and without PTSD. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2013;43:238–245. doi: 10.1016/j.pnpbp.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Glatt SJ, Tylee DS, Chandler SD, Pazol J, Nievergelt CM, Woelk CH, et al. Blood-based gene-expression predictors of PTSD risk and resilience among deployed marines: A pilot study. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2013;162.4:313–326. doi: 10.1002/ajmg.b.32167. [DOI] [PubMed] [Google Scholar]
- 13.Langfelder P, Horvath S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinformatics. 2008;9.1:559.13. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller JA, Oldham MC, Geschwind DH. A Systems Level Analysis of Transcriptional Changes in Alzheimer's Disease and Normal Aging. Journal of Neuroscience. 2008;28.6:1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saris C, Horvath S, van Vught PWJ, vanEs MA, Blaue HM, Fuller TF, et al. Weighted gene co-expression network analysis of the peripheral blood from Amyotrophic Lateral Sclerosis patients. BMC Genomics. 2009;10.1:405. doi: 10.1186/1471-2164-10-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474.7351:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang Y, Kim J, Shin JY, Kim JI, Seo JS, Webster MJ, et al. Gene expression profiling by mRNA sequencing reveals increased expression of immune/inflammation-related genes in the hippocampus of individuals with schizophrenia. Translational Psychiatry. 2013;3.10:e321. doi: 10.1038/tp.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, Cheng L, Grennan K, Pibiri F, Zhang C, Badner JA, et al. Two gene co-expression modules differentiate psychotics and controls. Molecular Psychiatry. 2012;18.12:1308–1314. doi: 10.1038/mp.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torkamani A, Dean B, Schork NJ, Thomas EA. Coexpression Network Analysis of Neural Tissue Reveals Perturbations in Developmental Processes in Schizophrenia. Genome Research. 2010;20.4:403–412. doi: 10.1101/gr.101956.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Gusman FD, Charney DS, et al. The development of a Clinician-Administered PTSD Scale. J. Trauma. Stress. 1995;8:75–90. doi: 10.1007/BF02105408. [DOI] [PubMed] [Google Scholar]
- 21.King DW, Leskin GA, King LA, Weathers FW. Confirmatory factor analysis of the Clinician-Administered PTSD Scale: Evidence for the dimensionality of posttraumatic stress disorder. Psychol. Assess. 1998;10:90–96. [Google Scholar]
- 22.Weathers FW, Keane TM, Davidson JR. Clinician-administered PTSD scale: a review of the first ten years of research. Depress. Anxiety. 2001;13:132–156. doi: 10.1002/da.1029. [DOI] [PubMed] [Google Scholar]
- 23.Weathers FW, Ruscio AM, Keane TM. Psychometric properties of nine scoring rules for the clinician-administered posttraumatic stress disorder scale. Psychol. Assess. 1999;11:124–133. [Google Scholar]
- 24.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th edition. Washington DC: American Psychiatric Press; 2000. [Google Scholar]
- 25.Blanchard EB, Hickling EJ, Taylor AE, Loos WR. Psychiatric morbidity associated with motor vehicle accidents. J. Nerv. Ment. Dis. 1995;183:495–504. doi: 10.1097/00005053-199508000-00001. [DOI] [PubMed] [Google Scholar]
- 26.Blanchard EB, Hickling EJ, Vollmer AJ, Loos WR, Buckley TC, Jaccard J. Short-term follow-up of post-traumatic stress symptoms in motor vehicle accident victims. Behaviour Research and Therapy. 1995;33:369–377. doi: 10.1016/0005-7967(94)00067-t. [DOI] [PubMed] [Google Scholar]
- 27.Blanchard EB, Hickling EJ, Barton KA, Taylor AE, Loos WR, Jones-Alexander J. One-year prospective follow-up of motor vehicle accident victims. Behaviour Research and Therapy. 1996;34:775–786. doi: 10.1016/0005-7967(96)00038-1. [DOI] [PubMed] [Google Scholar]
- 28.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth GK. Limma: linear models for microarray data. In: Gentlemen R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 30.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Miller JA, Cai C, Langfelder P, Geschwind DH, Kurian SM, Salomon DR, Horvath S. Strategies for aggregating gene expression data: the collapse Rows R function. BMC Bioinformatics. 2011;12:322. doi: 10.1186/1471-2105-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, et al. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shoemaker JE, Tiago L, Ghosh S, Matsuoka Y, Kawaoka Y, Kitano H. CTen: A Web-based Platform for Identifying Enriched Cell Types from Heterogeneous Microarray Data. BMC Genomics. 2012;13.1:460. doi: 10.1186/1471-2164-13-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zarbock A, Polanowska-Grabowska RK, Ley K. Platelet-neutrophil-interactions: Linking Hemostasis and Inflammation. Blood Reviews. 2007;21.2:99–111. doi: 10.1016/j.blre.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Beck F, Geiger J, Gambaryan S, Veit J, Vaudel M, Nollau P, et al. Time-resolved characterization of cAMP/PKA-dependent signaling reveals that platelet inhibition is a concerted process involving multiple signaling pathways. Blood. 2014;123(5):e1–e10. doi: 10.1182/blood-2013-07-512384. [DOI] [PubMed] [Google Scholar]
- 36.Daly ME. Determinants of Platelet Count in Humans. Haematologica. 2010;96.1:10–13. doi: 10.3324/haematol.2010.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raslova H, Kauffmann A, Sekkai D, Ripoche H, Larbret F, Robert T, et al. Interrelation between Polyploidization and Megakaryocyte Differentiation: A Gene Profiling Approach. Blood. 2007;109.8:3225–3234. doi: 10.1182/blood-2006-07-037838. [DOI] [PubMed] [Google Scholar]
- 38.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, et al. INTERFEROME V2.0: An Updated Database of Annotated Interferon-regulated Genes. Nucleic Acids Research. 2012;41.D1:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pacak K. Stressor Specificity of Central Neuroendocrine Responses: Implications for Stress-Related Disorders. Endocrine Reviews. 2001;22.4:502–548. doi: 10.1210/edrv.22.4.0436. [DOI] [PubMed] [Google Scholar]
- 40.Bray PF, Mckenzie SE, Edelstein LC, Nagalla S, Delgrosso K, Ertel A. The Complex Transcriptional Landscape of the Anucleate Human Platelet. BMC Genomics. 2013;4.1:1. doi: 10.1186/1471-2164-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Austin AW, Wissmann T, Von Kanel R. Stress and Hemostasis: An Update. Seminars in Thrombosis and Hemostasis. 2013;39.08:902–912. doi: 10.1055/s-0033-1357487. [DOI] [PubMed] [Google Scholar]
- 42.Walburn J, Vedhara K, Hankins M, Rixon L, Weinman J. Psychological stress and wound healing in humans: a systematic review and meta-analysis. J Psychosom Res. 2009;67(3):253–271. doi: 10.1016/j.jpsychores.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 43.Gouin J-P, Kiecolt-Glaser JK. The Impact of Psychological Stress on Wound Healing: Methods and Mechanisms. Immunology and Allergy Clinics of North America. 2011;31.1:81–93. doi: 10.1016/j.iac.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eraly SA, Nievergelt CM, Maihofer AX, Barkauskas DA, Nilima Biswas N, Agorastos A, et al. Assessment of Plasma C-Reactive Protein as a Biomarker of Posttraumatic Stress Disorder Risk. JAMA Psychiatry. 2014;71.4:423. doi: 10.1001/jamapsychiatry.2013.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maunder RG, Hunter JJ, Feinman SV. Interferon Treatment of Hepatitis C Associated With Symptoms of PTSD. Psychosomatics. 1998;39.5:461–464. doi: 10.1016/s0033-3182(98)71308-8. [DOI] [PubMed] [Google Scholar]
- 46.Dieperink E, Leskela J, Dieperink ME, Evans B, Thuras P, Ho SB. The Effect of Pegylated Interferon-α2b and Ribavirin on Posttraumatic Stress Disorder Symptoms. Psychosomatics. 2008;49.3:225–229. doi: 10.1176/appi.psy.49.3.225. [DOI] [PubMed] [Google Scholar]
- 47.Butcher SK, Lord JM. Stress Responses and Innate Immunity: Aging as a Contributory Factor. Aging Cell. 2004;3.4:151–160. doi: 10.1111/j.1474-9728.2004.00103.x. [DOI] [PubMed] [Google Scholar]
- 48.Clark SM, San J, Francis TC, Nagaraju A, Michael KC, Keegan AD, et al. Immune status influences fear and anxiety responses in mice after acute stress exposure. Brain, behavior, and immunity. 38:192–201. doi: 10.1016/j.bbi.2014.02.001. 20014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maes M, Kubera M, Leunis J. The gut-brain barrier in major depression: Intestinal mucosal dysfunction with an increased translocation of LPS from gram negative enterobacteria (leaky gut) plays a role in the inflammatory pathophysiology of depression. Nueroendocrinology Letters. 2008;29(1):117–124. [PubMed] [Google Scholar]
- 50.Aiboshi J, Moore EE, Ciesla CJ, Silliman CC. Blood transfusion and the two-insult model of post-injury multiple organ failure. Shock. 2001;15:302–306. doi: 10.1097/00024382-200115040-00009. [DOI] [PubMed] [Google Scholar]
- 51.Veldhuis TB, Floris T, van der Meide PH, Vos IM, de Vries HE, Dijkstra CD, et al. Interferon-beta prevents cytokine-induced neutrophil infiltration and attenuates blood–brain barrier disruption. J. Cerebral Blood Flow Metab. 2003;23:1060–1069. doi: 10.1097/01.WCB.0000080701.47016.24. [DOI] [PubMed] [Google Scholar]
- 52.Bhatia M, Moochhala S. Role of inflammatory mediators in the pathophysiology of acute respiratory distress syndrome. J. Pathol. 2004;202:145–156. doi: 10.1002/path.1491. [DOI] [PubMed] [Google Scholar]
- 53.Giannoudis PV. Current concepts of the inflammatory response after major trauma: an update. Injury. 2003;34:397–404. doi: 10.1016/s0020-1383(02)00416-3. [DOI] [PubMed] [Google Scholar]
- 54.Minagar A, Alexander JS. Blood-brain Barrier Disruption in Multiple Sclerosis. Multiple Sclerosis. 2003;9.6:540–549. doi: 10.1191/1352458503ms965oa. [DOI] [PubMed] [Google Scholar]
- 55.Carmeliet P, Strooper BD. Alzheimer's Disease: A Breach in the Blood–brain Barrier. Nature. 2012;485.7399:451–452. doi: 10.1038/485451a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differential gene expression analyses were performed using a moderated t statistic within Dataset 1 and Dataset 2. A cross-sectional analysis compared PTSD cases to controls post-deployment in Dataset 1 (47 PTSD cases vs. 47 controls) and subsequently in Dataset 2 (24 PTSD cases vs. 24 controls) to reveal 294 and 61 differentially expressed genes, respectively, with a significant overlap (a). The top 3 most significant biological processes (annotated with DAVID) based on over-expressed and under-expressed genes identified from Dataset 1 are reported (b). Subsequently, the same paired data were analyzed pre-deployment for Dataset 1 and Dataset 2 revealing 662 and 178 differentially expressed genes, respectively, with a significant overlap (c). The top 3 most significant biological processes from Dataset 1 are reported (d). Utilizing, the paired structure of the data, a longitudinal contrast analysis was applied to identify genes behaving differently across time within the PTSD and control groups. This analysis revealed a total of 177 genes in Dataset 1 and 110 genes in Dataset 2, with minimal overlap (e) and no significant functional annotation (f). Up and down symbols are relative to the PTSD group. Significance threshold for genes was set to a nominal p < 0.05 where as we used a more strict threshold for functional annotations with a Bonferroni corrected p-value < 0.05.
Module Significance (MS) across (a) pre- and (c) post-deployment modules in Dataset 2. WGCNA detected nine modules post-deployment from a combination of PTSD cases and control (a) and eleven modules at pre-deployment from a combination of PTSD risk cases and controls (b). The y axis indicates MS by calculating the average −log10 p-values, generated by a moderated t-test, for each gene within a particular module, when assessing differential expression between PTSD cases and controls. Here, a kruskal-wallis p-value was used only for descriptive purposes and not inferential. Modules denoted with an asterisk (*) have module eigengenes values (ME) significantly correlated to group status (i.e. PTSD cases or controls). Differences in ME expression were measured using a two-tailed student’s t-test on and a p-value < 0.05 is considered significant. Boxplots are displayed for each main group. Post-deployment module M3B displayed a moderate, yet insignificant, over-expression (b) and pre-deployment M4A displayed a significant over-expression (d).
Enrichment analysis and correlation networks for module M3A (a & b) identified post-deployment and module M4A (c & d) identified pre-deployment in Dataset 2. Enrichment analysis was used to identify the top 6 Reactome ontology terms (black bars), all DAVID ontology terms (grey bars) and the top cell-type signature (white bar) over-represented in the list of genes in each module. All terms were deemed significant as assessed by a hypergeometric test (FDR corrected p-value <0.05). Gene-networks were constructed selecting the top 150 most significant connections. Nodes represent genes and edges represent correlations. The top 5 hub genes with the highest correlation with ME are shown in larger sizes.
Heatmaps representation of the 51 overlapping genes from module M1A, M2A, M3A, and M4A. Heatmaps are divided into four main parts. Dataset 1 modules (a), M1A on the top row and M2A on the bottom row, are split into PTSD cases (left column) and controls (right column). Dataset 2 modules (b), M3A on the top row and M4A on the bottom row, are split into PTSD cases (left column) and controls (right column). All subjects have been sorted in an identical fashion in order to properly visualize differences in expression as occurring within each individual sample across the two time-points (i.e. Subject 1 post-deployment is directly above Subject 1 pre-deployment). Red represents over-expression, green represents under-expression and black represents the mean.
Venn Diagram (a) depicting significant overlap in hemostasis and blood coagulation genes belonging to modules M1B post-deployment and Module 16 pre-deployment in Dataset 1 as well as modules Module 7 post-deployment and Module 6 pre-deployment in Dataset 2. The overlap identified 46 genes found across all four analyses which are displayed in table format (b) along with the corresponding kME rank (i.e. rank of connectivity) for each gene within a particular module. Higher rank indicates hub gene status, the top 10 genes for each module are in bold. Heatmaps representing these 46 genes are shown across both datasets and are divided into four main parts. Dataset 1 modules (c), M1B on the top row and Module 16 on the bottom row, are split into PTSD cases (left column) and controls (right column). Dataset 2 modules (d), Module 7 on the top row and Module 6 on the bottom row, are split into PTSD cases (left column) and controls (right column). All subjects have been sorted in an identical fashion in order to properly visualize differences in expression as occurring within each individual sample across the two time-points (i.e. Subject 1 post-deployment is directly above Subject 1 pre-deployment). Red represents over-expression, green represents under-expression and black represents the mean according to the scale bar according to scale bar.
Data Availability Statement
RNA-Seq and microarray gene expression data are freely available at the Gene Expression Omnibus under the SuperSeries accession number GSE64814 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64814).
Full Methods and any associated references are available in Supplementary Methods.